Abstract
RNA-Seq provides an unbiased way to study a transcriptome, including both coding and non-coding genes. To date, most RNA-Seq studies have critically depended on existing annotations, and thus focused on expression levels and variation in known transcripts. Here, we present Scripture, a method to reconstruct the transcriptome of a mammalian cell using only RNA-Seq reads and the genome sequence. We apply it to mouse embryonic stem cells, neuronal precursor cells, and lung fibroblasts to accurately reconstruct the full-length gene structures for the vast majority of known expressed genes. We identify substantial variation in protein-coding genes, including thousands of novel 5′-start sites, 3′-ends, and internal coding exons. We then determine the gene structures of over a thousand lincRNA and antisense loci. Our results open the way to direct experimental manipulation of thousands of non-coding RNAs, and demonstrate the power of ab initio reconstruction to render a comprehensive picture of mammalian transcriptomes.
INTRODUCTION
A critical task in understanding mammalian biology is defining a precise map of all the transcripts encoded in a genome. While much is known about protein-coding genes in mammals, recent studies have suggested that the mammalian genome also encodes many thousands of large ncRNA genes1–3,4. Recently, we used a chromatin signature, combining Histone 3 Lysine 4 tri-methylation modifications (H3K4me3) that mark the promoter region and Histone 3 Lysine 36 tri-methylation modifications (H3K36me3) that mark the entire transcribed region (Supplementary Fig. 1), to discover the genomic regions encoding ~1600 large intergenic ncRNAs (lincRNAs) in four mouse cell types4, and ~3300 lincRNAs across 6 human cell types5.
Defining the complete gene structure of these lincRNAs is a pre-requisite for experimental and computational studies of their function. We previously gained initial insights by hybridizing total RNA to tiling microarrays defined across the K4-K36 region4. This provided a coarse list of putative exonic locations, but could not define the precise gene structures and exon connectivity.
Advances in massively-parallel cDNA sequencing (RNA-Seq) have opened the way to unbiased and efficient assays of the transcriptome of any mammalian cell6,7,8–10. Recent studies in mouse and human cells have mostly focused on using RNA-Seq to study known genes6,8,7,10,11, and depended on existing annotations. They were thus of limited utility for discovering the complete gene structure of lincRNAs or other non-coding transcripts.
An alternative strategy is to use an ab initio reconstruction approach9,12–14 to learn the complete transcriptome of an individual sample from only the unannotated genome sequence and millions of relatively short sequence reads. A complete ab initio transcriptome reconstruction of a sample will (1) identify all expressed exons; (2) enumerate all the splicing events that connect them; (3) combine them into transcriptional units; (4) determine all isoforms, including alternative ends, and (5) discover novel transcripts. A successful ab initio method should be applicable to large and complex mammalian genomes, and should be able to reconstruct transcripts of variable sizes, expression levels and protein-coding capacity.
Despite early successes in yeast9, ab initio reconstruction of a mammalian transcriptome has remained an elusive and substantial computational challenge. There has been important recent progress, including (1) efficient gapped aligners (e.g., TopHat13) that can map short reads that span splice junctions (‘spliced reads’); (2) use of such gapped alignments to identify novel splicing events9,13; (3) exon identification methods14; and (4) genome-independent assembly of unmapped reads to sequence contigs (e.g., Abyss12). Each of these methods provides an important component towards reconstruction, but none can reconstruct the complete transcriptome of a mammalian cell, due to scaling issues9, limitations in handling splicing14, or inability to identify transcripts with moderate coverage12.
Here, we present Scripture, a comprehensive method for ab initio reconstruction of the transcriptome of a mammalian cell that uses gapped alignments of reads across splice junctions (exploiting recent increases in read length) and reconstructs reads into statistically significant transcript structures. We apply Scripture to RNA-Seq data from mouse embryonic stem cells (ESC), neural progenitor cells (NPC), and mouse lung fibroblasts (MLF) and correctly identify the complete annotated full-length gene structures for the vast majority of expressed known protein coding genes. The reconstruction of the three transcriptomes reveals substantial variation in protein coding genes between cell types, including thousands of novel 5′-start sites, 3′ ends, or additional coding exons. Many of these variant structures are supported by independent data. We also discover the gene structure and expression level of over 2000 non-coding transcripts, including hundreds of transcripts from previously identified lincRNA loci, over a thousand additional lincRNAs with similar properties, and hundreds of multi-exonic antisense ncRNAs. We show that lincRNAs have no significant coding potential, and that they are evolutionary conserved. Our results open the way to direct experimental manipulation of this new class of genes and highlight the power of RNA-seq along with an ab initio reconstruction to provide a comprehensive picture of cell specific transcriptomes.
RESULTS
RNA-seq libraries
We used massively parallel (Illumina) sequencing to sequence cDNA libraries from polyA(+) mRNA from ESC, NPC and MLF cells, with 76 base paired-end reads. For the ESC library, we generated a total of 152 million paired-end reads. Using a gapped aligner13, 93 million of these were alignable (497Mb aligned bases, 262X average coverage of known protein coding genes expressed in ESC). We obtained similar numbers for the NPC and MLF libraries (Methods). In ESC, 76% of these reads map within the exonic regions of known protein-coding genes, 9% are in introns of known protein coding genes, and 15% map in intergenic regions. We found a strong correlation between expression levels of protein-coding genes as measured by RNA-Seq and Affymetrix expression arrays (r=0.88 for all genes, Supplementary Fig. 2).
Scripture: a statistical method for ab initio reconstruction of a mammalian transcriptome
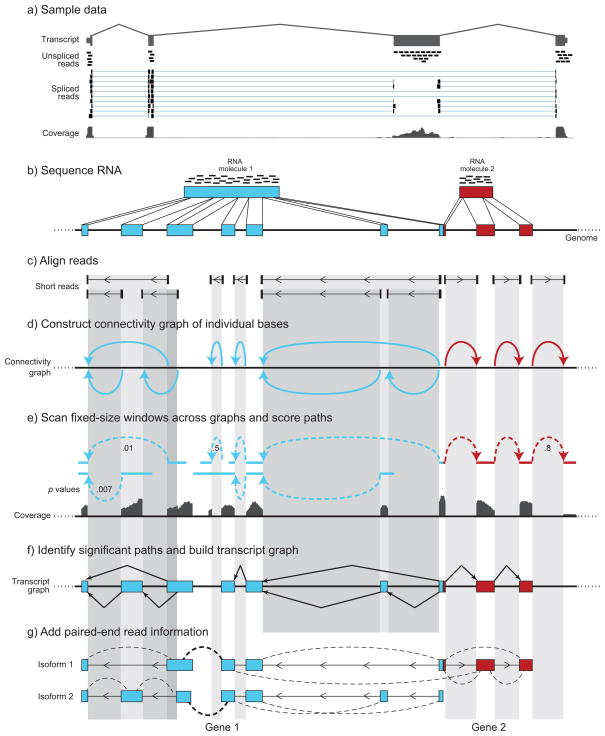
We next developed Scripture, a genome-guided method to reconstruct the transcriptome using only an RNA-Seq dataset and an (unannotated) reference genome sequence. Scripture consists of five steps (Fig. 1, Supplementary Note 1, Methods). First, we use reads aligned to the genome, including those with gapped alignments13 spanning exon-exon junctions (‘aligned spliced reads’, Fig. 1c). ‘Spliced’ reads provide direct information on the location of splice junctions within the transcript, and ~30% of 76 base reads are expected on average to span an exon-exon junction. From the aligned spliced reads, we construct a connectivity graph (Fig. 1d), where two bases in the genome are connected if they are immediate neighbours either in the genomic sequence itself or within a spliced read. We use agreement with splicing motifs at each putative junction to orient the connection (edge) in the connectivity graph9,13 (Fig. 1d). Second, to infer transcripts, we use a statistical segmentation approach4 and both spliced and non-spliced reads to identify paths in the connectivity graph with mapped read enrichment compared to the genomic background (Fig. 1e). This is done by scoring a sliding window using a test statistic for each region, computing a threshold for genome-wide significance, and using the significant windows to define intervals. Third, from the paths, we construct a transcript graph connecting each exon in the transcript (Fig. 1f). Each path through the graph is directed and represents one oriented (strand-specific) isoform of the gene (Fig. 1e). Alternative spliced isoforms are identified by considering all possible paths in the transcript graph. Fourth, we augment the transcript graph with connections based on paired-end reads and their distance constraints, allowing us to join transcripts or remove unlikely isoforms (Fig. 1g, below). Finally, we generate a catalogue of transcripts defined by the paths through the transcript graph.
Figure 1. Scripture: a method for ab initio transcriptome reconstruction from RNA-Seq data.
(a) Spliced and unspliced reads. Shown is a typical expressed 4-exon gene (1500032D16Rik, top, exons: grey boxes) with coverage from different type of reads. Unspliced reads (black bars) fall within a single exon, whereas splice reads (dumbbells) span exon-exon junctions (thin horizontal lines connect the alignment of a read to the exons it spans). The coverage track (bottom) shows the aggregate coverage of both spliced and unspliced reads. (b–g) A schematic description of Scripture. (b) A cartoon example. Reads (black bars) originate from sequencing a contiguous RNA molecule. Shown are transcripts from two different genes (blue and red boxes), one with seven exons (blue boxes) and one with three exons (red boxes), which are adjacent in the genome (black line). The grayscale vertical shading in subsequent panels is shown for visual tracking. (c) Spliced reads. Scripture is initiated with a genome sequence and spliced aligned reads (dumbbells) with gaps in their alignment (thin horizontal lines). Scripture uses splice site information to orient splice reads (arrow heads). (d) Connectivity graph construction. Scripture builds a connectivity graph by drawing an edge (curved arrow) between any two bases that are connected by a spliced read gap. (Edges are color coded to relate to the original RNA and eventual transcript). (e) Path scoring. Scripture scans the graph with fixed-sized windows and uses coverage from all reads (spliced and non-spliced, bottom track) to score each path for significance (p-values shown as edge labels). (f) Transcript graph construction. Scripture merges all significant windows and uses the connectivity graph to give significant segments a graph structure (three graphs in this example). (g) Refinement with paired-end data. Scripture uses paired-end (dashed curved lines) to join previously disconnected graphs (Gene 1, bold dashed line), find break point regions within contiguous segments (e.g. no dashed lines between Gene 1 and 2), and eliminate isoforms that result in paired-end reads mapping at a distance with low likelihood.
Paired-end reads in transcriptome reconstruction and resolution of alternative spliced isoforms
Paired-end information, consisting of reads that came from the two ends of the sequenced RNA fragment, provides valuable additional information in the reconstruction.
First, the presence of paired-ends linking two regions shows that they appear in the same transcript; such a connection might not otherwise be apparent because low expression levels or non-alignable sequence might prevent a continuous chain of overlapping sequence reads (spliced or unspliced) across the transcript. We thus augment the transcript graphs with paired-end information, where available, to (indirectly) link nodes in the graph. We use these indirect links (Fig. 1g) to add edges between disconnected graphs, add internal nodes (exons) that might have been missed within a path (transcript), and add extra support for existing edges. This refines the structure of our transcripts and increases our confidence in them, especially in lowly-expressed transcripts that are more likely to have coverage gaps.
Second, the distribution of library insert sizes constrains the distance between the paired end reads; these distance constraints can be used to infer the relative likelihood of some potential transcripts (for example, those in which the paired ends would be much closer or much further than expected). We infer the distribution of insert sizes for a given library from the position of read pairs on transcripts from those genes for which there is only a single transcript model (i.e., no detectable alternative splicing, Methods). For example, in the ESC library, this distribution matches well with the experimentally determined sizes. Using this distribution we assign likelihoods to each connection, filtering unlikely ones (Methods).
Reconstruction of full-length gene structures
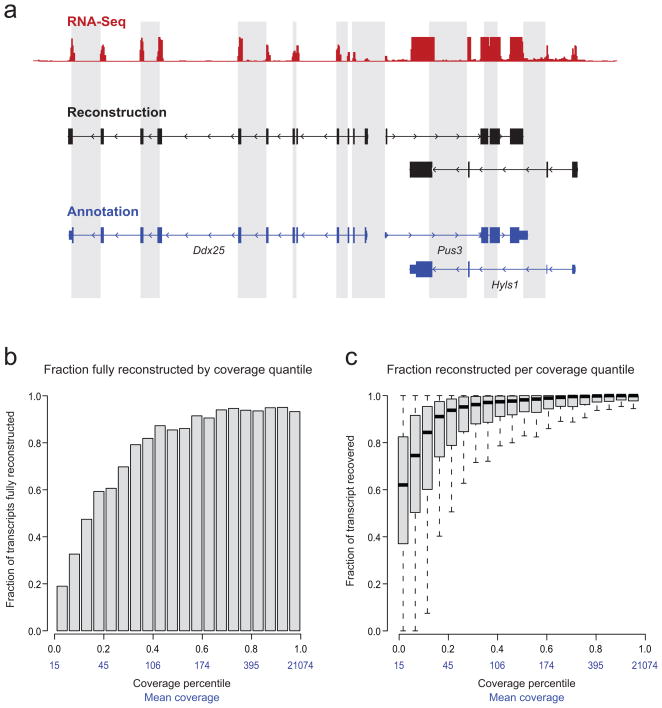
We applied Scripture to our mouse ES RNA-Seq dataset, and compared our reconstructions to protein-coding gene annotations15. Scripture identified 16,389 nonoverlapping, multi-exonic transcript graphs which correspond to 15,352 known multi-exonic genes (Methods). 88.4% of reconstructed genes are covered by a single graph (no fragmentation of the reconstructed transcript) and 8.0% are covered by two transcript graphs (fragmentation of the transcript to two separate pieces in the reconstruction). Focusing on the 13,362 genes with a significant expression level (P<0.05, Methods), Scripture reconstructed the full-length structure of the longest known splice isoform (from 5′ to 3′ end, including all exons and splice junctions, Fig. 2a) for 10,355 of them (~78%). All of our reconstructed transcripts for known multi-exonic transcripts also had the correct orientation (strand), allowing us to reconstruct genes that overlap one another on opposite strands (Fig. 2a).
Figure 2. Scripture correctly reconstructs full length transcripts for the majority of annotated protein coding genes.
(a) A typical Scripture reconstruction on mouse chr9. Top (red) – RNA-Seq read coverage (from both non-spliced and spliced reads); middle (black) – three transcripts reconstructed by Scripture, including exons (black boxes) and orientation (arrow heads); bottom (blue) –RefSeq annotations for this region. All three transcripts are fully reconstructed from 5′ to 3′ ends capturing all internal exons; notice that Scripture correctly reconstructed the overlapping transcripts Pus3 and Hyls1. (b) Fraction of genes fully reconstructed in different expression quantiles (5% increments) in ESC. Each bar represents a 5% quantile of read coverage for genes expressed (mean read coverage is noted in blue). The height of each bar is the fraction of genes in that quantile that were fully reconstructed. For example, ~20% of the transcripts at the bottom 5% of expression levels are fully reconstructed; ~94% of the genes at the top 95% of expression are fully reconstructed. (c) Portion of gene length reconstructed in different expression quantiles in ESC. Shown is a box plot of the portion of each transcript’s length that was covered by a Scripture reconstruction in each 5% coverage quantile. The black line in each box is at the median, the rectangle spans the 25% and 75% coverage quantiles; the whiskers depict the annotations in the quantile most and least covered by our reconstruction. For example, at the bottom 5% of expression, Scripture reconstruct a median length of 60% of the full length transcript.
Complete transcript structures are recovered across a very broad range of expression levels (Fig. 2b,c) for both single and multi-exonic genes. For example, Scripture accurately reconstructs the full-length transcript of ~73% of the known protein-coding genes at the second quintile of expression, and ~94% of the genes from the top quintile. Furthermore, the average proportion of bases constructed for each transcript was high (Fig. 2c). Even for the bottom 5% of expressed genes, we recover on average 62% of each of these transcripts’ bases (Fig. 2c). For single-exon genes, we recover on average 80% of the transcribed bases. We obtained similar results in the other two cell types (19,835 and 20,407 transcript graphs for 14,212 and 13,351 known genes in NPC and MLF, respectively). Most of the genes that are not fully reconstructed are those with low expression levels; it should be possible to reconstruct most of these by generating additional RNA-Seq data. The few highly expressed genes that are not fully reconstructed are either the result of alignment artifacts caused by recent processed pseudogenes or stem from novel transcriptome variations, missing from the current annotation (explored in detail below).
Novel transcriptome variations in annotated protein-coding genes
Given that the vast majority of the Scripture reconstructions of protein-coding genes are extremely accurate, we next investigated the differences between the reconstructed transcriptome and the known gene annotations (Supplementary Table 1). We focused on transcripts with (i) novel 5′ start sites; (ii) novel 3′ ends; and (iii) previously unidentified exons within the transcriptional units of known protein-coding genes. In each category, we first discuss below the reconstructed transcripts in ESC and then consider the results for the NPC and MLF.
(i) Alternative 5′ start sites are supported by H3K4me3 marks
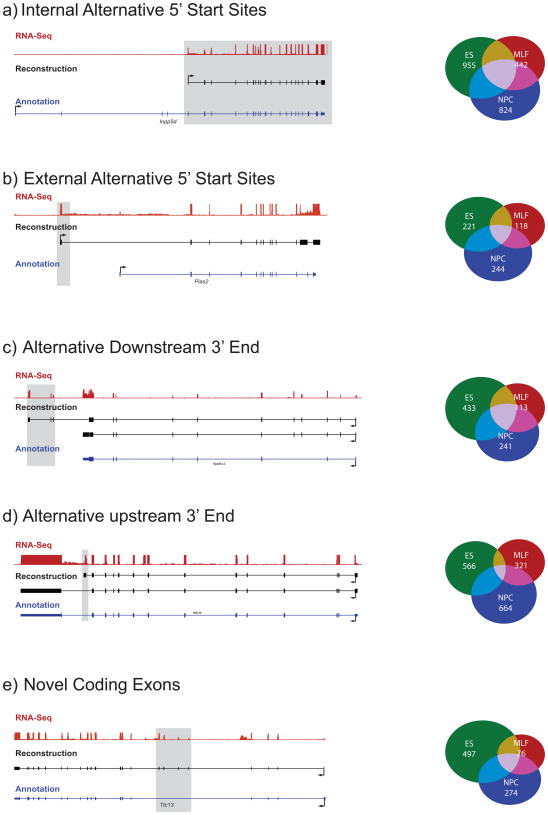
We found 1804 transcripts in ESC that match the annotated 3′-end but have an alternative 5′ start site, derived from an additional exon not overlapping the annotated first exon. We distinguish between internal alternative 5′ start sites (1397 cases, Fig. 3a) that occur downstream of the annotated start, and external alternative 5′ start sites (407 cases, Fig. 3b) that occur upstream of the annotated start. 90% of the internal 5′-start sites and 75% of the external 5′ start sites contain an H3K4me3 modification, a mark of the promoter region of genes16 (Supplementary Fig. 3). These alternative start sites are on average 21kb upstream of the annotated site, substantially revising the annotated promoters. Notably, ~60% of the transcripts with an alternative start site (internal or external) had no reconstructed isoform starting at the annotated 5′-start site.
Figure 3. Alternative 5′ ends, 3′ ends and novel coding exons in transcripts reconstructed by Scripture.
Shown are representative examples (tracks, left) and summary counts (Venn diagrams, right) of five categories of variations discovered in Scripture transcripts compared to the known annotations. In each representative example, shown is the coverage by RNA-Seq reads (top track, red), the reconstructed annotation (middle track, black), and the known annotation (bottom track, blue). The novel regions in the reconstruction are marked by gray shading. In each proportional Venn diagram we show the number of transcripts in this class in each cell type (ESC – green, NPC – blue, MLF – red) and their overlap. (a) Internal alternative 5′ start; (b) External alternative 5′ start; (c) Alternative downstream 3′ end (extended termination); (d) Alternative upstream 3′ end (early termination); (e) Novel coding exons.
We observed similar results from NPC and MLF (Fig, 3a,b, Venn diagrams, Supplementary Table 1). Altogether, we identified 2813 internal 5′ start sites (2302 are supported by K4me3 in their respective tissues), and 807 external 5′ start sites in at least one cell type. In particular, 33% of these novel 5′ ends are likely unique to ESC.
(ii) Alternative 3′ UTRs are supported by polyadenylation motifs
There are 551 (~4%) ESC-reconstructed transcripts with an alternative 3′-end downstream of the annotated 3′-end (mean distance 30 kb downstream, Fig. 3c). Of these, 275 (~50%) have evidence of a polyadenylation motif within the novel 3′ exon, which is only slightly lower than for annotated 3′ ends (60%), and much higher than for randomly chosen size-matched exons (6%). The frequency of the polyadenylation motif supports the accuracy of the reconstruction.
To conservatively distinguish between upstream (early) termination and incomplete reconstruction, we designated novel 3′ ends only in those cases that did not overlap any of the known exons in the annotated transcript and that contained complete 5′ start sites. We identified 759 transcripts with upstream 3′-ends in ESC (Fig. 3d), 44% of them containing a poly-adenylation motif, supporting their biological relevance. For the vast majority (90%) of these transcripts, Scripture also reconstructed an isoform that contained the annotated 3′ end.
We observed similar results for NPC and MLF (Fig. 3c,d, Venn diagrams, Supplementary Table 1). Altogether, we identified 940 downstream 3′ ends and 1850 upstream 3′ ends in at least one cell type.
(iii) Additional coding exons are highly conserved and preserve ORFs
We found 534 transcripts in ESC with at least one additional previously unannotated internal coding exon spliced into annotated protein-coding transcripts (Fig. 3e). These transcripts contained 588 novel internal exons, ranging in length from 6bp to 3.5kb (median 111bp, 60–224 20%–80% quantiles). Of these additional exons, 322 (54.5%) are present in all versions of the reconstructed transcript in ESC. The vast majority (83%) of these novel exons retain the reading frame of the transcript, and are as highly conserved as known coding exons (Supplementary Fig. 4), consistent with their coding capacity. We validated the presence of the novel exons within 5 of 5 tested transcripts, using RT-PCR followed by Sanger sequencing (Methods).
We observed similar results in MLF (124 transcripts, 144 exons) and NPC (325 transcripts, 363 exons) (Fig. 3e, Venn diagram). The majority (~70%) are present in all versions of the reconstructed transcript within a cell type. Altogether, we identified 960 novel internal exons in at least one cell type (Fig. 3e, Venn diagram).
Discovery of the complete gene structures of hundreds of previously identified lincRNA loci
We next turned to identifying the gene structures of transcripts expressed from known lincRNAs loci. We had previously identified 317 lincRNA loci based on K4-K36 domains in ESC cells4. When applied to ESC RNA-Seq data, Scripture reconstructed multi-exonic gene structures for 250 (78.8%) of them (Fig. 4a). This is comparable to the proportion (78.5%) reconstructed for protein-coding genes with K4-K36 domains in ESC. Scripture reconstructed 87% (160/183) of ESC lincRNAs for which we previously identified an RNA hybridization signal from tiling microarrays. We discuss possible reasons for the few remaining discrepancies in Supplementary Note 2.
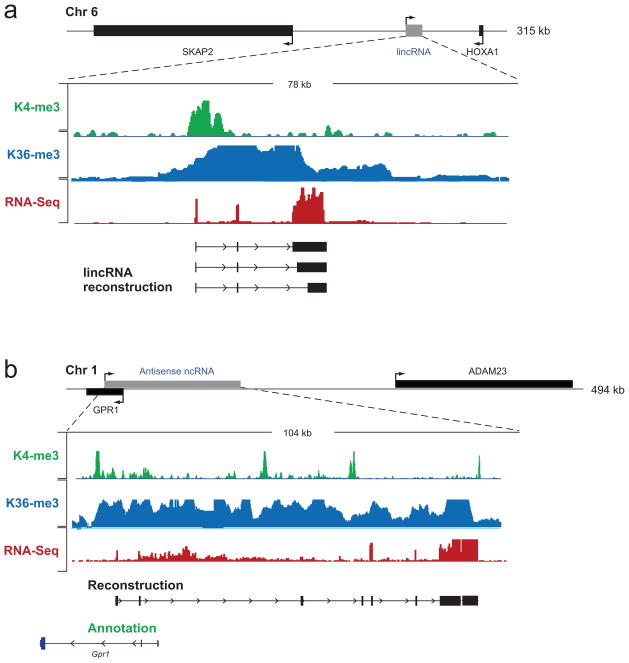
Figure 4. Non-coding transcripts reconstructed by Scripture.
(a) A representative example of a lincRNA expressed in ESC. Top panel – mouse genomic locus containing the lincRNA and its neighbouring protein coding genes. Bottom panel – zoom in on the lincRNA locus showing the coverage of H3K4me3 (green track), H3K36me3 (blue track), and RNA-Seq reads (red track) overlapping the transcribed lincRNA locus, as well as its Scripture reconstructed transcript isoforms (black). (b) A representative example of a multi-exonic antisense ncRNA expressed in ESC. Top panel – mouse genomic locus containing the antisense transcript. Bottom panel – zoom in on the antisense locus showing the coverage of H3K4me3 (green track), H3K36me3 (blue track), and RNA-Seq reads (red track) overlapping the transcribed antisense locus, as well as its Scripture reconstructed gene structure (black).
The reconstructed lincRNA transcripts in ESC have 3.7 exons on average, an average exon size of 350 bp, and an average mature spliced size of 3.2 kb (compared to 9.7 exons, exon length of 291 bp, and average length of 2.9kb for protein coding genes). The Scripture-identified strand information for each lincRNA is consistent with that inferred from the location of K4me3 modification, and with the orientation determined from a strand-specific RNA-Seq library which we generated independently (Methods). The majority of lincRNAs likely represent 5′ complete transcripts based on overlap with H3K4me3 (82%) and 3′ complete transcripts based on presence of a polyadenylation motif (~50%, comparable to 60% for protein-coding genes and far above background of 6%).
Similarly, Scripture successfully reconstructed lincRNA gene structures for K4-K36 lincRNA loci in MLF and NPC (232 of 289 in MLF and 224 of 270 in NPC). Most are likely 5′ complete (69% in MLF and 81% in NPC based on overlap with H3K4me3) and many may be 3′ complete based on detectable 3′ polyadenylation sites (18% in MLF and 37% in NPC). In addition, we successfully reconstructed another 116 lincRNAs previously identified only in mouse embryonic fibroblasts but which were now reconstructed in at least one of the other three cell types. Altogether, we identified gene structures for 609 previously defined lincRNA loci in at least one of the three cell types.
Discovery of novel lincRNAs
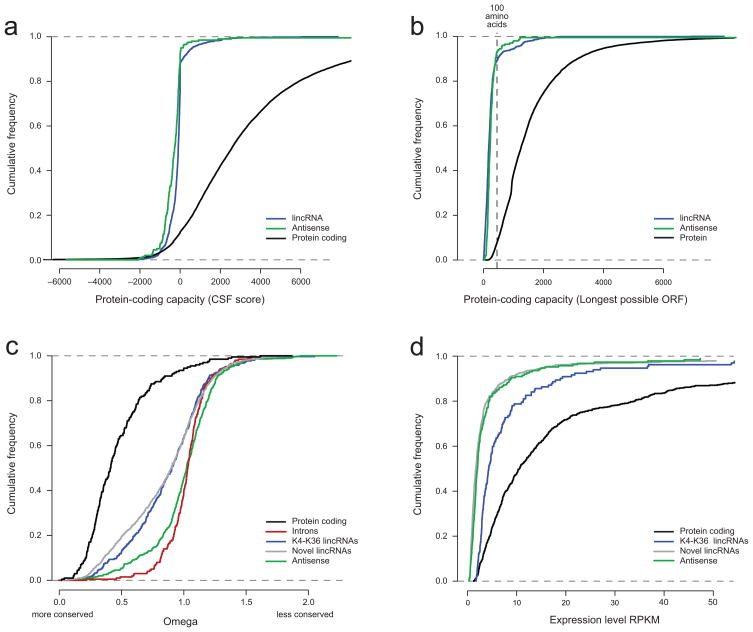
In addition to the previously identified lincRNAs, we found another 1140 multi-exonic transcripts that map to intergenic regions (591 in ESCs, 318 in MLF, and 528 in NPC). The majority of these transcripts do not appear to encode proteins, and are designated as non-coding, based on their Codon Substitution Frequency (CSF) scores17–18 (Methods) across the mature (spliced) RNA transcript (88%, Fig. 5a), and the lack of an open reading frame (ORF) larger than 100 amino acids (80%, Fig. 5b). Careful review of the remaining ~12%, reveals 66 loci that are likely to be novel protein coding genes (high CSF score, ORF >200 amino acids, and very high evolutionary conservation, Supplementary Fig. 5).
Figure 5. Protein coding capacity, conservation levels and expression of lincRNAs and multi-exonic antisense transcripts.
(a–b) Coding capacity of protein coding, lincRNAs and multi-exonic antisense transcripts. Shown is the cumulative distribution of CSF scores (a) and maximal ORF length (b) for protein coding transcripts (black), lincRNAs (blue) and multi-exonic anti-sense transcripts (green). (c) Conservation levels for exons from protein coding transcripts, lincRNAs, multi-exonic antisense transcripts and introns. Shown is the cumulative distribution of sequence conservation across 29 mammals for exons from protein-coding exons (black), introns (red), exons from previously annotated lincRNA loci (blue), exons from newly annotated lincRNA transcripts (grey), and exons from multi-exonic antisense transcripts (green). (d) Expression levels of protein coding, lincRNAs and multi-exonic antisense transcripts. Shown is the cumulative distribution of expression levels (RPKM) in ESC for protein coding transcripts (black), transcripts from previously annotated lincRNA loci (blue), transcripts from newly annotated lincRNA loci (gray), and multi-exonic antisense transcripts (green).
Most of the novel lincRNA loci were not identified in our previous study due to the stringent criteria we imposed when using chromatin maps to identify lincRNAs. Specifically, we required that a K4-K36 domain extend over at least 5 Kb and be well-separated from the nearest known gene locus4. Indeed, the vast majority of novel intergenic transcripts (76%) were enriched for a K4-K36 domain (a comparable proportion as for expressed protein-coding genes) but failed to meet the other two criteria or were too weak to be identified at a genome-wide significance (without knowing their locus a priori). On average, the genomic loci of the novel lincRNAs are closer to neighboring genes, have smaller genomic sizes (~3.5Kb average) and shorter transcript lengths (859bp). Of the lincRNAs that did not have a chromatin signature that reached genome-wide significance, ~40% showed chromatin modifications enriched at a nominal significance level (compared to 57% for protein coding genes).
On average, the lincRNAs are expressed at readily detectable levels, albeit somewhat lower than those of protein-coding genes. The median expression level of the reconstructed lincRNAs (as estimated by RPKM, Methods) is approximately 3-fold lower than that of protein-coding genes (Fig. 5d), with ~25% of lincRNAs having expression levels higher than the median level for protein-coding genes (Fig. 5d). The novel lincRNAs identified in this study are expressed at somewhat lower levels than those from chromatin identified loci, consistent with the fact that chromatin enrichment is positively correlated with expression levels (Fig. 5d).
We compared the novel lincRNA genes to a collection of ~35,000 mouse cDNA and found evidence that ~43% of our lincRNAs were present in this collection1. This is comparable to the reported fraction (40%) of known transcripts covered by the same cDNA catalogue1. The remaining lincRNAs are unique to this study. These were likely previously missed due to the different cell types and limited coverage of the previous study1.
Most lincRNAs are evolutionarily conserved, with 22% of bases under purifying selection
The reconstructed full-length gene structures of lincRNAs allow us to accurately assess their evolutionary sequence conservation in each exon and in small windows. To this end, we identified the orthologous sequences for each lincRNA across 29 mammals and estimated conservation by a metric (ω, Methods) reflecting the total contraction of the branch length of the evolutionary tree connecting them19. We calculated ω over the entire lincRNA transcript, as well as over individual exons.
Based on our high resolution gene structures, the lincRNA sequences show significantly greater conservation than random genomic regions or introns (Fig. 5c), comparable to 8 known functional lincRNAs20,21,22, and lower than protein-coding exons. The results are consistent with our previous estimates of conservation4. Interestingly, conservation levels are indistinguishable between the chromatin defined lincRNAs4 and the novel ones identified only in this study (Fig. 5c), consistent with membership in the same class of functional large ncRNA genes. These conservation levels are considerably higher than those reported for a previous catalogue of large non-coding RNAs1.
We also determined the specific regions within each lincRNA that are under purifying selection and thus likely to be functional, by computing ω within short windows (Methods). On average, 22% of the bases within the lincRNAs lie within conserved patches (comparable to 25% for the 8 known functional lincRNAs, much higher than 7% for intronic bases and lower than 77% of protein coding bases, Supplementary Fig. 6). These conserved patches provide a critical starting point for functional studies23.
Variations in lincRNA expression and isoforms
A substantial fraction (~41%) of the novel lincRNAs reconstructed in at least one cell type shows evidence for expression in at least two of the three cell types. This is comparable to the 45% of the previously identified lincRNAs present in at least 2 out of the 3 cell types. In contrast, 80% of expressed protein coding genes are expressed across two of the three cell types. This is not merely a result of the lower overall expression of lincRNAs, since the fraction of cell-type specific lincRNAs is higher than that of tissue specific protein-coding genes in every expression quantile (Supplementary Fig. 7). Thus, lincRNAs are likely to be more tissue-specific than protein coding genes.
A substantial portion of lincRNA loci also produce alternative spliced isoforms. For example, within ESC we identified two or more alternative spliced isoforms for 25% of lincRNA genes, comparable for 30% of protein coding genes (15% of lincRNAs in MLF have alternative spliced isoforms, and 14.7% in NPC). Altogether, 28.8% of the 1749 lincRNA loci have evidence for alternative isoforms in any of the three cell types.
Identification of hundreds of large antisense transcripts
Scripture reconstructed hundreds of transcripts that overlap known protein-coding gene loci but are transcribed in the opposite orientation and likely represent anti-sense transcripts. To determine orientation, we required that any identified antisense transcript be multi-exonic (Methods).
Using these criteria, we identified 201 antisense multi-exonic transcripts in ESC (Fig. 4b); these transcripts have an average 5 exons per transcript and an average transcript size of 1.7Kb. On average, the antisense transcripts overlap the genomic locus of the sense protein coding gene by 1023 bp (83% of the transcript length), and most (64%) overlap at least one sense exon, but this overlap is substantially lower (766 bp, 48%). Some of these antisense transcripts (79, ~40%) were identified by a previous cDNA sequencing study1,24, but the majority (122, ~60%) were previously unidentified. Most (~85%) of anti-sense transcripts are non-protein coding by both ORF analysis (Fig. 5b) and CSF scores (Fig. 5a). Four of the newly identified antisense transcripts had a large, conserved open reading frame and are likely novel, previously unannotated protein coding genes.
We validated the reconstructed ESC anti-sense transcripts by three independent sets of experimental data. First, the majority of the anti-sense loci carry an H3K4me3 mark at their 5′-end (Fig. 4b), consistent with their independent and antisense transcription (e.g., 64% of the 164 transcripts where it is possible to detect an independent H3K4me3 mark, because the 5′-end of the anti-sense transcript does not overlap the 5′-ends of the sense gene). Second, we generated and sequenced a strand-specific library in ESC (17.5M reads, Illumina, Methods), and found a significant number of reads on the anti-sense strand in >90% of cases (the remaining are likely missed in this limited sequencing due to lower expression). Finally, we confirmed 5 of 5 tested anti-sense transcripts using RT-PCR to unique exons of the antisense transcript (Methods) followed by Sanger sequencing.
We obtained similar results for anti-sense transcripts in MLF and NPC (112 and 202 multi-exonic antisense transcripts, respectively). Altogether, we identified 469 antisense transcripts expressed in at least one cell type, only 125 of which (27%) were previously identified in large scale sequencing of mouse cDNAs24. The remaining 344 (73%) were previously unidentified by this study, likely reflecting the distinct cell types used in this study, and the limited coverage of previous catalogues.
The 469 anti-sense transcripts are expressed at comparable levels to the novel lincRNAs (Fig. 5d), but show substantially lower sequence conservation. Indeed, the antisense ncRNAs showed very little evolutionary conservation as estimated by the ω metric for the portions that do not overlap protein-coding exons on the sense strand, suggesting that the antisense ncRNAs are a distinct class from the lincRNAs (Fig. 5c).
DISCUSSION
Despite the availability of the genome sequence of many mammals, a comprehensive understanding of the mammalian transcriptome has been an elusive goal. In particular, the computational tools needed to reconstruct all full-length transcripts from the wealth of short read data were largely missing. A recent study proposed to overcome this limitation experimentally by using very long reads (e.g.. 454 sequencing), as a scaffold for short read reconstruction25. This is applicable, albeit at a substantial cost, for highly expressed genes, but would require extraordinary depth to cover more lowly expressed ones.
Here, we present Scripture, a novel computational method to reconstruct a mammalian transcriptome with no prior knowledge of gene annotations. Scripture relies on longer reads that span splice junctions to connect discontiguous (spliced) segments, resolve multiple splice isoforms, and leverages paired-end information to refine these transcripts. Scripture can identify short but strongly expressed transcripts as well as much lower expressed transcripts for which there is aggregate evidence along the entire transcript length. While Scripture does rely on a reference genome sequence, many of its components can also be used in the development of methods for assembly of transcripts from read data only.
We applied Scripture to RNA-Seq data from pluripotent ES cells and differentiated lineages and showed that we can accurately reconstruct the majority of expressed annotated protein coding genes, at a broad range of expression levels, as well as uncover a large number of novel isoforms in the protein-coding transcriptome. This variation may play key regulatory roles, defining new cell-type specific promoters, UTRs and protein-coding exons. We leveraged Scripture’s sensitivity and resolution to reconstruct the gene structures and strand information of hundreds of lincRNAs and multi-exonic antisense transcripts, many of whom are only moderately expressed.
Scripture identified over a thousand lincRNAs across the three cell types studied. The substantial majority of the lincRNAs identified were not previously found by classical large-scale cDNA sequencing1. Many of these lincRNAs could not be reliably identified solely on the basis of chromatin structure, owing to their proximity to protein-coding genes or their short genomic lengths. Overall, we find that the ratio of expressed protein-coding to non-coding genes in these cell types is ~10:1, but that the total number of RNA molecules is more heavily biased toward the protein-coding fraction (~30:1), similar to previous observations26.
Scripture identifies precise gene structures for the majority of previously found lincRNA loci (as well as for the newly discovered ones), a pre-requisite for further studies. For example, we used these to identify the specific regions within each lincRNA that are under purifying selection (conservation), a starting point for experimental and computational investigation.
Taken together our results highlight the power of ab initio reconstructions to discover novel transcriptional variation within known protein coding genes, and provide a rich catalog of precise gene structures for novel non-coding RNAs. The next step is clearly to apply this approach to a wide range of mammalian cell types, to obtain a comprehensive picture of the mammalian transcriptome.
Data Availability
The sequencing data in this study is available at the NCBI Gene Expression Omnibus (GEO) under accession number GSE20851. The Scripture method is implemented as a stand-alone Java application and is available at www.broadinstitute.org/software/Scripture/, along with all assembled transcripts in both GFF and BED file formats. Additionally, all transcript graphs are available in the dot graph language.
MATERIALS AND METHODS
Cell culture
Mouse embryonic stem cells (V6.5) were co-cultured with irradiated MEFs (GlobalStem; GSC-6002C) on 0.2% gelatin coated plates in a culture media consisting of Knockout DMEM (Invitrogen; 10829018) containing 10% FBS (GlobalStem; GSM-6002), 1% pen-strep 1% Non-essential amino acids, 1% L-glutamine, 4ul Beta-mercaptoethanol, and .01% LIF (Millipore; ESG1106). ESC were passaged once on gelatin without MEFs before RNA extraction. V6.5 ES cells were differentiated into neural progenitor cells (NPC) through embryoid body formation for 4 days and selection in ITSFn media for 5–7 days, and maintained in FGF2 and EGF2 (R&D Systems) as described27. The cells uniformly express Nestin and Sox2 and can differentiate into neurons, astrocytes and oligodendrocytes. Mouse lung fibroblasts (ATCC), were grown in DMEM with 10% fetal bovine serum and penicillin/streptomycin at 37°, 5% CO2.
RNA Extraction & Library Preparation
RNA was extracted using the protocol outlined in the RNeasy kit (Qiagen). Extracts were treated with DNase (Ambion 2238). Polyadenylated RNAs was selected using Ambion’s MicroPoly(A)Purist kit (AM1919M) and RNA integrity confirmed using Bioanalyzer (Agilent). We used a cDNA preparation procedure that combines a random priming step with a shearing step8–9,28 and results in fragments of ~700 bp in size. We previously found9,28 that this protocol provides relatively uniform coverage of the whole tanscript, thus assisting in ab initio reconstruction. Specifically, a ‘regular’ RNA sequencing library (non strand specific) was created as previously described28, with the following modifications. 250 ng of polyA+ RNA was fragmented by heating at 98°C for 33 minutes in 0.2 mM sodium citrate, pH 6.4 (Ambion). Fragmented RNA was mixed with 3 μg random hexamers, incubated at 70°C for 10 minutes, and placed on ice briefly before starting cDNA synthesis. First strand cDNA synthesis was performed using Superscript III (Invitrogen) for 1 hour at 55°C, and second strand using E. coli DNA polymerase and E. coli DNA ligase at 16°C for 2 hours. cDNA was eluted using Qiagen MiniElute kit with 30ul EB buffer. DNA ends were repaired using dNTPs and T4 polymerase, (NEB) followed by purification using the MiniElute kit. Adenine was added to the 3′ end of the DNA fragments to allow adaptor ligation using dATP and Klenow exonuclease (NEB; M0212S) and purified using MiniElute. Adaptors were ligated and incubated for 15 minutes at room temperature. Phenol/choloform/isoamyl alcohol (Invitrogen 15593-031) extraction followed to remove the DNA ligase. The pellet was then resuspend in 10ul EB Buffer. The sample was run on a 3% Agarose gel (Nusieve 3:1 Agarose) and a 160 – 380 base pair fragment was cut out and extracted. PCR was performed with Phusion High-Fidelity DNA Polymerase with GC buffer (New England Biolabs) and 2M Betaine (Sigma). [PCR conditions: 30 sec at 98°C, (10 sec at 98°C, 30 sec at 65°C, 30 sec at 72°C -16 cycles) 5 min at 72°C, forever at 4°C], and products were run on a poly-acrylamide gel for 60 minutes at 120 volts. The PCR products were cleaned up with Agencourt AMPure XP magnetic beads (A63880) to completely remove primers and product was submitted for Illumina sequencing.
The “strand-specific” library was created from 100 ng of polyA+ RNA using the previously published RNA ligation method29 with modifications from the manufacturer (Illumina, manuscript in preparation). The insert size was 110 to 170 bp.
RNA-Seq library sequencing
All libraries were sequenced using the Illumina Genome Analyzer (GAII). We sequenced 3 lanes for ESC corresponding to 152 million reads, 2 lanes for MLF corresponding to 161 million reads, and 2 lanes for NPC corresponding to 180 million reads.
Alignments of reads to the genome
All reads were aligned to the mouse reference genome (NCBI 37, MM9) using the TopHat aligner13. Briefly, TopHat uses a two-step mapping process, first uses Bowtie30 to align all reads that map directly to the genome (with no gaps), and then maps all the reads that were not aligned in the first step using gapped alignment. TopHat uses canonical and non-canonical splice sites to determine possible locations for gaps in the alignment.
Generation of connectivity graph
Given a set of reads aligned to the genome, we first identified all spliced reads, as those whose alignment to the reference genome contains a gap. These reads and the reference genome are used to construct connectivity graphs. Each connectivity graph contains all bases from a single chromosome. The nodes in the graph are bases and the edges connect each base to the next base in the genome as well as to all bases to which it is connected through a ‘spliced’ read (Fig. 1). In the analysis presented, we defined an edge between any two bases in the chromosome that were connected by two or more spliced reads. The connectivity graph thus represents the contiguity that exists in the RNA but that is interrupted by intron sequences in the reference genome.
Identification of splice site motifs and directionality
We restricted our analysis to splice reads that mapped connecting donor/acceptor splice sites, either canonical (GT/AG) or non-canonical (GC/AG and AT/AC). We oriented each mapped spliced read using the orientation of the donor/acceptor sites it connected.
Construction of transcript graphs
The ‘spliced’ edges in the connectivity graph reflect bases that were connected in the original RNA but are not contiguous in the genome. To construct a transcript graph, we ‘thread’ the connectivity graph (which was constructed only from the genome and spliced reads) with the non-spliced (contiguous) reads, to provide a quantitative measure of the reads supporting each base and edge. We then use a statistical segmentation strategy to traverse the graph topology directly and determine paths through the connectivity graph that represent a contiguous path of significant enrichment over the background distribution (below). In this segmentation process, we scan variable sized windows across the graph and assign significance to each window. We then merge significant paths into a transcript graph. Specifically, for a window of fixed size, we slide the window across each base in the connectivity graph (after augmenting it with the non-spliced reads). If a window contains only contiguous non-spliced reads, then it represents a non-spliced part of the transcript. However, if the window hits an edge in the connectivity graph connecting two separate parts of the genome (based on two or more spliced reads), then the path follows this edge to a non-contiguous part of the genome, denoting a splicing event. Similarly, when alternative splice isoforms are present, if a base connects to multiple possible places, then all windows across these alternative paths are computed. Using a simple recursive procedure we can compute all paths of a fixed size across the graph.
Identification of significant segments
To assess the significance of each path, we first define a background distribution. We estimate a genomic defined background distribution by permuting the read alignments in the genome and counting the number of reads that overlap each region and the frequency by which they each occur. Specifically, if we are interested in computing the probability of observing alignment a (of length r) at position i (out of a total genome size of L) we can permute the alignments and ask how often read a overlaps position i. Under this uniform permutation model, the probability that read a overlaps position i is simply r/L. Extending this reasoning, we can compute the probability of observing k reads (of average length r) at position i as the binomial probability. Given the large number of reads and the large genome size, the binomial formula can be well approximated by a Poisson distribution where λ=np (or the number of reads/number of possible positions).
Given a distribution for the real number of counts over each position we scan the genome for regions that deviate from the expected background distribution. First consider a fixed window size w. We slide this window across each position (allowing for overlapping windows), and compute the probability of each observed window based on a Poisson distribution with λ=wnp. Since we are sliding this window across a genome of size L, we correct our nominal significance for multiple testing, by computing the maximum value observed for a window size (w) across a number of permutations of the data. This distribution controls the family-wise error rate, defined as the probability of observing at least one such value in the null distribution31. Notably, we can estimate this maximum permutation distribution well by a distribution known as the scan statistic distribution32, which depends on the size of the genome that we scan, the window size used, and our estimate of the Poisson λ parameter. This method provides us with a general strategy to determine a multiple testing corrected P-value for a specified region of the genome in any given sample. We use this method to compute a corrected significance cutoff for any given region.
Finally, to identify significant intervals, we scan the genome using variable sized windows, computing significance values for each and filtering by a 5% significance threshold. For each window size, we merge the significant regions that passed this cutoff into consecutive intervals. We trim the ends of the intervals as needed, since we are computing significant windows (rather than regions) and it is possible that an interval need not be fully contained within a significant region. Trimming is performed by computing a normalized read count for each base in the interval compared to the average number of reads in the genome. We then trim the interval to the maximum contiguous subsequence of this value. We test this trimmed interval using the scan procedure and retain it only if it passes our defined significance level.
We work with a range of different window sizes in order to detect paths (intervals) with variable support, Small windows have power to identify short regions of strong enrichment (e.g. short exon which is highly expressed), whereas long windows capture long contiguous regions with often lower and more ‘diffuse’ enrichment levels (e.g. a longer lower expression transcript, whose ‘moderate evidence’ aggregates along its entire length).
Estimation of library insert size
We estimated the insert size distribution by taking all reconstructed transcripts for which we only reconstructed a single isoform and computing the distribution of distances between the paired-end reads that aligned to them.
Weighting of isoforms using paired end edges
Using the size constraints imposed by the length of the paired ends, we assigned weights to each path in the transcript graph. We classified all paired ends overlapping a given path and assigned them to all possible paths that they overlapped. We then assigned a probability to each paired end of the likelihood that it was observed from this transcript given the inferred insert size for the pair in that path. We used an empirically determined distribution of insert sizes, estimated from single isoform graphs. We then scaled each value by the average insert size. We refer to this scaled value as our insert distribution. For each paired end in a path, we computed I, the inferred insert size (the distance between nodes following along the full path) minus the average insert size. We then determined the probability of I as the area in our insert distribution between −I, I. This value is the probability of obtaining the observed paired end insert distance given this distribution of paired end reads. To aggregate these into weights for each path, we simply weight each paired end by its probability of observing to the given path. Paired ends that equally support multiple isoforms will count equally for all, but paired ends with biases toward some isoforms and against others will provide weighted evidence for each isoform. We assign this weight to each isoform path. This score is normalized by the number of paired ends overlapping the path. We filter paths with little support (normalized score<0.1) of paired reads supporting it.
Determination of expression levels from RNA-Seq data
Expression levels are computed as previously described8. Briefly, the expression of a transcript is computed in Reads Per Kilobase of exonic sequence per Million aligned reads (RPKM) defined as: , where r is the number of reads mapped to the exonic region of the transcript, t is the total exonic length of the transcript, and R is the total number of reads mapped in the experiment.
Array expression profiling in ESC cells
Microarray hybridization data was obtained from our previous studies including ESC, NPC16 and MLF4.
Comparisons to known annotation
The reconstructed transcripts were compared to the RefSeq genome annotation15 (NCBI Release 39). To determine whether a known annotation of a protein coding gene from RefSeq was fully reconstructed, we first compared the 5′ and 3′ ends of the reconstructed vs the annotated transcript. If these overlapped, we further verified that all exons in the annotated transcript matched those in the reconstructed version. To score the portion of an annotated transcript covered by our reconstructions, we found the reconstructed transcript whose exons covered the largest fraction of the annotated transcript, and reported the portion of the annotation that it covered.
ChIP-seq profiles in ESC cells and determination of K4 and K36 regions
To determine regions enriched in chromatin marks from ChIP-seq data we used our previously described method4, applied to ESC, MLF, and NPC data4,16.
Determination of external and internal 5′ start sites
We identified alternative 5′ start sites by comparing the 5′ exon of our reconstructed transcripts to the location of the 5′ exon of the annotated gene overlapping it. If the reconstructed 5′ start site resided upstream to the annotated 5′ we termed it ‘external start site’. For the novel 5′ ends that are downstream of the annotated 5′ end (internal) we required a few additional criteria to avoid reconstruction biases due to low coverage. First, we required that the novel internal 5′ end do not overlap any of the known exons within the known gene. Second, we required that the reconstructed gene contains a completed 3′ end. To determine the presence of H3K4me3 modifications overlapping the promoter regions defined by these novel start sites, we computed regions of enriched K4me3 genome-wide (as previously described) and intersected the location of the novel 5′ exon (both internal and external) with the location of a K4me3 peak.
Determination of premature/extended 3′ end
To determine novel 3′ ends, we compared the locations of the 3′ exon of our reconstructed 3′ ends and those of annotated genes. If the reconstruction extended past the annotated 3′ end, we classified it as an extended 3′ end. If the reconstruction ended before the annotated 3′ end we required that it not overlap any known exon and have a fully reconstructed 5′ start site.
Determination of sequence conservation levels
We used the SiPhy19 algorithm and software package (http://www.broadinstitute.org/genome_bio/siphy/) to estimate ω, the deviation (‘contraction’ or ‘extension’) of the branch length compared to the neutral tree based on the total number of substitutions estimated from the alignment of the region of interest across 20 placental mammals (build MM9, http://hgdownload.cse.ucsc.edu/goldenPath/mm9/multiz30way/). For global (whole transcript) conservation, we estimated ω for each protein coding, lincRNA and antisense transcript exon and compared it to similarly sized regions within introns. To identify local regions of conservation within a transcript, we computed ω for all 12-mers within the transcript sequence, and assigned a p-value for each 12-mer based on the chi-square distribution, as previously described19. We then took all 12-mers showing significance at p< 0.05, collapsed overlapping 12-mers, and identified constrained regions within the transcript (e.g. Supplementary Fig. 6).
ORF determination
We estimated maximal supported open reading frames (ORFs) for each transcript built by scanning for start codons and computing the length (in nucleotides) until the first stop codon was reached.
CSF Scores
To further estimate the coding potential of novel transcripts, we evaluated whether evolutionary sequence substitutions were consistent with the preservation of the reading frame of any detected peptide. In a nutshell, if a transcript encodes a protein, we expect a reduction in frame shifting indels, non synonymous changes and, in general, any substitution that affects the encoded protein. To assess this, we used Codon Substitution Frequency (CSF) method as previously described17–18.
RT-PCR validations
Primers were obtained for a randomly selected set of predicted lincRNA, protein coding genes, antisense transcripts, and intron primers (Supplementary Table 2); all begining with M13 primer sequence. RNA from ESC cells was extracted using Qiagen’s RNeasy kit (74106). A one-step cDNA/RT-PCR reaction was run using Invitrogen’s one-step RT-PCR kit (12574-018), following the manufacturer’s instructions, with the following PCR protocol: 55°C for 30 minutes, 94°C for 2 minutes (94°C for 15 seconds, 64°C for 30 seconds, 68°C for 1 minute – 40 cycles) 68°C for 5 minutes, 4°C forever. Samples were separated on a 3% agarose gel, and all bands were cut out and gel extracted using the QIAquick Gel Extraction Kit 28706. 30ng of DNA were mixed with 3.2pmol M13 forward or M13 reverse primer for sequencing.
Supplementary Material
Supplementary Figure 1. Comparison of RNAseq and tiling array gene structures. Shown is a genomic locus for an example lincRNA. Shown are K4me3 (green) and K36me3 (blue) chromatin marks, hybridization signal from a tiling array (grey), RNA-Seq read coverage (red) and Scripture reconstructions. The reconstruction is at substantially higher resolution than the tiling array information, and it identified exons and connectivity missed by the tiling arrays.
Supplementary Figure 2. Correlation in expression levels between our RNA-Seq sample and Affymetrix arrays. Shown is a density scatter plot, showing the estimated expression ranks for annotated protein coding genes from RNA-seq data (by RPKM) and Affymetrix arrays (by MAS 5.0 estimation). Darker shades indicate more genes and lighter shades indicated fewer genes.
Supplementary Figure 3. H3K4me3 support for an alternative 5′ end. Shown is the genomic locus at chr18:77,304,429-77,392,539, along with known annotated transcripts (blue, bottom), the reconstructed transcript from ESC (black) with an alternative external 5′ end, and K4me3 data (green tracks, top) for four cell types. The alternative 5′ end in ESC is associated with an ESC-specific K4me3 mark.
Supplementary Figure 4. Conservation of novel protein coding exons. Shown is the cumulative distribution of sequence conservation across 29 mammals (ω) for novel protein coding exons (blue), annotated protein coding exons (green), and introns (red). The novel exons are conserved at comparable levels to annotated ones.
Supplementary Figure 5. Conservation of novel protein coding genes. Shown is the cumulative distribution of sequence conservation across mammals (ω) for exons from novel protein coding genes (blue), annotated protein coding genes (green), and introns (red). The novel genes are conserved at comparable levels to annotated ones.
Supplementary Figure 6. Patches of conservation in lincRNAs. Shown are two examples of lincRNAs and the conservation patterns across their exons. In each panel; shown is the relevant genomic locus (top) with a zoom in (bottom), listing ω (top, green), RefSeq gene anontations (middle, blue tracks), and 30-way multiz alignment tracks (bottom, black). (a) Known functional lincRNA, XIST. Zoom in on its 5′ exon illustrates the conservation patterns within the exon. (b) A novel lincRNA. Zoom in on one of its exons illustrates the conservation patterns within the exons, comparable to those in (a).
Supplementary Figure 7. LincRNAs are more cell type specific across different expression levels. Shown are the fractions of ES specific transcripts for lincRNAs (dark bars) and protein coding genes (light bars) at different ESC expression quantiles. In each quantile, the fraction of lincRNAs with ES specific expression is higher than the fraction of protein coding transcripts with ES specific expression.
Acknowledgments
We thank Marius Wernig for providing NPC, Mike Lin and Manolis Kellis for CSF code, the Broad Sequencing Platform for sample sequencing, Leslie Gaffney for assistance with graphics, Chris Burge, Jason Merkin, Rob Bradley and members of Lander and Regev labs, in particular Moran Yassour, Tarjei Mikkelsen, and Ido Amit for helpful discussions. AR and JLR are fellows of the Merkin Family Foundation for Stem Cell Research at the Broad Institute. M. Guttman is a Vertex scholar. Work was supported by a Burroughs Wellcome Fund Career Award at the Scientific Interface, an NIH PIONEER award, an NHGRI R01, and the Howard Hughes Medical Institute (AR), and NHGRI and the Broad Institute of MIT and Harvard (ESL).
References
- 1.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (New York, NY) 2007;316:1484–1488. doi: 10.1126/science.1138341. 1138341 [pii] 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science (New York, NY) 2004;306:2242–2246. doi: 10.1126/science.1103388. 1103388 [pii] 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. nature07672 [pii] 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America; 2009. pp. 11667–11672. 0904715106 [pii] 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. nmeth.1223 [pii] 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 7.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. nature07509 [pii] 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. nmeth.1226 [pii] 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 9.Yassour M, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci U S A. 2009;106:3264–3269. doi: 10.1073/pnas.0812841106. 0812841106 [pii] 10.1073/pnas.0812841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. ng.259 [pii] 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 11.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. nature07638 [pii] 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birol I, et al. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25:2872–2877. doi: 10.1093/bioinformatics/btp367. btp367 [pii] 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. btp120 [pii] 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denoeud F, et al. Annotating genomes with massive-scale RNA sequencing. Genome Biol. 2008;9:R175. doi: 10.1186/gb-2008-9-12-r175. gb-2008-9-12-r175 [pii] 10.1186/gb-2008-9-12-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. gkl842 [pii] 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. nature06008 [pii] 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin MF, Deoras AN, Rasmussen MD, Kellis M. Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PLoS Comput Biol. 2008;4:e1000067. doi: 10.1371/journal.pcbi.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin MF, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. gr.6679507 [pii] 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garber M, et al. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics (Oxford, England) 2009;25:i54–62. doi: 10.1093/bioinformatics/btp190. btp190 [pii] 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 21.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. 309/5740/1570 [pii] 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 25.Wu JQ, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0914114107. 0914114107 [pii] 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. 04-PLBI-RA-0934R2 [pii] 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger MF, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010 doi: 10.1101/gr.103697.109. gr.103697.109 [pii] 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. S0092-8674(08)00448-0 [pii] 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. gb-2009-10-3-r25 [pii] 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewens WJ, Grant GR. Statis[t]ical methods in bioinformatics: an introduction. 2. Springer; 2005. [Google Scholar]
- 32.Glaz J, Naus JI, Wallenstein S. Scan statistics. Springer; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Comparison of RNAseq and tiling array gene structures. Shown is a genomic locus for an example lincRNA. Shown are K4me3 (green) and K36me3 (blue) chromatin marks, hybridization signal from a tiling array (grey), RNA-Seq read coverage (red) and Scripture reconstructions. The reconstruction is at substantially higher resolution than the tiling array information, and it identified exons and connectivity missed by the tiling arrays.
Supplementary Figure 2. Correlation in expression levels between our RNA-Seq sample and Affymetrix arrays. Shown is a density scatter plot, showing the estimated expression ranks for annotated protein coding genes from RNA-seq data (by RPKM) and Affymetrix arrays (by MAS 5.0 estimation). Darker shades indicate more genes and lighter shades indicated fewer genes.
Supplementary Figure 3. H3K4me3 support for an alternative 5′ end. Shown is the genomic locus at chr18:77,304,429-77,392,539, along with known annotated transcripts (blue, bottom), the reconstructed transcript from ESC (black) with an alternative external 5′ end, and K4me3 data (green tracks, top) for four cell types. The alternative 5′ end in ESC is associated with an ESC-specific K4me3 mark.
Supplementary Figure 4. Conservation of novel protein coding exons. Shown is the cumulative distribution of sequence conservation across 29 mammals (ω) for novel protein coding exons (blue), annotated protein coding exons (green), and introns (red). The novel exons are conserved at comparable levels to annotated ones.
Supplementary Figure 5. Conservation of novel protein coding genes. Shown is the cumulative distribution of sequence conservation across mammals (ω) for exons from novel protein coding genes (blue), annotated protein coding genes (green), and introns (red). The novel genes are conserved at comparable levels to annotated ones.
Supplementary Figure 6. Patches of conservation in lincRNAs. Shown are two examples of lincRNAs and the conservation patterns across their exons. In each panel; shown is the relevant genomic locus (top) with a zoom in (bottom), listing ω (top, green), RefSeq gene anontations (middle, blue tracks), and 30-way multiz alignment tracks (bottom, black). (a) Known functional lincRNA, XIST. Zoom in on its 5′ exon illustrates the conservation patterns within the exon. (b) A novel lincRNA. Zoom in on one of its exons illustrates the conservation patterns within the exons, comparable to those in (a).
Supplementary Figure 7. LincRNAs are more cell type specific across different expression levels. Shown are the fractions of ES specific transcripts for lincRNAs (dark bars) and protein coding genes (light bars) at different ESC expression quantiles. In each quantile, the fraction of lincRNAs with ES specific expression is higher than the fraction of protein coding transcripts with ES specific expression.
Data Availability Statement
The sequencing data in this study is available at the NCBI Gene Expression Omnibus (GEO) under accession number GSE20851. The Scripture method is implemented as a stand-alone Java application and is available at www.broadinstitute.org/software/Scripture/, along with all assembled transcripts in both GFF and BED file formats. Additionally, all transcript graphs are available in the dot graph language.