Abstract
The quantitative analysis of biochemical reactions and metabolites is at frontier of biological sciences. The recent availability of high-throughput technology data sets in biology has paved the way for new modelling approaches at various levels of complexity including the metabolome of a cell or an organism. Understanding the metabolism of a single cell and multi-cell organism will provide the knowledge for the rational design of growth conditions to produce commercially valuable reagents in biotechnology. Here, we demonstrate how equations representing steady state mass conservation, energy conservation, the second law of thermodynamics, and reversible enzyme kinetics can be formulated as a single system of linear equalities and inequalities, in addition to linear equalities on exponential variables. Even though the feasible set is non-convex, the reformulation is exact and amenable to large-scale numerical analysis, a prerequisite for computationally feasible genome scale modelling. Integrating flux, concentration and kinetic variables in a unified constraint-based formulation is aimed at increasing the quantitative predictive capacity of flux balance analysis. Incorporation of experimental and theoretical bounds on thermodynamic and kinetic variables ensures that the predicted steady state fluxes are both thermodynamically and biochemically feasible. The resulting in silico predictions are tested against fluxomic data for central metabolism in E. coli and compare favourably with in silico prediction by flux balance analysis.
Keywords: systems biology, constraint-based modelling, linear polytope, logarithmic polytope, algebraic geometry
1 Introduction
It has long been appreciated that biological systems are subject to physico-chemical laws. These laws impose certain constraints on the feasible set of enzymatic reaction rates and metabolite concentrations. Within the physico-chemically feasible set, the actual values of evolved kinetic parameters will determine the functional state of the cell. In the absence of experimentally determined kinetic parameters, it is still possible to mathematically formulate and computationally implement systems of equations, the solutions to which represent a physico-chemically feasible set of functional states. Ideally, one seeks to incorporate all relevant physico-chemical constraints in a single framework. This brings a model closer to our conception of reality, reduces the size of the feasible set, and is aimed at increasing the predictive capacity of such models. However, it is not always computationally tractable to apply all known physical laws since the resulting equations are difficult to solve. This is especially so for genome scale models where there are typically a large number of equations and variables.
Flux balance analysis is a standard constraint-based modelling approach, based the assumption that mass is conserved in a biochemical system of reactions (40). In addition, within the timescale of interest, often one can reasonably assume that the concentrations of metabolites are time invariant, that is, the system is assumed to be at a steady state. These two constraints define a convex polytope of feasible reaction fluxes, even though absolute metabolite concentrations are not explicitly represented. Herein we derive a formalism where reaction flux to obeys mass conservation, energy conservation, the second law of thermodynamics, and mass action kinetics. The resulting constraint equations define a non-convex feasible set, with metabolite concentration and kinetic parameters are explicitly represented. We also demonstrate the application of this modelling approach with an example using a model of central metabolism in E. coli.
In addition, theoretically we illustrate how experimental knowledge of physiologically feasible ranges and theoretical biochemical predictions of feasible ranges of other pertinent variables may be incorporated into this new modelling framework (21). Natural selection has to operate within the constraints enforced by physical laws. By integration of genome scale stoichiometric, thermodynamic and kinetic constraints, in a framework amenable to numerical analysis, we aim to investigate in silico, the mechanisms responsible for the results of adaptive evolution experiments. This is the ultimate goal, but there are significant mathematical and computational modelling challenges which need to be addressed before these biologically more pertinent issues can be addressed.
Stoichiometric models of metabolism continue to grow in size due to new models of higher organisms (51) and incremental extension of existing models (20, 53). As the dimensions of the stoichiometric matrix increases, computational complexity theory dictates that some computational analysis techniques no longer become practical due to memory or time constraints (55). This problem is exemplified by vertex enumeration of convex polytope where no known polynomial time algorithm exists (3). Vertex enumeration is the key step in elementary mode, or extreme pathway analysis (40). Unless significant advances are made in reducing the complexity of vertex enumeration, one cannot hope to utilise this analysis method for the latest genome scale models (20). An additional problem is that existing methods of low complexity may be numerically unstable for large matrices with coefficients spread over many orders of magnitude. The application of less conventional linear algebra to design a more numerically stable, yet low complexity algorithm can help to alleviate such problems (47).
In tandem with growth of stoichiometric matrices, there are efforts to increase their predictive capacity by incorporating additional constraints, beyond mass conservation with a steady state assumption. The addition of Boolean genetic regulatory constraints, to flux balance models of E. coli metabolism, increases their qualitative predictive capacity when compared to experimental data of mutant growth capability on various media (14). Predictions of qualitative growth capability or quantitative growth rate (28) are interpretations of the numerical values of exchange fluxes from flux balance analysis of stoichiometric models. It is not possible to reliably predict internal fluxes in genome scale metabolic models without additional thermodynamic constraints. The simple fact is that stoichiometrically balanced loops are invariably present in such networks and, with only stoichiometric constraints, they exhibit cyclic net fluxes entirely independent of exchange fluxes. Energy conservation, and the second law of thermodynamics, can be combined with stoichiometric constraints to retain only mass balanced and thermodynamically feasible fluxes (6). However, although such fluxes obey mass conservation, energy conservation and the second law of thermodynamics, additional constraints are required to make them biochemically feasible.
Standard chemical potential forms a bridge between metabolite concentration and metabolic flux in thermodynamically constrained stoichiometric models. Experimentally, one can measure standard chemical potential and combine this data with stoichiometric models. This approach has shown promise in constraining viable fluxes and assisting interpretation of metabolome data (32). Theoretically, one can estimate standard chemical potential using group contribution methods (35, 24, 31). The group contribution method uses knowledge of the chemical energy within recurring chemical bonds and enumerates the prevalence of these bonds, within a compound of known structure, to estimate the entire standard chemical potential. Using this method, it is possible to estimate the standard chemical potential of almost all compounds in E. coli (20). These estimated results compare favourably with experimental measures, provided that the associated uncertainty in estimation is acknowledged (23, 31). Experimental and theoretical approaches are desirable and complimentary, since it is laborious and difficult to measure all the values experimentally whereas estimates need experimental confirmation.
Using standard chemical potential and metabolite concentration, one can place upper and lower bounds on the in vivo change in chemical potential of almost all reactions in genome scale stoichiometric models of metabolism (20). Given a stoichiometric model where all reactions were initially reversible, quantitative bounds on change in chemical potential, in combination with mass conservation, energy conservation and second law of thermodynamics, constrain the feasible steady state fluxes to thermodynamically feasible and physiologically relevant solutions. Without such bounds on chemical potential, the set of thermodynamically feasible solutions could include degradation of biomass into external nutrient metabolites, the opposite of the more probable biomass synthesis direction. Maximising bacterial growth, constrained by mass balance, energy conservation and the second law of thermodynamics, integrated with standard chemical potential and metabolite concentration estimates, has recently been shown to significantly improve the correspondence between computed and experimental internal flux rates in E. coli (26).
With the size of stoichiometric models increasing and simultaneously the scope of constraints expanding a pressing problem is to find a mathematical formalism which encompasses all current constraints in a concise form yet lends itself to robust, scalable numerical analysis. In addition, it is desirable to use a form which is invariant to the addition of new, possibly nonlinear, constraints. In particular, efforts are under way to collate the kinetic equations and parameters of enzymes in model organisms (42), including E. coli. Building on earlier work (41), herein we present a mathematical formulation which includes steady state mass conservation, energy conservation, the second law of thermodynamics, and extends to encompass a system of reversible kinetic equations where bounds on standard chemical potential, metabolite or enzyme concentration and kinetic variables can be incorporated. We refer to kinetic variables rather than kinetic parameters to emphasise that this is a constraint-based approach to steady state kinetics where the variables are not exactly specified, but constrained to a bounded interval. The resulting system of simultaneous linear equations in linear variables, combined with linear equations in logarithmic variables, shows a strong mathematical similarity with the system of equations studied in chemical reaction equilibrium analysis (50, 46). The outline of the mathematical description below mimics the order of the aforementioned constraints. During the description we highlight salient mathematical features and use simple toy metabolic examples for illustration of key concepts. Initial efforts to apply this modelling approach to a genome scale model of E. coli metabolism is also illustrated.
2 Mass conservation at steady state
The set of chemical reactions in a metabolic network can be represented as a set of chemical equations. Embedded in these chemical equations is information about reaction stoichiometry. Stoichiometry represents the number of molecules of reactant consumed, and product produced, in a single chemical reaction. Since elements, Carbon, Nitrogen, Oxygen etc, are neither created nor destroyed in a biochemical reaction, the total number of each element, or moiety, is conserved even though their relative arrangements may change as old chemical bonds are broken and new ones are formed. All of the stoichiometric information about a metabolic network can be represented within a matrix, the stoichiometric matrix, S ∈ ℤm,n, i ∈ 1…m, j ∈ 1…n, where m is the number of metabolites and n is the number of reactions (e.g. Supplementary Material A). Each elementary reversible chemical transformation is represented by a pair of conjugate columns of the stoichiometric matrix, one forward elementary reaction and one reverse elementary reaction, odd columns, j, and even columns, j + 1, respectively. Each row represents the participation of a single metabolite in all possible reactions. An integer coefficient in a column of a stoichiometric matrix gives the number of molecules of a single metabolite consumed or produced in that reaction. The stoichiometric matrix linearly transforms a flux vector, , into a vector of metabolite concentration time derivatives, giving the fundamental equation of dynamic mass conservation (27), which, with the assumption of steady state concentration, gives
| (1) |
The steady state assumption asserts that the concentration of metabolites is time invariant. Since Eq. 1 is generally under-determined, rank(S) < m < n, there are many flux vectors which satisfy the steady state mass conservation constraint.
3 Energy conservation and the second law of thermodynamics
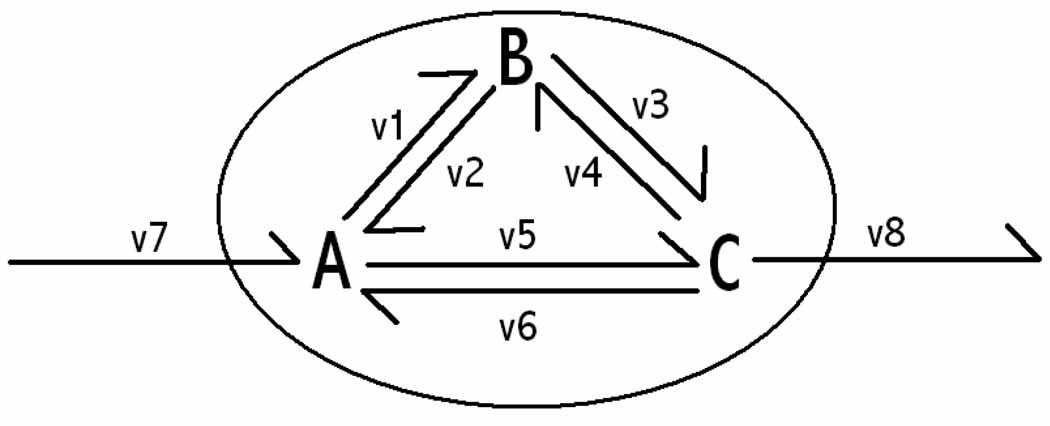
Theoretical application of thermodynamic principles to chemical networks in terms of differential geometry, algebraic topology and circuit theory were considered by Oster and Perelson in the 1970’s (38, 39). This work remains to fully impact systems biology, most likely because of a dearth of advanced mathematical training amongst the systems biology community. When applied to biochemical networks, energy conservation, and the second law of thermodynamics, prevent net flux around stoichiometrically balanced cycles within metabolic networks, as emphasised by Beard, Liang & Qian (6). A finer point to note here is the implicit assumption that thermodynamically unfavourable biochemical transformations cannot be driven by a gradient of temperature, this is may not be the case for endothermic chemotrophes (33). Given the entire set of elementary reactions in a metabolic network, a stoichiometrically balanced cycle is a subset of contiguous reactions that form a subnetwork of chemical transformations that are perfectly mass balanced, without a member of the subset being an exchange reaction. An example of such a stoichiometrically balanced cycle is given in Figure 1. By the second law of thermodynamics, net reaction flux from reactants to products requires a concomitant thermodynamic driving force, provided by a drop in chemical potential from reactants to products. In Figure 1, if A has a greater chemical potential than B, and B greater than C, then by the principle of conservation of energy, C must have lower chemical potential than A. By the second law of thermodynamics, the net flux is in the direction of decreasing chemical potential, therefore there must be net flux in the direction A → C. Therefore, there is no net flux around the cycle A → B → C → A. Many more complicated stoichiometrically balanced cycles exist in genome scale models, to the extent that they become unidentifiable by sight alone and innumerable using vertex enumeration algorithms (3) in the latest genome scale models for E. coli (20).
Figure 1. A toy metabolic network.
An artificial metabolic network consisting of one source exchange reaction, one sink exchange reaction and a three reversible isomerisations in a stoichiometrically balanced cycle, {υ1, υ3, υ5}, or equivalently {υ2, υ4, υ6}. The oval represents the boundary between the internal organism and external environment.
Quantitative thermodynamic constraints prevent net flux around stoichiometrically balanced loops but it is possible to have no net flux around a loop yet still violate thermodynamic constraints. Consider Figure 1, if A is in equilibrium with B and B is in equilibrium with C then by the second law of thermodynamics, there is no net flux A → B → C and hence no net flux around the cycle. In flux balance analysis models, net flux between A → C is still possible even if there is no net flux A → B → C. However, net flux A → C violates energy conservation since A must be in equilibrium with C, when A is in equilibrium with B and B is in equilibrium with C. Conservation principles and model consistency from different perspectives are two sides of the same coin.
Let us consider the first ñ columns of the stoichiometric matrix corresponding exclusively to internal reactions, denoted S̃ ∈ ℤm,ñ. If one computes a rational basis, K ∈ ℤñ,k, for the null space of S̃, then K spans the vector space of stoichiometrically balanced loops at steady state and, by the definition of a null space, S̃ · K ≡ 0. Multiplying across by a vector of metabolite chemical potentials, μ ∈ ℝ1,m, gives μ · S̃ · K = 0. Since μ · S̃ = Δμ̃, the vector of change in chemical potential for internal reactions, by substitution then transposing, one arrives at
| (2) |
This is an invariant, first derived by Beard and Qian (6), on total change in chemical potential at steady state for each stoichiometrically balanced loop and linear combinations thereof. Biochemically it enforces conservation of chemical potential energy by internal reactions of a stoichiometric network. By the second law of thermodynamics, net flux is with the direction of drop in chemical potential so this constraint is required in addition to energy conservation, in order to eliminate flux around these loops.
The net steady state change in chemical potential with respect to the forward reaction, Δμ̃j, is given by the logarithm of the ratio of reverse divided by forward flux, scaled by temperature, T, and the gas constant, R,
| (3) |
Here we assign the odd index j to the forward reaction, and even index j + 1 to the reverse reaction. The change in chemical potential with respect to the forward direction is equal to the change in chemical potential with respect to the reverse reaction if one changes its sign. Eq. 3 ensures that net flux is in the direction of a drop in chemical potential energy, in accordance with the second law of thermodynamics. Assuming the existence of a chemical potential for each metabolite, mass action kinetics, constant temperature and pressure, within a finite volume element, this thermodynamic relationship applies to elementary reversible reactions arbitrarily far from equilibrium (43). Obviously, at equilibrium, the rate of the forward and reverse reactions is equal so there is no change in chemical potential. The connection between Eq. 3 and the work of Hill on catalytic cycle fluxes (25), Ussing, Hodgkin and Huxley on ion transport, and Crooks on entropy production in microscopically reversible systems (15) has recently been discussed (7). It is fair to say that the exact validity of this relation, beyond the assumption of an elementary reaction, is still an open issue.
By considering all chemical transformations to be reversible, rather than approximating certain steps with ’irreversible’ reactions, with Eq. 3, we see that assigning zero flux to a reverse reaction dictates that the change in chemical potential, for the ’irreversible’ forward reaction, should be negative infinity. Such a situation is not physically possible as all changes in chemical potential are finite. This is a difference between the kinetic studies which routinely assume irreversibility and thermodynamic studies which can not. Given Eq. 2, by substituting an appropriately signed vector of equations 3 in place of Δμ̃, after some algebraic manipulation we derived the novel equation
| (4) |
where P ∈ ℤk,ñ and ln(ṽ) denotes the component-wise natural logarithm of each flux in the vector of fluxes. (See Supplementary Material B for details an example). Eq. 4 simultaneously applies energy conservation and the second law of thermodynamics as constraints on logarithmic elementary flux. One advantage of this novel reformulation is that it eliminates thermodynamically infeasible fluxes without dependence on a priori knowledge of change in chemical potential, cf. (6). This is important since standard chemical potential is unknown experimentally for certain metabolites and accurate theoretical estimates are difficult because the metabolites contain substructures for which no group contribution estimate is known (31). Another advantage is that it explicitly considers elementary forward and reverse flux, rather than net flux, which is important in isotopomer analysis (49).
4 Kinetic constraints
The vast majority of kinetic parameters available in the literature are inappropriate for models of in vivo metabolism because they are byproducts of enzyme mechanistic studies carried out at temperature and pH far from in vivo conditions (13). However, mechanistic enzyme classification also indicates a specific mathematical form of kinetic equation for an enzyme in accordance with its mechanism. Different enzyme classes, with distinct kinetic equations, specify different potential relationships between their variables independent of actual numerical values for kinetic variables. For example, consider two bisubstrate reactions, one with a sequential ordered mechanism versus one with a ping pong mechanism (11). Since their rate equations are different, the relationship between the possible values of their variables are different. Just as the stoichiometric matrix constrains flux distributions, kinetic equations constrain distributions of kinetic variables, metabolite and enzyme concentrations at steady state. Since flux is a function of kinetic variables, metabolite and enzyme concentration, then kinetic equations implicitly constrain flux distributions. The kinetic equations for many E. coli enzymes are available in the literature and, as prerequisite for genome scale modelling, are in the process of being collated in a manually curated database (42). Even in cases where the rate equation is unknown, one can approximate the kinetics with a simple mass action rate law.
4.1 Phenomenological versus elementary kinetic variables
Reversible kinetic equations typically appear in one of two mathematical forms depending on whether phenomenological or elementary kinetic variables are used. Phenomenological variables are microscopic properties of enzymes (18) but are more easily measured in macroscopic experiments with ensembles of enzymes (12), whereas elementary kinetic variables correspond to discrete approximations of major conformational transitions and are much more difficult to measure experimentally. Consider one of the isomerase reactions from Figure 1 modelled as a three step kinetic mechanism
| (5) |
with associated elementary reaction kinetic variables, k. Reactions are modelled stoichiometrically as a one step mechanism as it is the net stoichiometry of the entire reaction which acts as a constraint. The phenomenological, reversible, Michaelis-Menten kinetic equation for mechanism 5 is
| (6) |
where υnet is the net flux through the reaction, a and b are substrate and product concentrations, and eAB is the total enzyme concentration. Specificity constants, kA and kB, reflect the specificity of the enzyme for A or B in the presence of competing substrates. Michaelis constants, KmA and KmB, are apparent dissociation constants. Phenomenological kinetic variables are functions of overlapping sets of elementary kinetic variables so they are not independent. In the case of reaction 5
| (7) |
These functions can be viewed as implicit polynomial equations relating experimentally measurable quantities to quantitatively modelled kinetic variables, see Supplementary Material B. This facilitates the incorporation of appropriate experimental kinetic data (42) or theoretical predictions on optimal enzyme characteristics (21).
4.2 Thermodynamic Haldane equation
For a reversible reactions, with ordered binding and dissociation of substrates and products, the thermodynamic Haldane equation (11) relates standard change in chemical potential, , to a ratio of forward and reverse elementary rate constants
| (8) |
For reversible reactions where the kinetic mechanism is not ordered, such as the ’Ping Pong Bi Bi’ mechanism (11), or random, such as the ’Rapid Equilibrium Random Uni Bi Bi’ mechanism, the thermodynamic Haldane is no longer simply the ratio of two monomials formed from the forward and reverse rate constants (11). Instead, the thermodynamic Haldane is a more complicated polynomial. In such a situation one may use a transformation from polynomial to linear, linear-logarithmic equations in order to incorporate the thermodynamic Haldane equation into the system of constraint equations. The transformation from polynomial to linear, linear-logarithmic equations is discussed in section 4.3.
For ordered kinetic mechanisms, given the logarithmic identity
| (9) |
the relation between standard change in chemical potential for an elementary reaction, and a single row of the stoichiometric matrix, , one can rewrite Eq. 8 in linear homogeneous form in terms of logarithmic elementary kinetic parameters
| (10) |
Thus, standard chemical potential, 0 < μo ∈ ℝ1,m, can be used to linearly constrain the relationship between logarithmic elementary kinetic constants. Generalised to a system of reactions, in matrix form, we have
| (11) |
where I is the identity matrix, , and k+ ∈ ℝñ and k− ∈ ℝñ are vectors of elementary kinetic constants, ordered appropriately according to each internal reaction. An identity matrix is a square matrix with all zero coefficients, except for the diagonal which consists of unitary coefficients. Eq. 11 constrains the kinetic parameters of the model to be consistent with thermodynamics, a constraint often overlooked in kinetic models (17) and likewise in numerical sampling of candidate values for kinetic parameters (19).
4.3 Kinetic rate equations
For reversible reactions, elementary kinetic variable rate equations express net flux as a function of elementary kinetic variables, metabolite concentrations and enzyme concentration. By splitting a reversible rate equation, into separate unidirectional forward and reverse flux equations, we have two polynomial equations relating elementary flux, metabolite and enzyme concentrations. For an entire metabolic network, all elementary unidirectional rate equations, and the relations between phenomenological and elementary kinetic variables, form a system of implicit homogeneous polynomials. We demonstrate how any system of polynomial equations can be algebraically transformed into a conjugate set of linear equalities, logarithmic equalities and logarithmic inequalities. This conjugate linear-logarithmic system has the same mathematical form as the aforementioned linear-logarithmic flux constraints representing mass balance and thermodynamic feasibility.
Consider a d dimensional system of m homogeneous polynomials, with n distinct monomials. Here m is the number of metabolites + enzymes in a system, whilst d is the number of unidirectional reactions + kinetic constants + metabolites + enzymes. A distinct monomial is a single term in a polynomial ignoring coefficients, e.g. 6x2y3 and −4x2y3 are indistinct monomials. The number of distinct monomials depends on the type of polynomial kinetic expression used to model each reaction (see Supplementary Material D for workthrough of toy example). Any system of polynomials, i ∈ 1…m, can be represented, without algebraic manipulation, in the form
| (12) |
where Ai,j is the coefficient of monomial j in polynomial i, yr is a variable and Qj,r is a an exponent of variable yr within distinct monomial j. The coefficients are integers in the matrix A ∈ ℤm,n. 0 < y ∈ ℝd since it is non-physical to consider variables or concentrations of zero or less. Each row of Q ∈ ℕn,d corresponds to one distinct monomial and each column of Q corresponds to one variable.
Now we arrive at the first key algebraic step. Let yr ≡ exp (zr), then, by the exponential identities, (ea)b = eab and eaeb = ea+b, we have
If we consider exp (Qj ·z), j ∈ 1…n, as one component of an n dimensional vector then Eq. 12 condenses to become
| (13) |
where z ∈ ℝd. This prepares the way for the second key step.
In the field of algebraic geometry, in particular, the study of algebraic varieties, which are geometric manifestations of solution sets of systems of polynomial equations, the matrix Q composed of exponents of distinct monomials, leads to the study of mathematical structures known as Newton polytopes (22). A polytope is a generalisation of a polygon to arbitrary dimension. To construct a Newton polytope, one considers the exponents of a single distinct monomial, a row in Q, as a vertex of this polytope. A vertex is a generalisation of a corner, to arbitrary dimension. The set of rows j ∈ 1…n of Q form such a set of vertices that define a polytope. It is a fundamental result from convex geometry that there are two equivalent ways to represent a polytope, in terms of a set of vertices, a vertex representation, or terms of a set of linear equations and inequalities, a half-space representation. Facet enumeration, using a convex hull algorithm, can transform a vertex representation into a half space representation. The Newton polytope of a polynomial system is the convex hull of Q when one considers each row as a vertex. We invite the interest of the algebraic geometry community at this stage as we depart from the usual perspective and instead consider each column of Q as the vertex of a polytope and use a vertex enumeration algorithm to compute the ’dual Newton polytope’.
Since the polynomials we consider are homogeneous, no entire row of Q is devoid of nonzero coefficients. Qj,r = 0 when a monomial indexed j has a variable indexed r with a zero exponent since . Since Qj,r ∈ ℕ we consider each column of Q, which is formed from the exponents of a single variable yr across all n distinct monomials of the polynomial system, as a vertex of a polytope in the positive quadrant. Using facet enumeration we can compute the convex hull of Q and derive an equivalent half-space representation of the dual Newton polytope as a system of affinely independent inhomogeneous linear equalities and inequalities with new matrices B ∈ ℤb,n, C ∈ ℤc,n, and vectors b ∈ ℝb, c ∈ ℝc, which by the definition of Q as a set of vertices of this ’dual Newton polytope’ satisfy the equations
| (14) |
| (15) |
i.e. the new matrices B and C, with respective vectors b and c, are defined to be the halfspace representation of the same polytope with vertex representation defined by the set of vertices in Q. These are equivalent representations of the same convex polytope. Now, let Q · z ≡ ln (x), then Eq. 13 becomes
| (16) |
and equations 14 and 15 become
| (17) |
| (18) |
Intuitively, if one considers Eq. 16 as a polytope in linear scale, and Eqs. 17 and 18 as a polytope in logarithmic scale, then the simultaneous solution to equations 16, 17 and 18 is the intersection of a polytope in linear scale with the exponential of a polytope on a logarithmic scale (see Supplementary Figure 3).
Since we let yr ≡ exp (zr) and Q · z ≡ ln (x), it is consistent to relate the new vector x to the original vector y with Q · ln(y) = ln(x). This relation can be equivalently expressed as
| (19) |
where I ∈ ℤn,n is an identity matrix. If the number of different monomials is greater than the number of dimensions of the polynomial system, Q ∈ ℕn,d, n > d, then Q · ln(y) = ln(x) is overdetermined. In this case, not every x will necessarily correspond to a solution y. However, since Eq. 19 is the of same mathematical form as 17 we can incorporate it into the system of constraints to guarantee a solution to y for each x. The output of the transformation from a system of polynomial equations in y, using exponential identities and facet enumeration, is a set of linear equations in linear variables, plus a set of equalities and inequalities in ln(x) and ln(y). In the case of a polynomial system of kinetic equations, y encompasses external fluxes, v̅, internal fluxes, ṽ, forward and reverse elementary kinetic variables, k+ and k−, metabolite concentrations, m, and enzyme concentrations, e. i.e. . Here we assume a unique enzyme for each reaction, so the dimension of y will be lower if the same enzyme catalyses multiple reactions. Thus we have the combined system of m linear equalities on linear variables, b + n linear equalities on logarithmic variables, and c linear inequalities on logarithmic variables.
| (20) |
| (21) |
| (22) |
where the various subscripts to Q denote the columns appropriate to the variables indicated. A detailed example of the transformation is given in Supplementary Material D. In the case of (pseudoelementary) mass action kinetics, where flux is a function of a single monomial, then vertex enumeration is not required to exactly linearise the equations in terms of logarithmic variables. In section 7 we provide such an example, with an application to central E. coli metabolism, moreover, see Supplementary Material E for an example of this exact linearisation for a few reversible reactionsE.
5 Thermodynamic and kinetic inequalities
Broad bounds on metabolite or enzyme concentrations, fluxes or phenomenological kinetic variables, are available from theoretical consideration of intracellular kinetics (21) guided in specific cases by experimental data. In all cases, whether due to lack of knowledge or inherent errors in experimental determination, such data results in a pair of bounding inequalities on the variable of interest. For example, estimates of standard chemical potential come with a margin of uncertainty. The effect of this uncertainty on prediction of reaction directionality is discussed elsewhere (31, 52). This uncertainty may be represented by inequalities on the allowed range of standard chemical potential
| (23) |
Likewise, broad estimates on physiologically realistic intracellular metabolite concentrations (20) lead to similar bounding inequalities. When experimental metabolite concentrations are available this data leads to tighter bounds on the model. Model representation of inter-compartmental concentration gradients is essential to permit net diffusive flux of transport reactions. Such considerations, beyond the realm of flux balance analysis, are vital to permit in silico growth of more realistic models which include potential gradients, in addition to fluxes. Of particular importance is the difference in concentration of H+ ions between separate compartments of the electrochemical chain (23). This electron-motive force can be represented in compartmentalised, charge balanced, constraint based models (20) when experimental data on intra- and periplasmic pH is included.
Establishing tight bounds on genome wide enzyme concentrations is a significant problem. Given the high level of noise in both microarray (29) and quantitative proteomics data (16) it is difficult to make conclusions on the relationship between transcript and protein abundance. If one assumes enzyme concentration is proportional to transcript abundance in global gene expression studies(14), then enzyme concentration can be quantitatively estimated through protein molecular weight, estimates of total dry weight of protein per cell and thermodynamically grounded preprocessing of raw microarray data (9). At best, such effort with experimental data gives rise to loose bounds on intracellular enzyme concentration for a particular nutrient medium appropriate to the microarray data (8). However, this may be important if the upper bound is so close to zero as to render flux through that particular enzyme inconsequential with respect to the flux of active pathways. As such, detecting the presence versus absence of a transcript is a crucial output of microarray preprocessing (10). As experimental systems biology continues its rapid progress one can expect tightening of these inequalities.
6 Integrated stoichiometric, thermodynamic and kinetic constraint-based modeling
The rationale for the reformulation of constraints becomes apparent when one compares the mathematical form of the resulting equations. Eq. 1, the steady state mass conservation invariant and Eq. 20, the linear portion of the reformulated kinetic constraints are of the same mathematical form and together can be represented by
| (24) |
where S̅ and S̃ denote the external and internal columns of the stoichiometric matrix. Equation 4, the novel compact reformulation of energy conservation and the second law of thermodynamics, Eq. 11, the linear reformulation of the thermodynamic Haldane in terms of logarithmic kinetic variables and Eq. 21, the logarithmic equality portion of the reformulated kinetic constraints, are all linear forms in logarithms and together can be represented by
| (25) |
Experimental data or theoretical estimates of kinetic or thermodynamic variables give rise to bounding inequalities such as 23. By taking the logarithm, splitting paired bounding inequalities into two inequalities and reversing the sign of the upper bound inequality, we have the same form as Eq. 22, the logarithmic inequality portion of the reformulated kinetic constraints. Combined they are of the form
| (26) |
Observing equations 24, 25 and 26, biologically, they represent the combination of many different constraints, but mathematically, they are in the same form as equations 20, 21 and 22, derived from a system of polynomial equations. If we let w ≡ ln(x) then equations 20, 21 and 22 are mathematically equivalent to
| (27) |
but this has the algorithmic advantage that a wide dynamic range of variable is facilitated in logarithmic scale. In fact, all constraints formulated thus far could be expressed as a system of polynomial equations by applying the inverse of the transformation sequence outlined in subsection 4.3.
7 Application to Escherichia coli
We tested the predictive capacity our approach by implementing mass conservation, energy conservation and the second law of thermodynamics for a core metabolic model of E. coli metabolism (m = 72 metabolites, 76 pairs of internal reactions and ñ = 19 net exchange reactions) (37). We assumed that each overall reaction followed pseudo-elementary mass action kinetics and thus, applied thermodynamic Haldane constraints to the ratio of forward and reverse pseudo-elementary rate constants. The equality constraints took the general form
| (28) |
| (29) |
where the boundary condition on the mass conservation constraints, a ≡ −S̅ · v̅, was derived from the optimal exchange fluxes obtained using flux balance analysis. For the latter, we fixed uptake/secretion rates for D-glucose, oxygen, ethanol, acetate, D-lactate, succinate, pyruvate and formate corresponding to aerobic growth on glucose minimal medium in a chemostat (Sample ID: GR04, (30)) . As described previously (52), for all metabolites, we used known standard (Legendre) transformed chemical potentials, uo′, back-calculated from experimentally measured equilibrium constants.
We used a bespoke solver to satisfy the non-linear, non-convex feasibility problem posed by Eq. 28 and 29. Due to uncertainty in the absolute values of in vivo kinetic parameters, and the nature of available fluxomic data, we focused on comparing net flux vectors to evaluate the predictive capacity. Also, where two reactions occur sequentially in an unbranched pathway, we compare only one predicted flux with fluxomic data. Figure 2 compares linear, linear-logarithmic prediction of net flux rates with fluxomic data, and also with net flux rates predicted by flux balance analysis. With flux balance analysis, typically there exist alternative optimal net flux vectors satisfying the same optimal value of the linear objective function, in this case, the biomass production rate. Therefore, we used flux variability analysis (34) to predict the maximum and minimum net flux rates consistent with mass conservation at an optimal biomass production rate and the aforementioned fixed exchange reaction rates.
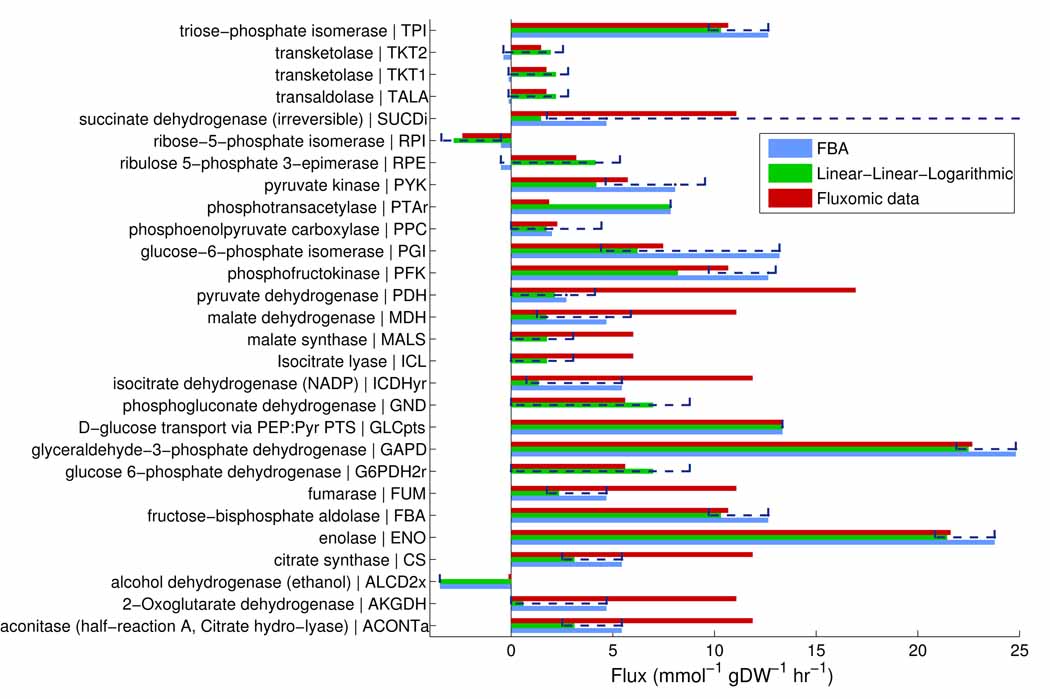
Figure 2. Fluxomic data compared with in silico predictions.
Comparison of in vivo fluxomic data, chemostat grown E. coli in an aerobic glucose-minimal medium (red, (30)), with in silico prediction by flux balance analysis (FBA, blue, (48)) and our equality constrained linear, linear logarithmic approach (green). A unique flux vector with minimum Euclidean norm, represents a single flux balance analysis prediction of net flux (blue). The fluxes predicted with the linear, linear logarithmic approach may extend outwidth the range of flux variablity for optimal flux balance analysis solutions (dashed bars) due to the omission of strict inequality constraints on reaction direction. As discussed in section 7, our approach provides a more refined prediction of pentose phosphate flux (e.g. RPE), and flux through the glycoxylate bypass (e.g. ICL), but does not capture the in vivo distribution of flux towards different fermentative end products (e.g. PTAr). Reaction abbreviations correspond to those in the map of central metabolism (Supplementary Figure 4).
In comparison with flux balance analysis, our linear, linear-logarithmic method does provide a more accurate prediction of the flux split between the decarboxylating oxidative branch of the pentose phosphate pathway and the proximal reactions of glycolysis. Also, our method improves the flux prediction through the glycoxylate bypass of the distal, oxidative part of the tricarboxylic cycle. Overall our flux prediction is superior to that by flux balance analysis; however, the prediction of flux through other reactions, such as the balance between different fermentative end products, does still not agree well with the fluxomic data. This preliminary comparison with experimental data indicates that the inclusion of thermodynamic constraints does refine our ability to predict in vivo flux. Further comparison with fluxomic data from various experimental conditions will be required for a definitive conclusion of predictive capacity.
We also tested our approach with a genome scale model of E. coli metabolism, iAF1260 (20) (data not shown). However, without additional inequality constraints, it is not possible to represent the qualitatively assigned local reaction directionality which accompany the iAF1260 model (52). Such constraints seem essential for a reliable prediction using flux balance analysis but their inclusion into the current formulation presents a significant numerical analysis challenge. The main difficulty is that local reaction directionality constraints may give rise to a feasible flux balance analysis problem, but an infeasible problem when additional thermodynamic constraints are required to be satisfied. Moreover, when a solver fails to converge to the solution of a non-convex feasibility problem, it is presently difficult to differentiate between a limitation in the ’solving’ capability of the algorithm versus an infeasible problem to begin with. This problem is an inherent feature of many non-convex feasibility problems. We continue efforts to add the capability to reliably solve such larger scale problems with inequality constraints.
8 Discussion
We have demonstrated how the mathematical constraints representing steady state mass conservation, energy conservation, the second law of thermodynamics, reversible polynomial kinetics, experimental and theoretical estimates of thermodynamic and kinetic variables can all be integrated into the composite mathematical form of equations 27. This reformulation is mathematically exact in the sense that it uses no approximations around arbitrary reference states, as with the ’linlog kinetics’ approach (45). The thermodynamic reformulation relies on a fundamental equation in non-equilibrium steady state thermodynamics (5) complementing existing work of Beard et. al. (6). The reformulation of reversible polynomial kinetic equations relies on judicious application of exponential identities, their logarithmic analogues, and a novel application of a well known principle from convex geometry. This principle is the representational equivalence of a polytope, as a set of vertices, a vertex representation, or a set of linear equations and inequalities, a half-space representation. The mathematical incorporation of numerical bounds on experimental and theoretical estimates of thermodynamic and kinetic variables, detailed in table 1 follows trivially in this constraint-based approach.
Table 1.
Input data for integrated modelling. Reaction stoichiometry is essential input data whereas experimentally measured fluxes or concentrations are optional. Experimentally derived or computationally estimated bounds on standard chemical potential are not essential but serve to quantitatively dictate the direction of net flux for kinetically irreversible reactions in the absence of manually assigned metabolic reconstruction (54) reaction directions. Since standard chemical potential is on a logarithmic scale, with respect to concentration, bounds on the former are more significant for determining the feasible set of reaction directions. In an iterative model development approach, initially, one may assume mass action kinetics for all reactions then subsequently incorporate more specific polynomial rate laws, such as Michaelis-Menten kinetics, as the model becomes more sophisticated(42). Kinetic rate laws dictate the magnitude of net reaction flux given a particular metabolite concentration, assuming the reaction is thermodynamically feasible.
| Data | Comments |
|---|---|
| Stoichiometry | Essential |
| Standard Chemical Potentials |
Experimentally measured(2, 1) or group contribution estimates(24, 23). |
| Kinetic Rate Equations | Mass action assumption or enzyme specific polynomial kinetic rate laws(11) |
| Kinetic parameters | Optional, upper & lower bounds from experiments |
| Fluxes | Optional, upper & lower bounds from experiments |
| Concentrations | Optional, upper & lower bounds from experiments |
The mathematical generality of the integrated system of linear, linear-logarithmic constraint equations 27, lies in the possibility to reformulate any system of polynomial equations in such a form. For constraint-based modelling of biological networks, this means that, theoretically, any new nonlinear constraints, formalised as an arbitrary system of implicit polynomials, can be integrated with the existing constraint-based modelling of steady state metabolism. Practically, computational complexity may limit the size of polynomial system that can be transformed in reasonable time. In systems biology, variants of vertex enumeration algorithms are used to compute a convex basis for the stoichiometric matrix (44). These convex bases correspond directly to stoichiometrically balanced pathways of contiguous reactions. Facet enumeration is dual to vertex enumeration, so the worst case computational complexity of vertex enumeration is identical to facet enumeration. Given Q ∈ ℕn,d, the latest algorithms (4) find all facets in worst case time of 𝒪(nd2) per facet (3).
The number of facets contributes to the size of the constraint satisfaction problem through the matrix A in Eq. 24 and matrix B in Eq. 25. To our knowledge, there is no closed form for the number of facets corresponding to a given a set of vertices. However, it is possible to estimate the number of facets and hence the computation time, prior to facet enumeration (4). Assuming mass action kinetics for each reaction, the integration of kinetic constraints does not require facet enumeration, see Supplementary Material E. In this case the numerical constraint feasibility problem requires a simultaneously solving m linear mass conservation constraints (one for each metabolite) Eq. 24, linear thermodynamic & kinetic equalities on logarithmic variables (for reversible elementary reactions) Eq. 25, and m linear inequalities on the estimated range of in vivo standard chemical potential.
The motivation for this work arose out of a requirement to integrate various constraints into a concise mathematical form, yet preserve desirable characteristics of the equations such that they are amenable to numerical analysis. Even though the feasible set is non-convex, and therefore non-linear, the near-linear structure of equations 27, even with a combination of linear and logarithmic variables, indicates potential for scalable numerical analysis. Moreover, with a central metabolic model of E. coli metabolism, we demonstrated that our method can be readily applied to improve the prediction of in vivo fluxes, in comparison with flux balance analysis. Nevertheless, future biological applications will be required in order to establish the generality of such conclusions for growth of other in a range of different environments.
With regard to genome scale models metabolism, the current numerical challenge lies in solving this non-convex feasibility problem when there are non-trivial inequality constraints. It practice, the current difficulty is distinguishing between failure of the current solver to converge due to (i) problem infeasibility due to over tight inequality constraints, versus (ii) failure to converge due to a shortcoming in algorithm design. It is important to realise that this same issue arises with any system of constraint equations and inequalities, defining a non-convex feasible set (36). It is not particular to our refomulation, and can occur with large systems of polynomial rate equations without inequalities.
The numerical solution of large systems of polynomial rate equations, even without inequality constraints, is as yet an open research question (36). Further work needs to be done to either, develop an algorithmic test for infeasibility of linear, linear-logarithmic equations with non-trivial inequalities, or, develop an algorithmic approach which is guaranteed to converge to a solution, if a solution exists. It is interesting to note that systems of linear, linear-logarithmic equations also arise in the classical problem of predicting metabolite concentrations at equilibrium (50, 46). There, the requirement for equilibrium concentration to be positive requires a trivial inequality constraint and the algorithms for solving such systems can be proven to converge (50). It remains to be seen if the sucessful algorithmic approaches from chemical reaction equilibrium analysis can be adapted to solve our novel non-equilibrium formulation with non-trivial inequality constraints.
Supplementary Material
Acknowledgements
The authors would like to thank Neema Jamshidi & Hong Qian for critical reading of the manuscript. R.M.T.F. was supported by a National University of Ireland, Galway, Science Faculty Fellowship. I.T. was supported by NIH grant Grant 5R01GM057089-11.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
R.M.T. Fleming, Center for Chromosome Biology, School of Natural Sciences, National University of Ireland, Galway.
I. Thiele, Center for Systems Biology, University of Iceland, Reykjavik, Iceland.
G. Provan, Department of Computer Science, University College Cork
H.P. Nasheuer, Center for Chromosome Biology, School of Natural Sciences, National University of Ireland, Galway, Ireland, and Systems Biology Ireland, Galway, Ireland
References
- 1.Alberty RA. Biochemical Thermodynamics: Applications of Mathematica. Hoboken, NJ: Wiley-Interscience; 2006. [PubMed] [Google Scholar]
- 2.Alberty RA. Thermodynamics of Biochemical Reactions. Hoboken, NJ: Wiley-Interscience; 2003. [Google Scholar]
- 3.Avis D. Polytopes - Combinatorics and Computation, Birkhauser-Verlag, chapter lrs: A Revised Implementation of the Reverse Search Vertex Enumeration Algorithm. 2000. pp. 177–198. [Google Scholar]
- 4.Avis D. Computational experience with the reverse search vertex enumeration algorithm. Optimization Methods and Software. 1998;10:107–124. [Google Scholar]
- 5.Beard DA, Babson E, Curtis E, Qian H. Thermodynamic constraints for biochemical networks. Journal of theoretical biology. 2004;228:327–333. doi: 10.1016/j.jtbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Beard DA, Liang S, Qian H. Energy balance for analysis of complex metabolic networks. Biophysical journal. 2002;83:79–86. doi: 10.1016/S0006-3495(02)75150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beard DA, Qian H. Relationship between thermodynamic driving force and one-way fluxes in reversible processes. Public Library of Science - One. 2007;2:e144. doi: 10.1371/journal.pone.0000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyer A, Hollunder J, Nasheuer HP, Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Molecular & cellular proteomics. 2004;3:1083–1092. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Binder H. Thermodynamics of competitive surface adsorption on DNA microarrays. Journal of Physics: Condensed Matter. 2006;18:S491–S523. [Google Scholar]
- 10.Binder H, Preibisch S. GeneChip microarrays-signal intensities, RNA concentrations and probe sequences. Journal of Physics: Condensed Matter. 2006;18:S537–S566. [Google Scholar]
- 11.Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. London: Taylor & Francis Group; 2007. [Google Scholar]
- 12.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. 3rd edition. London: Portland Press; 2004. [Google Scholar]
- 13.Cornish-Bowden A, Hofmeyr JHS. Enzymes in context: Kinetic characterization of enzymes for systems biology. The Biochemist. 2005;27:11–13. [Google Scholar]
- 14.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BØ. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 15.Crooks GE. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Physical Review E. 1999;60:2721–2726. doi: 10.1103/physreve.60.2721. [DOI] [PubMed] [Google Scholar]
- 16.Deeds EJ, Ashenberg O, Shakhnovich EI. A simple physical model for scaling in protein-protein interaction networks. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:311–316. doi: 10.1073/pnas.0509715102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ederer M, Gilles E. Thermodynamically feasible kinetic models of reaction networks. Biophysical journal. 2007;92:1846–1857. doi: 10.1529/biophysj.106.094094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nature chemical biology. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 19.Famili I, Mahadevan R, Palsson BØ. k-Cone analysis: Determining all candidate values for kinetic parameters on a network scale. Biophysical journal. 2005;88:1616–1625. doi: 10.1529/biophysj.104.050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular systems biology. 2007;3:e121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fersht A. Structure and mechanism in protein science: A guide to enzyme catalysis and protein folding. New York: W. H. Freeman; 1999. [Google Scholar]
- 22.Gel’fand I, Zelevinskij A. Discriminants, Resultants, and Multidimensional Determinants. New York: Birkhauser; 1994. [Google Scholar]
- 23.Henry CS, Broadbelt LJ, Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophysical journal. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophysical journal. 2006;90:1453–1461. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill TL. Free Energy Transduction in Biology. New York: Academic Press; 1977. [Google Scholar]
- 26.Hoppe A, Hoffmann S, Holzhutter HG. Including metabolite concentrations into flux balance analysis: Thermodynamic realizability as a constraint on flux distributions in metabolic networks. BioMed Central Systems Biology. 2007;1:23. doi: 10.1186/1752-0509-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn F, Jackson R. General mass action kinetics. Archive for Rational Mechanics and Analysis. 1972;47:81–116. [Google Scholar]
- 28.Ibarra RU, Edwards JS, Palsson BØ. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y, Yugi K, Arakawa K, Iwata N, Toya Y, Nakayama Y, Nishioka T, Shimizu K, Mori H, Tomita M. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science (New York, N.Y.) 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 31.Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophysical journal. 2008;95:1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kümmel A, Panke S, Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Molecular systems biology. 2006;2:e34. doi: 10.1038/msb4100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu JS, Marison IW, von Stockar U. Microbial growth by a net heat up-take: a calorimetric and thermodynamic study on acetotrophic methanogenesis by Methanosarcina barkeri. Biotechnology and bioengineering. 2001;75:170–180. doi: 10.1002/bit.1176. [DOI] [PubMed] [Google Scholar]
- 34.Mahadevan R, Schilling C. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metabolic engineering. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Mavrovouniotis ML. Estimation of standard Gibbs energy changes of biotransformations. The Journal of biological chemistry. 1991;266:14440–14445. [PubMed] [Google Scholar]
- 36.Neumaier A. Acta Numerica. Cambridge University Press; 2004. Complete Search in Continuous Global Optimization and Constraint Satisfaction. [Google Scholar]
- 37.Orth J, Fleming R, Palsson BØ. Escherichia coli and Salmonella: Cellular and Molecular Biology, ASM Press, chapter Reconstruction and use of microbial metabolic networks: the core Escherichia coli metabolic model as an educational guide. 2009 doi: 10.1128/ecosalplus.10.2.1. (in press). [DOI] [PubMed] [Google Scholar]
- 38.Oster GF, Perelson AS. Chemical reaction dynamics. Archive for Rational Mechanics and Analysis. 1974;55:230–274. [Google Scholar]
- 39.Oster GF, Perelson AS. Chemical reaction dynamics, part II: Reaction networks. Archive for Rational Mechanics and Analysis. 1974;57:31–98. [Google Scholar]
- 40.Palsson BØ. Systems Biology: Properties of Reconstructed Networks. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 41.Qian H, Beard DA. Thermodynamics of stoichiometric biochemical networks in living systems far from equilibrium. Biophysical chemistry. 2005;114:213–220. doi: 10.1016/j.bpc.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Rojas I, Golebiewski M, Kania R, Krebs O, Mir S, Weidemann A, Wittig U. Storing and annotating of kinetic data. In silico biology. 2007;7:37–44. [PubMed] [Google Scholar]
- 43.Ross J. Thermodynamics and fluctuations far from equilibrium, volume 80 of Springer Series in Chemical Physics. New York: Springer; 2008. [Google Scholar]
- 44.Schilling CH, Letscher D, Palsson BØ. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. Journal of theoretical biology. 2000;203:229–248. doi: 10.1006/jtbi.2000.1073. [DOI] [PubMed] [Google Scholar]
- 45.Smallbone K, Simeonidis E, Broomhead DS, Kell DB. Something from nothing: Bridging the gap between constraint-based and kinetic modelling. Federation of European Biochemical Societies Journal. 2007;274:5576–5585. doi: 10.1111/j.1742-4658.2007.06076.x. [DOI] [PubMed] [Google Scholar]
- 46.Smith WR, Missen RW. Chemical Reaction Equilibrium Analysis: Theory and Algorithms. New York: Wiley; 1982. [Google Scholar]
- 47.Vallabhajosyula RR, Chickarmane V, Sauro HM. Conservation analysis of large biochemical networks. Bioinformatics (Oxford, England) 2006;22:346–353. doi: 10.1093/bioinformatics/bti800. [DOI] [PubMed] [Google Scholar]
- 48.Varma A, Palsson BØ. Metabolic capabilities of Escherichia coli: I. Synthesis of biosynthetic precursors and cofactors. Journal of theoretical biology. 1993;165:477–502. doi: 10.1006/jtbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 49.Wiechert W. The thermodynamic meaning of metabolic exchange fluxes. Biophysical journal. 2007;93:2255. doi: 10.1529/biophysj.106.099895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeleznik F, Gordon S. Calculation of complex chemical equilibria. Industrial & Engineering Chemistry. 1968;60:27–57. [Google Scholar]
- 51.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleming R, Thiele I, Nasheuer H. Quantitative assignment of reaction directionality in constraint-based models of metabolism: application to Escherichia coli. Biophysical chemistry. 2009;145:47–56. doi: 10.1016/j.bpc.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiele I, Jamshidi N, Fleming R, Palsson BØ. Genome-scale reconstruction of E. coli ’s transcriptional and translational machinery: A knowledge-base, its mathematical formulation, and its functional characterization. PLoS computational biology. 2009;5:e1000312. doi: 10.1371/journal.pcbi.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nature protocols. 2009;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeung M, Thiele I, Palsson B. Estimation of the number of extreme pathways for metabolic networks. BioMed Central Bioinformatics. 2007;8:363. doi: 10.1186/1471-2105-8-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.