Abstract
Methylation of CpG islands in promoter regions is often associated with gene silencing and aberrant DNA methylation occurs in most cancers, leading to the silencing of some tumor suppressor genes. Reversal of this abnormal hypermethylation by DNA methylation inhibitors is effective in re-activating methylation-silenced tumor-suppressor genes both in vitro and in vivo. Several DNA methylation inhibitors have been well studied; the most potent among them is 5-aza-2’-deoxycytidine (5-Aza-CdR), but it can induce myelosuppression in patients. S110 is a dinucleotide consisting of 5-Aza-CdR followed by a deoxyguanosine which we previously showed to be effective in vitro as a DNA methylation inhibitor while being less prone to deamination by cytidine deaminase, making it a promising alternative to 5-Aza-CdR. Here we show that S110 is better tolerated than 5-Aza-CdR in mice, and is as effective in vivo in inducing p16 expression, reducing DNA methylation at the p16 promoter region, and retarding tumor growth in human xenograft. We also demonstrate that S110 is effective by both intraperitoneal (IP) and subcutaneous (SQ) deliveries. S110 therefore is a promising new agent that acts similarly to 5-Aza-CdR and has better stability and less toxicity.
Introduction
DNA methylation is employed by mammalian cells in maintaining a normal expression pattern; it is involved in the regulation of imprinted gene expression and X-chromosome inactivation, among others [1–3]. Methylation of CpG islands in promoter regions is often associated with gene silencing, and aberrant DNA methylation occurs in most cancers, leading to the silencing of some tumor suppressor genes [4, 5]. Reversal of this abnormal hypermethylation by DNA methylation inhibitors has been shown to be effective in re-activating methylation-silenced tumor-suppressor genes both in vitro and in vivo [6].
DNA methylation inhibitors can be further dividied into two groups—nucleoside analogs and non-nucleoside analogs—and most have been well studied for their mechanisms of actions and clinical potentials [6]. Nucleoside analogs are converted into nucleotides and incorporated into DNA, and there they can trap DNMTs by forming covalent complexes [7–9]. 5-Azacytidine (5-Aza-CR) and 5-Aza-CdR are two well-known DNA methylation inhibitors, and have been approved by the Food and Drug Administration for the treatment of myelodysplastic syndrome [6, 10, 11]. Unlike traditional chemotherapy agents, DNA methylation inhibitors do not induce immediate cell death at their optimal dosage, although cytotoxicity can occur at high concentrations. Cells need to be proliferating for effective incorporation of drugs into DNA and reactivate methylation-silenced tumor-suppressor genes that in turn make the cells more responsive to apoptotic or cell-cycle regulating signals [6]. Given their potential in clinical applications, much effort has been invested to develop more stable forms of these known DNA methylation inhibitors that can be effectively delivered to cancer cells.
Since Laird et al. [12] showed that 5-Aza-CdR was effective in reducing the occurence of intestinal adenomas in ApcMin/+ mice, there has been many animal studies that examine the actions of epigenetic drugs. Zebularine, another promising DNA methylation inhibitor, has been shown to be effective at reducing tumor growth in vivo [13, 14]. Karam et al. [15] reported that HDAC inhibitor FK228 can inhibit transitional cell carcinoma xenograft growth with minimal undesirable side effects. Moreover, 5-Aza-CdR and zebularine have been shown to decrease vessel formation, a necessary step for tumor formation, in mouse tumor models [16]. Many studies have examined the combinatorial effects of different epigenetic drugs in mouse tumor models [17–19].
We examined the in vivo effect of the dinucleotide S110, which consists of 5-Aza-CdR followed by a deoxyguanosine. S110 has been shown to be effective in vitro in inducing p16 expression and is more stable than 5-Aza-CdR due to decreased deamination by cytidine deaminase [20]. We now show that S110 is better tolerated than 5-Aza-CdR in tumor-free mice, and is as effective in vivo in inducing p16 expression and reducing DNA methylation at the p16 promoter region. We demonstrate that S110 is effective at retarding tumor growth in a xenograft model, and we also demonstrate that the effect can be achieved by both IP and SQ deliveries. S110 therefore serves as a promising new agent that acts similarly to 5-Aza-CdR and has better stability and less toxicity.
MATERIALS AND METHODS
In Vivo Drug Tolerability Study
Non-tumor-bearing athymic nu/nu mice (Charles River, Wilmington, MA) were divided into six treatment groups with six animals per group. Treatments of S110 and 5-Aza-CdR were prepared in PBS and administered intravenously (IV) through tail vein injections. Doses and dosing schedules were designed so that after seven days each group received molar equivalents of either S110 or 5-Aza-CdR. Animals were treated on the following schedules for three weeks: Group 1 received 36.6 mg/kg S110 once weekly (Mon.) and Group 2 was administered 15 mg/kg 5-Aza-CdR once weekly. Group 3 was dosed with 18.3 mg/kg S110 twice weekly (Tues. and Thurs.) and group 4 received 7.5 mg/kg 5-Aza-CdR twice weekly. Finally, groups 5 and 6 received 12.2 and 5.0 mg/kg of S110 and 5-Aza-CdR, respectively administered three times weekly (Mon., Wed., and Fri.). Tolerability was grossly evaluated by body weight measurements and morbidity. Body weight measurements were recorded twice weekly.
In vivo xenograft drug efficacy studies with intraperitoneal delivery
The EJ6 human bladder cancer cell was used for this study, and experiments were done similarly to previously described [13]. EJ6 cells (5 × 105/injection) suspended in PBS were inoculated subcutaneously (SQ) into the right and left back (along the midaxillary lines) of 4- to 6-week-old female BALB/c athymic nude-Foxn1nu mice (Harlan, San Diego, CA). Mice were randomly divided into 3 groups. After 2–3 weeks and after macroscopic tumors (50–200 mm3) had formed, treatments were initiated. Tumors were measured with calipers, and tumor volumes (TVs) were calculated with the following formula: TV = LD2/2 (where L is the longest diameter and D is the shortest diameter). The fold differences in tumor growth among the various mice groups were calculated using relative TVs (RTVs), which are calculated as follows: RTV = TVn/TV0, where TVn is the tumor volume in mm3 at a given day n and TV0 is the tumor volume in mm3 at day 0 (initial treatment). Mice were weighed at the beginning and end of treatment to determine toxicity. The percent weight change for each mouse was calculated with the following formula: [(W6−W0)/W0] × 100% (where Wn is the mouse weight on day n).
5-Aza-CdR was used as the positive control and 0.45% PBS was used as the negative control. PBS, 5-Aza-CdR (dose of 5 mg/kg in PBS), and S110 (dose of 10 mg/kg in PBS) were administered daily by intraperitoneal (IP) injection over a period of 6 days.
All mice were sacrificed 24 hours after the last treatment. At this time, tumors were removed and each tumor was divided into two separate portions. One portion was immediately homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA) for RNA extraction, and the other portion was immediately frozen in liquid nitrogen for DNA extraction later. Genomic DNA and RNA would be used for analysis of the methylation status of p16 promoter by Ms-SNuPE and of gene expression by real time RT–PCR, respectively. All experimental protocols were approved by the Institutional Animal Care and Use Committee, in compliance with the Guide for the Care and Use of Laboratory Animals, University of Southern California.
In vivo xenograft drug efficacy studies with subcutaneous delivery
Athymic nu/nu mice (Charles River) were inoculated subcutaneously in the right hind flank with 107 EJ6 bladder cancer cells. After tumors reached 0.5 cm in diameter, animals were stratified into three groups with eight animals per group to begin treatments. Doses and dosing schedules were designed so that each group received molar equivalents of either S110 or 5-Aza-CdR. The agents were administered SQ once weekly at a dose of 12.2 mg/kg for S110 and 5.0 mg/kg for 5-Aza-CdR for three weeks. The study included an appropriate PBS control group. Tumor sizes by caliper and body weight measurements were taken twice weekly to monitor tumor growth inhibition and tolerability.
Nucleic acid extraction
RNA was extracted by first homogenizing tumor samples in TRIzol reagent (Invitrogen, Carlsbad, CA), and then following the manufacturer’s instructions. Genomic DNA was extracted by dissolving each tumor sample in 500µl of lysis buffer (100mM Tris HCl pH8.5, 5mM EDTA pH8.0, 0.2% SDS, 400mM NaCl, 100µg/ml proteinase K, and 10ug/ml RNase) overnight at 55°C, and then followed by phenol-chloroform extraction [21, 22].
Quantitative Real-Time Reverse-Transcription PCR
Total RNA was extracted from cells with the Invitrogen Trizol reagent (Carlsbad, CA). Reverse transcription (RT) was performed with M-MLV reverse transcriptase and random hexamers from Promega (Madison, WI). We performed quantitative real-time RT-PCR analysis as previously described [23] with DNA Engine Opticon System (MJ Research, Hercules, CA). The primers used are listed below: p16 sense, 5’-AGC CTT CGG CTG ACT GGC TGG-3’; p16 antisense, 5’-CTG CCC ATC ATC ATG ACC TGG A-3’; p16 fluorogenic probe, 5’-TGG ATC GGC CTC CGA CCG TAA CT-3’. The real-time RT-PCR conditions for both genes were as follows: 95°C for 9min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 1min.
Quantitative DNA Methylation Analysis by Methylation-Specific Single Nucleotide Extension (Ms-SNuPE)
Two µg of each genomic DNA sample was converted with sodium bisulfite as previously described [24], and each region of interest was amplified by PCR. The PCR conditions for p16 were as follows: 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 62°C for 1 min, and extension at 72°C for 1 min, and a final extension at 72°C for 10min. The bisulfite specific-PCR primer sequences are as follows: p16 sense, 5’- GTA GGT GGG GAG GAG TTT AGT T-3’, p16 antisense, 5’- TCT AAT AAC CAA CCA ACC CCT CCT-3’. The Ms-SNuPE conditions for p16 were as follows: 95°C for 2 min, 50°C for 2 min, and 72°C for 1 min. The p16 SNuPE primers are as follows: 5’-TTT TAG GGG TGT TAT ATT-3’, 5’-TTT TTT TGT TTG GAA AGA TAT-3’, and 5’-TTT GAG GGA TAG GGT-3’. The PCR amplicons were extracted with the Qiagen Gel Extraction Kit, and Ms-SNuPE analysis was performed to examine the methylation level changes as previously described [25].
RESULTS
S110 is better tolerated than 5-Aza-CdR in vivo in tumor-free nude mice
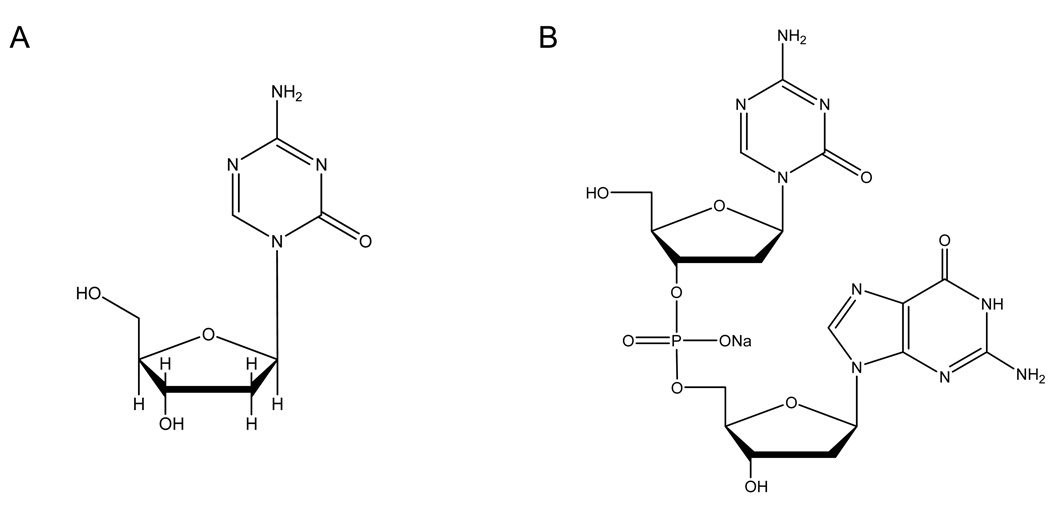
While 5-Aza-CdR is an effective DNA methylation inhibitor both in vitro and in vivo, it suffers from the drawback of having toxicity such as myelosuppression in patients [26]. S110 is a dinucleotide consisting of 5-Aza-CdR followed by a deoxyguanosine (Figure 1), and has been previously shown by Yoo et al. [20] to be effective in vitro as a DNA methylation inhibitor and comparable to 5-Aza-CdR in decreasing DNA methylation, reactivating p16 expression, depleting DNA methyltransferases and reducing cancer cell growth in vitro. Moreover, it has the advantage of being less prone to deamination by cytidine deaminase, making it a promising alternative to 5-Aza-CdR.
Figure 1.
Chemical structures of (A) 5-Aza-CdR and (B) S110
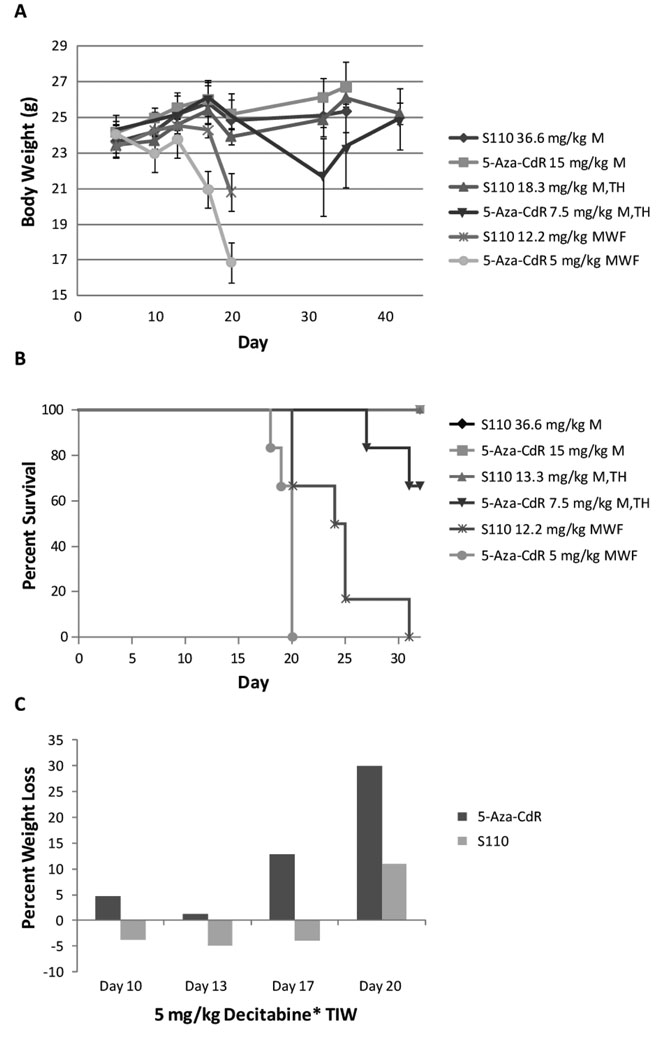
To compare the in vivo tolerability of S110 to that of 5-Aza-CdR, six groups of six tumor-free female nude mice each were randomly assigned to varying schedules according to Table 1. The groups were paired up so that S-110 was administered at molar equivalents of 5-Aza-CdR at the same frequencies in groups 1 and 2, groups 3 and 4, and groups 5 and 6. We first compared the tolerability by examining the relative body weights throughout the study (Figure 2A). Animals in groups 1 and 2 maintained or steadily gained body weight throughout the course of the study and were euthanized after 36 days. In groups 3 and 4, animals receiving S-110 also maintained or steadily gained weight, whereas the average body weight of those receiving 5-Aza-CdR began to decline after the first week of dosing. While the average body weight of both groups 5 and 6 rapidly decreased after the first week of dosing, eventually leading to death, those receiving S-110 maintained a healthy body weight slightly longer than animals receiving 5-Aza-CdR. It is of interest that despite the same total weekly dosage of 5-Aza-CdR or S110, both drugs cause more toxicity when given in smaller doses and higher frequency. In all three paired comparisons, S110 was better tolerated than 5-Aza-CdR in terms of weight change.
Table 1.
3 week tolerability study of S-110 and 5-Aza-CdR with varying schedules.
| Group | n | Treatment | Dose | Route | Schedule |
|---|---|---|---|---|---|
| 1 | 6 | S-110 | 36.6 mg/kg | IV | 3x Q7D |
| 2 | 6 | 5-Aza-CdR | 15 mg/kg | IV | 3x Q7D |
| 3 | 6 | S-110 | 18.3 mg/kg | IV | 3x BIW |
| 4 | 6 | 5-Aza-CdR | 7.5 mg/kg | IV | 3x BIW |
| 5 | 6 | S-110 | 12.2 mg/kg | IV | 3x TIW |
| 6 | 6 | 5-Aza-CdR | 5.0 mg/kg | IV | 3x TIW |
The purpose of this study was to compare the tolerability of S-110 to 5-Aza-CdR when administered intravenously on the same schedules. S-110 was administered as molar equivalents of 5-Aza-CdR. Female nude mice were used in this study.
IV, intravenous; Q7D, every 7 days; BIW, twice a week; TIW, three times a week.
Figure 2.
S110 is better tolerated than 5-Aza-CdR as demonstrated by relative body weight (A), percent survival (B), and percent weight loss (C) in normal tumor-free mice. Animals were treated according to the dosages and schedules listed in Table 1. A: Six animals per group treated with S110 maintained body weights better and survived longer (B) compared to those treated with 5-Aza-CdR. C: Histogram illustrating the effects of S-110 and 5-Aza-CdR treatment on body weight for groups 5 and 6 only, which received 5 mg/kg 5-Aza-CdR or a molar equivalent of S110 three times a week. Graphed is the average percent weight loss, therefore negative values are indicative of weight gain. Animals receiving 5-Aza-CdR began losing weight more rapidly than those receiving S-110.
We then examined the percent survival (Figure 2B) of the treated mice. There were no deaths in groups 1 and 2; however, two animals in group 4 died. All animals in group 3 survived, where as all animals in groups 5 and 6, which received molar equivalents of S110 or 5-Aza-CdR three times a week respectively, died before the completion of the study. However, animals receiving S-110 (group 5) survived 5–10 days longer on average than those receiving 5-Aza-CdR (group 6). Lastly, we compared the percent weight loss in groups 5 and 6 (Figure 2C). Graphed is the average percent weight loss, therefore negative values are indicative of weight gain. Animals receiving 5-Aza-CdR began losing weight faster than those receiving S-110; the later did not lose weight until ~ day 20 of the study. Taken together, S110 induced less weight loss and mortality than 5-Aza-CdR and is therefore more favorably tolerated in tumor-free mice.
S110 is effective at slowing tumor growth and inhibiting DNA methylation when delivered IP
EJ6 cells were inoculated SQ into the right and left back of female BALB/c athymic nude-Foxn1nu mice. Mice were randomly divided into 3 groups and after 2–3 weeks treatments were initiated. Five mice were injected IP with PBS, and 16 mice each were injected with 5-Aza-CdR at 5mg/kg of mouse weight and S110 at 10mg/kg of mouse weight. One mouse treated with 5-Aza-CdR died on day 6 before the harvest. Our previous experiment showed that S110 at 10mg/kg was an effective dose at reducing DNA methylation and retarding tumor growth, and caused roughly the same level of toxicity as 5-Aza-CdR (data not shown).
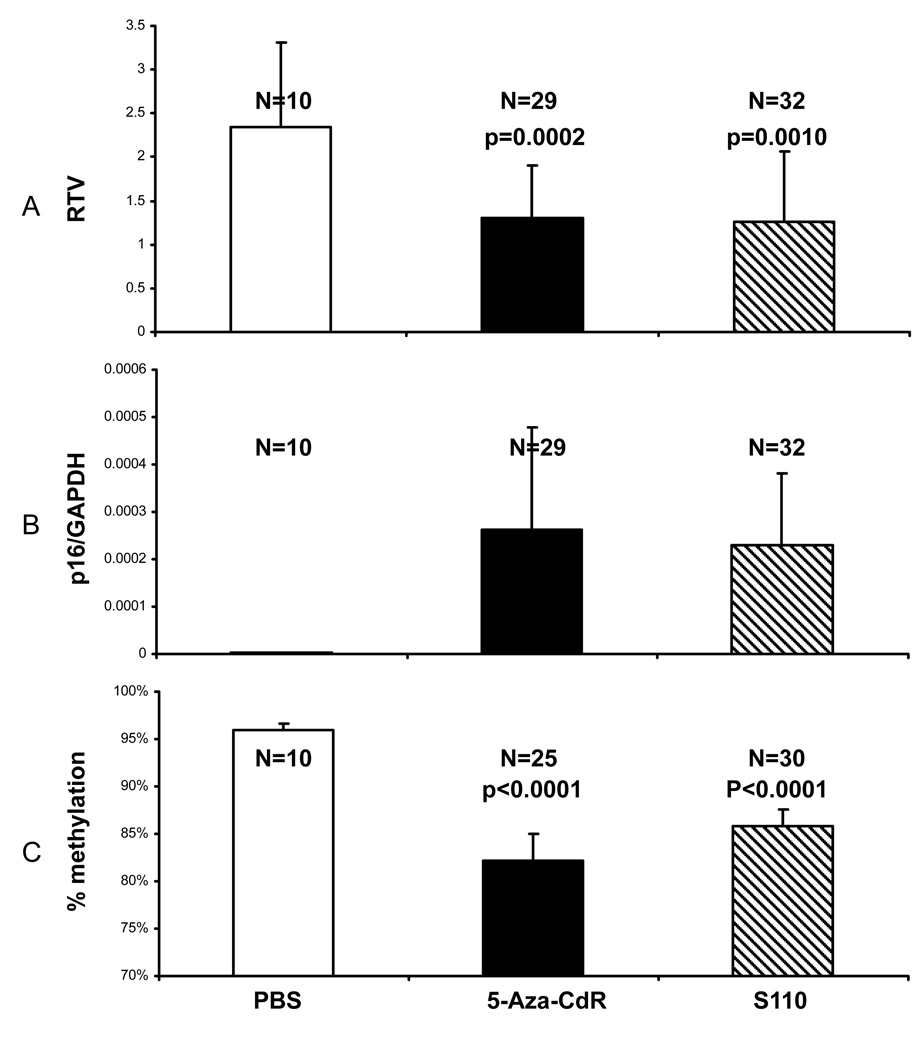
We studied the potential action of S110 on inhibiting tumor growth. We found that while neither 5-Aza-CdR nor S110 reduced the tumor sizes by the end of the treatment compared to the original sizes on day 0, they both retarded the growth rate of tumors on average compared to the PBS treatment (Figure 3A).
Figure 3.
S110 is effective at slowing EJ6 tumor growth in vivo (A), inducing p16 expression (B), and decreasing methylation (C) by IP administration. Mice were treated by IP injections of PBS, 5-Aza-CdR at 5mg/kg, or S110 at 10mg/kg daily for 6 days. A: Tumors were measured with calipers, and tumor volumes (TVs) were calculated by the following formula: TV = LD2/2 (where L is the longest diameter and D is the shortest diameter). The fold differences in tumor growth among the various mice groups were calculated using RTVs, which are calculated as follows: RTV = TV6/TV0, where TV6 is the tumor volume in mm3 at day 6 and TV0 is the tumor volume in mm3 at day 0 (initial treatment). B: Tumors were harvested on day 6, and total RNA extracted for RT-PCR analysis. Quantitative real time RT-PCR analysis for the p16 gene was done with normalization against the GAPDH reference gene. C: Tumors were harvested on day 6, and genomic DNA extracted for Ms-SNuPE analysis. Both 5-Aza-CdR and S110 are effective at reducing DNA methylation at the p16 gene promoter in the xenografts. The numbers of tumors and p values in each group are labeled above the histograms. The changes in percent methylation level in 5-Aza-CdR- and S110-treated groups are statistically significant to the PBS control treatments by the Student t-test. Results are shown as means + one standard deviation.
Real time RT-PCR analysis of the tumor tissues revealed that S110 was effective in vivo at reactivating the expression of the p16 gene, which is heavily methylated in the parent EJ6 cells (Figure 3B). In addition, S110 at 10mg/kg, which was at roughly the same molar concentration as 5-Aza-CdR at 5mg/kg, induced similar levels of p16 expression as 5-Aza-CdR at 5mg/kg. We also noticed that unlike in vitro studies, there was greater variation within treatment groups. This could be attributed to the different levels of response among different mice, or even between the two different tumors on the same mouse. Overall, S110 was effective in vivo at reactivating silenced genes, and its activity was comparable to 5-Aza-CdR at roughly the same molar concentrations.
In addition, S110 was effective in reducing the level of DNA methylation in vivo at the p16 promoter region (Figure 3C). While the PBS group showed consistent levels of high methylation, variations among different tumors were much more obvious in the 5-Aza-CdR and the S110 groups. On average, the DNA methylation level was roughly 96% in the PBS group, 82% in the 5-Aza-CdR group, and 86% in the 10mg/kg S110 group (Figure 3C). Though the changes in magnitude were small, they were statistically significant as determined by the Student t-test (Figure 3C) when compared to the PBS control group. Because Ms-SNuPE only examined the methylation changes at a few CpG sites, a different method such as genomic bisulfite sequencing might reveal more substantial methylation changes than was determined by Ms-SNuPE.
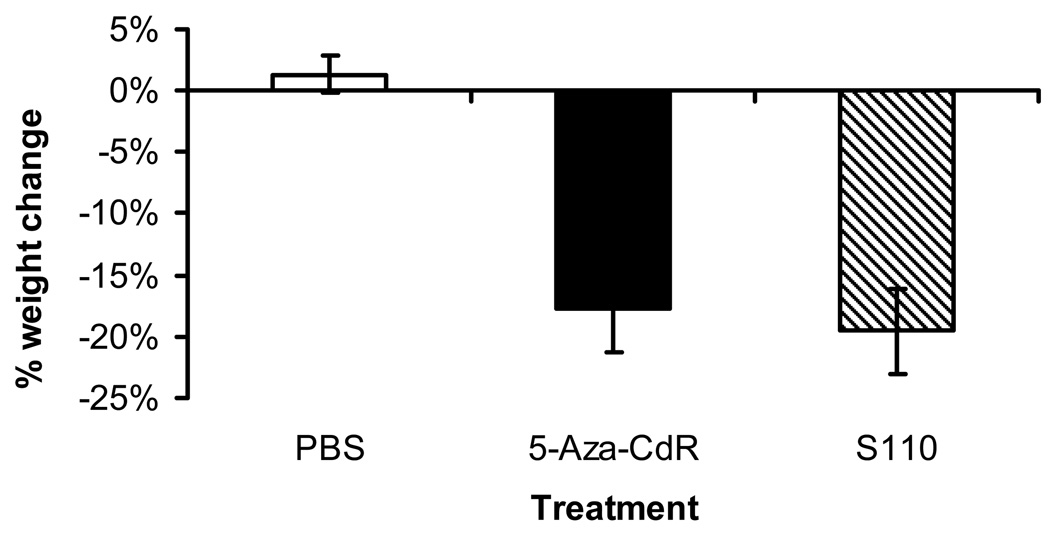
We then estimated the toxicities from the treatments by measuring the weights of the mice at the beginning and the end of the treatment (Figure 4). Mice treated with PBS did not experience much change in weight, and 5-Aza-CdR and S110 induced similar weight loss in mice. However, the mice treated with S110 appeared healthier compared to the mice treated with 5-Aza-CdR, both by appearance and by their normal level of activity. On the contrary, the mice treated with 5-Aza-CdR appeared cachectic and were not active. These data suggested that S110 could be a promising alternative to 5-Aza-CdR because, at roughly the same molar concentration, the former caused equitoxicity in mice as estimated by mouse weight while remaining comparable in terms of reactivating genes, reducing DNA methylation, and retarding tumor growth in a xenograft model.
Figure 4.
S110 is equitoxic to 5-Aza-CdR in vivo by IP administration Mice were weighed at the beginning and the end of treatment to determine toxicity. The percent weight change for each mouse was calculated with the following formula: [(W6-W0)/W0] × 100% (where Wn is the mouse weight on day n). The average percent weight change ± one standard deviation is plotted, and the number of mice in each group is labeled accordingly.
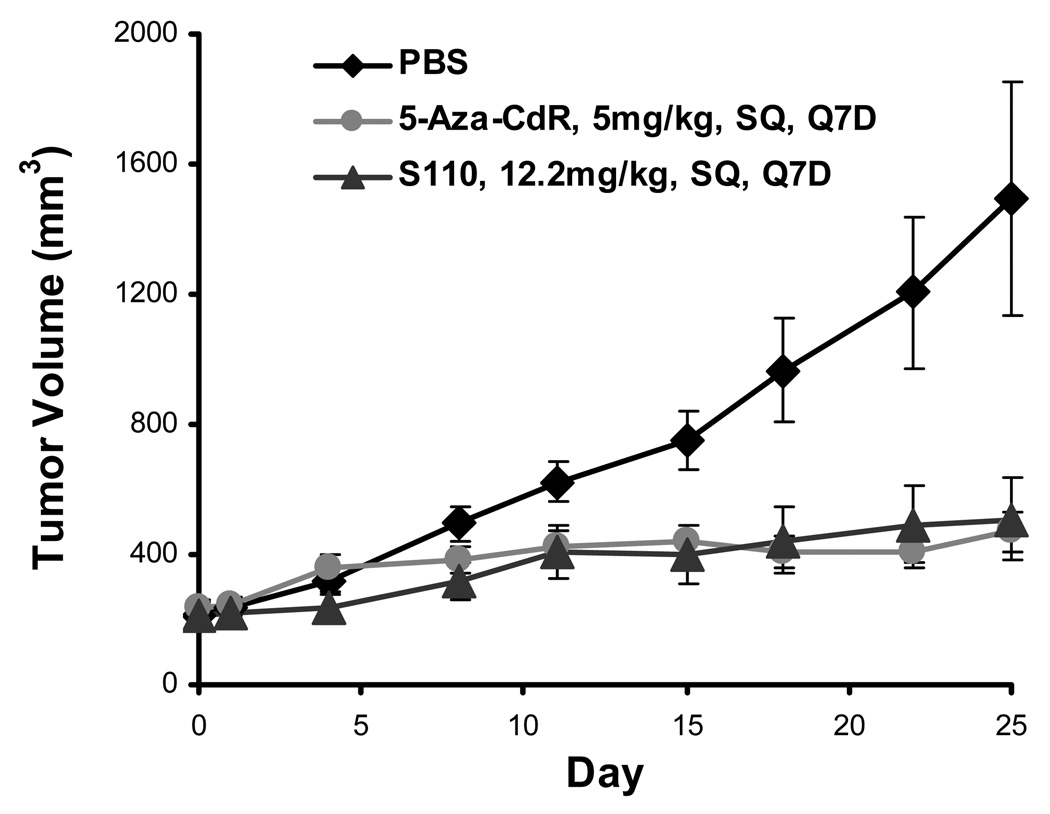
S110 is also effective at retarding tumor growth when delivered SQ
To further study the pharmacodynamic action of S110 in vivo, we again generated EJ6-xenografted nude mice and divided them randomly into three groups of eight to receive SQ PBS, 5-Aza-CdR at 5mg per kg of mouse weight, or S110 at 12.2mg per kg of mouse weight (equimolar to 5-Aza-CdR). Tumor volumnes were measured periodically for the total of 25 days, and the average tumor sizes for each group are shown in Figure 5. Both 5-Aza-CdR and S110 were effective at retarding tumor growth, and they were very comparable to each other in their efficacy. While the tumors did not shrink in size with either the 5-Aza-CdR or S110 treatment, they experienced very minimal growth while the tumors treated with PBS only showed substantial growth. Our results demonstrated that S110 can also be effective when given SQ. In addition, both 5-Aza-CdR and S110 induced much less toxicity as determined by mouse weight changes when given SQ compared to that with IP injections (data not shown), thus SQ injection could serve as a different way of delivery that would induce less toxicity.
Figure 5.
S110 is effective at slowing EJ6 tumor growth in vivo by subcutaneous (SQ) administration every seven days (Q7D). Eight mice each were treated by SQ injections of PBS, 5-Aza-CdR at 5mg/kg, or S110 at 12.2mg/kg every 7 days for 25 days. Results are given as the average of tumor volumes with one standard deviation.
DISCUSSION
Our results with S110 show that it is an effective DNA methylation inhibitor in vivo. It is effective at reducing DNA methylation as determined by Ms-SNuPE, and is effective in reactivating p16 expression as determined by RT-PCR. We also show that it can retard tumor growth in xenografts, and that it can be effectively delivered by both IP and SQ routes. Lastly, our data show that S110 is better tolerated than 5-Aza-CdR in vivo, suggesting that it can be an attractive alternative for potential clinical use.
Our results show that p16 is up-regulated to different extents in individual mice by the treatments. We notice that while S110 is effective in most tumors, there are individual variations in response to S110, as are also observed with 5-Aza-CdR. It would be of interest to determine which factors contribute to the differences in response. Having this knowledge would also aid us in the future to determine which patients would respond well to DNA methylation inhibitors clinically.
The individual variations we observe also suggest that 5-Aza-CdR and S110 could have secondary effects. Although both drugs have been shown to be effective DNA methylation inhibitors, they could have actions in addition to inhibiting DNA methyltransferases. Given that at the same level of efficacy, 5-Aza-CdR causes more toxicity in mice by their appearance than S110 in xenograft mice, it is reasonable to hypothesize that either or both drugs can cause changes in addition to affecting DNA methylation. Further studies should be conducted to investigate the actions of these drugs by examining global gene expression changes by microarray analysis. Such studies would illuminate on the global actions of these drugs and allow us to compare the actions of the different DNA methylation inhibitors side by side.
The more favorable tolerability of S110 compared to 5-Aza-CdR could be due to several possibilities. There could be differential uptake of the two drugs by different organs. Yoo et al. [27] showed that zebularine, another nucleoside-analog DNA methylation inhibitor, affected different organs in female mice differentially, with the greatest decrease in methylation level in the small intestine and colon. It is therefore reasonable to hypothesize that there are also differential effects of S110 and 5-Aza-CdR on different organs and that could lead to the better tolerability of S110. Until we know the exact mechanisms by which 5-Aza-CdR causes toxicity in vivo, we cannot fully answer the question why S110 is better tolerated. The answer is likely contributed by mechanisms at molecular, cellular, and tissue and systemic levels. It would also be of great interest to study the effects of the drugs on other mouse tissues to examine if the drugs have any adverse effects, and also to see if different tissues have preferential responses to the drugs.
Our previous study by Yoo et al. [20] showed that S110 is only effective at inhibiting DNA methylation after being cleaved into individual nucleotides, and this finding poses the question whether S110 is cleaved first in the circulation before entering individual cells, or cleaved later intracellularly after its uptake. From our data that show IV injections of S110 at equimolar concentrations to 5-Aza-CdR lead to less toxicity in tumor-free mice, one could argue that it is unlikely that S110 was completely cleaved in the circulation before reaching cells given the difference in tolerability. However, this remains a speculation, and more studies are needed in the future to elucidate the exact pharmacodynamic and pharmacokinetic mechanisms of S110.
Given the promising results from our S110 study, further experiments should be conducted to investigate the chemotherapeutic potential and toxicity of S110. In addition to comparing S110 to 5-Aza-CdR at equal molar concentrations, it would be informative to also compare the two drugs at equal toxicity levels because most clinical studies compare the efficacies of different drugs at equal toxicity levels. Having more knowledge of S110 would help us develop more effective DNA methylation inhibitors for chemotherapy in the future.
Acknowledgements
The authors would like to thank Dr. Yvonne Tsai for her invaluable assistance throughout this study, Dr. Hsiang-lin Chang and Dr. Erlinda Kirkman for their technical guidance, and Dr. Gangning Liang and Jeremy Lamb for editing the manuscript. This work is supported by the AACR Henry Shepard Bladder Cancer Research Grant 119993, NIH 5R37CA82422, the NIH CBM student training grant (5 T32 GM067587), and the China Scholarship Counsil.
BIBLIOGRAPHY
- 1.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 2.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 3.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153(4):773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 7.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33(1):9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 8.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984;81(22):6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou L, et al. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321(4):591–599. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaminskas E, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11(10):3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 11.Ruter B, Wijermans PW, Lubbert M. Superiority of prolonged low-dose azanucleoside administration? Results of 5-aza-2'-deoxycytidine retreatment in high-risk myelodysplasia patients. Cancer. 2006;106(8):1744–1750. doi: 10.1002/cncr.21796. [DOI] [PubMed] [Google Scholar]
- 12.Laird PW, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81(2):197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 13.Cheng JC, et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95(5):399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- 14.Herranz M, et al. The novel DNA methylation inhibitor zebularine is effective against the development of murine T-cell lymphoma. Blood. 2006;107(3):1174–1177. doi: 10.1182/blood-2005-05-2033. [DOI] [PubMed] [Google Scholar]
- 15.Karam JA, et al. The use of histone deacetylase inhibitor FK228 and DNA hypomethylation agent 5-azacytidine in human bladder cancer therapy. Int J Cancer. 2007;120(8):1795–1802. doi: 10.1002/ijc.22405. [DOI] [PubMed] [Google Scholar]
- 16.Hellebrekers DM, et al. Angiostatic activity of DNA methyltransferase inhibitors. Mol Cancer Ther. 2006;5(2):467–475. doi: 10.1158/1535-7163.MCT-05-0417. [DOI] [PubMed] [Google Scholar]
- 17.Venturelli S, et al. Epigenetic combination therapy as a tumor-selective treatment approach for hepatocellular carcinoma. Cancer. 2007;109(10):2132–2141. doi: 10.1002/cncr.22652. [DOI] [PubMed] [Google Scholar]
- 18.Morita S, et al. The synergistic effect of 5-aza-2'-deoxycytidine and 5-fluorouracil on drug-resistant tumors. Oncology. 2006;71(5–6):437–445. doi: 10.1159/000107110. [DOI] [PubMed] [Google Scholar]
- 19.Cantor JP, et al. Epigenetic modulation of endogenous tumor suppressor expression in lung cancer xenografts suppresses tumorigenicity. Int J Cancer. 2007;120(1):24–31. doi: 10.1002/ijc.22073. [DOI] [PubMed] [Google Scholar]
- 20.Yoo CB, et al. Delivery of 5-aza-2'-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 2007;67(13):6400–6408. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 21.Laird PW, et al. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19(15):4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q, et al. A simple, rapid method for isolation of high quality genomic DNA from animal tissues. Nucleic Acids Res. 1995;23(24):5087–5088. doi: 10.1093/nar/23.24.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heid CA, et al. Real time quantitative PCR. Genome Res. 1996;6(10):986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 24.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalgo ML, Jones PA. Quantitative methylation analysis using methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) Methods. 2002;27(2):128–133. doi: 10.1016/s1046-2023(02)00064-6. [DOI] [PubMed] [Google Scholar]
- 26.Issa JP, et al. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol. 2005;23(17):3948–3956. doi: 10.1200/JCO.2005.11.981. [DOI] [PubMed] [Google Scholar]
- 27.Yoo CB, et al. Long-term epigenetic therapy with oral zebularine has minimal side effects and prevents intestinal tumors in mice. Cancer Prev Res (Phila Pa) 2008;1(4):233–240. doi: 10.1158/1940-6207.CAPR-07-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]