Abstract
Seventy Escherichia coli isolates recovered from diseased chickens diagnosed with colibacillosis in Henan Province, China, between 2004 and 2005 were characterized for antimicrobial susceptibility profiles via a broth doubling dilution method. Overall, the isolates displayed resistance to trimethoprim-sulfamethoxazole (100%), oxytetracycline (100%), ampicillin (83%), enrofloxacin (83%), and ciprofloxacin (81%), respectively. Among the phenicols, resistance was approximately 79% and 29% for chloramphenicol and florfenicol, respectively. Molecular detection revealed that the incidence rates of the floR, cmlA, cat1, cat2 and cat3 were 29, 31, 16, 13, and 0%, respectively. Additionally, 10% of the isolates were positive for both floR and cmlA. As these antimicrobial agents may potentially induce cross-resistance between animal and human bacterial pathogens, their prudent use in veterinary medicine is highly recommended.
Keywords: antimicrobial resistance, Escherichia coli, florfenicol
Introduction
Diseases resulting from Escherichia coli (E. coli) infections, including colibacillosis, air sacculitis, and cellulitis, are responsible for high morbidity and mortality in poultry, and these diseases exert a significant economic influence on the poultry industry [1,6].
Antimicrobials are valuable tools for the treatment of clinical disease and for the maintenance of healthy, productive animals. However, recent reports have discovered increased resistance to the antimicrobial agents commonly utilized for treatment [1,4,25,27].
Florfenicol, a broad-spectrum antibiotic, belongs to the family of agents including thiamphenicol and chloramphenicol, which has played a hugely important role in reducing the enormous losses in the poultry industry resulting from certain bacterial diseases, including avian colibacillosis. However, Kim et al. [21] identified a novel plasmid-encoded gene (pp-flo) from Photobacterium piscicda in a study conducted in Japan [21]. More recently, florfenicol resistance conferred by the floR genes, referred to in the published literature as pp-flo, cmlA-like, floSt, flo, or floR, has also been detected in the Salmonella enteria serovar Typhimurium definitive phage type (DT) 104 [2,3,8-10,20], Salmonella enterica serovar Agona [9,14], E. coli [5,7,13,15,16,19,26], Klebsiella pneumoniae [12], Vibrio cholerae [17] and Pasteurella multocida [18], which mediate combined resistance to florfenicol and chloramphenicol.
Currently, very little data is available regarding the epidemiology and prevalence of antimicrobial-resistant veterinary pathogens in domestic animals, particularly in developing countries, including China, where antimicrobials are overused in veterinary medicine and domestic animals. Thus, the principal objective of the present study was to determine the antimicrobial susceptibility profiles among a collection of E. coli isolates collected from diseased chickens that were diagnosed with colibacillosis in China between 2004 and 2005. In addition, due to the high incidence of emerging florfenicol resistance in tested E. coli isolates 5 to 6 years after its introduction into veterinary clinics and the limited information regarding phenicol resistance in China, the resistance determinants for the phenotypes of phenicol resistance observed in these isolates were identified. The results presented herein may provide surveillance information for this specific region.
Materials and Methods
Bacterial strains
70 E. coli isolates were recovered from the livers of diseased chickens raised on 12 different poultry farms in Henan Province, China, from January 2004 to September 2005. All E. coli organisms were isolated and purified on MacConkey agar and verified as E. coli using the Vitek system (BioMerieux, USA). The strains were maintained at -86℃ until analysis. The CVM1841 and CVM827 strains, which were used as positive controls for the amplification of the floR and cmlA genes, were kindly donated by Dr. David White of the FDA, USA. The positive strains harboring the cat-1, cat-2 or cat-3 genes were obtained from the microbiology lab at Henan Agricultural University and were designated as strain C258, strain C337, and the strain C151, respectively.
Antimicrobial susceptibility determination
Antimicrobial minimum inhibitory concentrations (MIC) of the E. coli isolates were determined via the standard broth doubling dilution method on Muller-Hinton medium, and were interpreted in accordance with CLSI standards [11]. According to the suggestions provided in a previous report by Singer et al. [23], florfenicol resistance breakpoints in E. coli might be defined by an MIC of 32 µg/ml. The following antimicrobials were tested: ampicillin, ceftiofur, chloramphenicol, florfenicol, dihydrostreptomycin, gentamicin, amikacin, kanamycin, enrofloxacin, ciprofloxacin, oxytetracycline, and trimethoprim/sulfamethoxazole (China Institute of Veterinary Control, China). E. coli ATCC 25922 was used as a control in all of the MIC determinations.
Detection of florfenicol and chloramphenicol-resistance determinants
Genes encoding for florfenicol and chloramphenicol resistance determinants (floR, cmlA, cat-1, cat-2, and cat-3) were detected via PCR. Templates of total DNA from each isolate were prepared as previously described [5]. For the detection of the floR genes, one pair of the forward primer (flo1: 5'-GTGTCGTCACATCTACGGCCTTT-3') and the reverse primer (flo2: 5'-CAGACAGGATACCGACATTC AC-3') was designed using Oligo 6.0 software on the basis of the published floR gene sequence [26]. Between the two primers, the sequence region predicted an 882-bp fragment. For the positive control, the CVM1841 strain, which harbors the floR gene, was utilized. PCR was conducted in a final volume of 50 µl containing 1 µg of template DNA, 100 pmol of each primer (flo1/flo2), 1 × PCR buffer, 0.2 mM of each dNTP (dATP, dCTP, dGTP, dTTP) and 2.5 U of Ex Taq polymerase (Takara, Japan). A total of 32 cycles were conducted in the PCR Express (Thermo Hybaid, UK), under the following conditions: denaturation at 94℃ for 45 sec, annealing at 62℃ for 45 sec, and an extension step at 72℃ for 1 min.
The primer sets employed in the amplification of cmlA, cat-1, cat-2, and cat-3 were identical to those previously described [19,24]. For the amplification of the different amplicons, the appropriate program parameters were utilized. The predicted amplicons for the cmlA, cat-1, cat-2, and cat-3 genes were 699, 585, 495, and 508 bp, respectively. The CVM827, C258, C337, and C151 strains were utilized as positive controls for the amplification of the cmlA, cat-1, cat-2, and cat-3 genes, respectively.
Results
Antimicrobial susceptibility patterns of chicken E. coli isolates
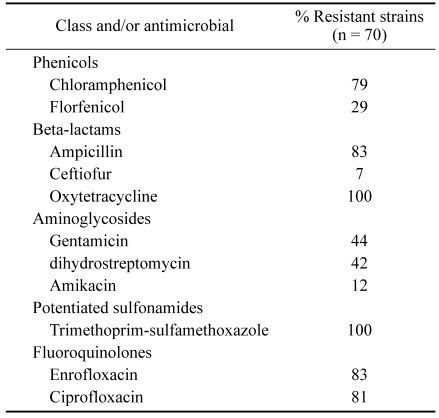
Seventy E. coli isolates recovered from diseased chickens diagnosed with colibacillosis were tested for their resistance to 11 antimicrobial agents of human and veterinary therapeutic significance. The rates of resistance, as determined via measurements of the MIC and comparisons to the resistance breakpoints established by CLSI, are listed in Table 1. The highest rates of resistance were to trimethoprim-sulfamethoxazole (100%), oxytetracycline (100%), ampicillin (83%), enrofloxacin (83%), ciprofloxacin (81%), and chloramphenicol (79%), respectively.
Table 1.
Antimicrobial resistance phenotypes of chicken E. coli isolates
With regard to multidrug resistance profiles, all of the isolates recovered from the diseased chickens proved resistant to more than 4 of the 11 tested antimicrobials, 93% were resistant to more than 8 antimicrobials, and 1% were resistant to all 11 of the antimicrobials. The majority of E. coli isolates from this study proved susceptible to ceftiofur (93%) and amikacin (88%).
Molecular detection of the florfenicol and chloramphenicol resistant deteminants
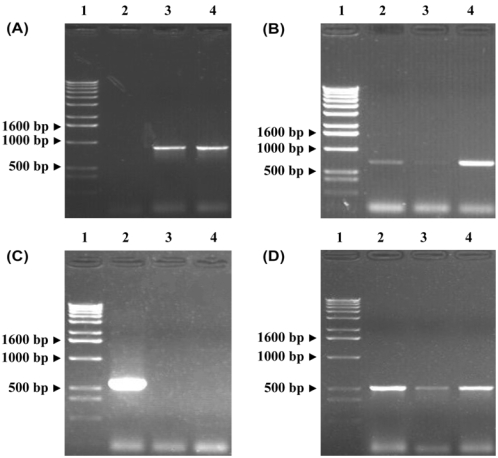
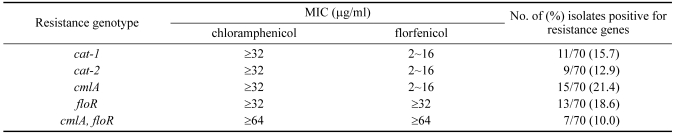
The genetic mechanisms relevant to chloramphenicol and florfenicol resistance in the chicken E. coli isolates were evaluated for the presence of 5 genes recognized to confer resistance to these antimicrobials: cmlA, cat-1, cat-2, cat-3, and floR. The different amplification products for the genes encoding for florfenicol and chloramphenicol resistance determinants are provided in Fig. 1. Using the total genomic DNA from each of the seventy chloramphenicol-resistant isolates as the PCR template, fifteen E. coli isolates (21.4%) were found to be positive for the cmlA gene and thirteen E. coli isolates (18.6%) were positive for the floR gene, with 20 of these isolates (28.6%) also harboring one of the chloramphenicol acetyltransferase genes (cat-1 or cat-2) (Table 2). Additionally, seven E. coli isolates (10%) were found to be positive for both the floR and cmlA genes, and both florfenicol and chloramphenicol MICs for these isolates were elevated (≥64 µg/ml) (Table 2).
Fig. 1.
Amplification of genes encoding for florfenicol and chloramphenicol resistance determinants. (A) The amplification of the floR genes. Lane 1: Marker; Lane 2: The negative isolate; Lane 3: The positive isolate; Lane 4: The positive control (B) The amplification of the cmlA genes. Lane 1: Marker; Lane 2-3: The positive isolate; Lane 4: The positive control. (C) The amplification of the cat-1 genes. Lane 1: Marker; Lane 2: The positive isolate; Lane 3-4: The negative isolates. (D) The amplification of the cat-2 genes. Lane 1: Marker; Lane 2-3: The positive isolate; Lane 4: The positive control.
Table 2.
Prevalence of cmlA, cat-1, cat-2 and floR genes in chloramphenicol-resistant chicken E. coli
Discussion
The bacterial isolates assessed in this study displayed similar levels of resistance to oxytetracycline, ampicillin, norfloxacin, and ciprofloxacin as were previously reported for E. coli strains isolated from diseased chickens in China by Yang et al. [27]. However, the chloramphenicol resistance rate (79%) determined in this study was significantly higher than that reported in the same country (24%), and this may reflect different patterns of antimicrobial use in different regions. In addition, as compared to the data from the only other report concerning florfenicol resistance in chicken E. coli isolates by Keyes et al. [19], the chicken E. coli isolates evaluated in this study displayed elevated resistance levels (Table 1).
Due to the wide use of oxytetracycline, sulfonamides, chloramphenicol, and fluoroquinolones for the treatment and prevention of diseases in chickens over the past decade, it was expected that the E. coli strains recovered from diseased chickens diagnosed with colibacillosis in this study would displayed a high rate of resistance to these drugs, and that was indeed the case. However, the high incidence of florfenicol resistance in the E. coli isolates tested herein was somewhat unexpected, as this drug was introduced into veterinary clinics for use in China only 6 to 7 years ago. This finding suggests that the selection pressure of chloramphenicol, as well as the other antimicrobials, may perform a relevant role in the emergence and dissemination of florfenicol resistance in E. coli.
Antimicrobials are useful therapeutic agents only if the drug concentrations achieved in the serum and tissue exceed the MIC of the drug. Based on this principle, Keyes et al. [19] suggested that florfenicol may not be therapeutically successful in some cases, as pharmacokinetic studies have demonstrated that the peak plasma florfenicol concentration in broiler chickens following oral administration of 15 mg/kg body weight is approximately 4 µg/ml. The florfenicol-resistant chicken E. coli isolates observed in both that study and the present study displayed florfenicol MICs in excess of that amount [19,22]. In this study, we also noted that 29 percent of the total isolates displayed florfenicol MICs in excess of 32 µg/ml. In order to ensure the rational and effective use of this drug, the expanded veterinary use of this drug in the treatment of E. coli-related chicken diseases can not be recommended at this time.
Acknowledgments
This work was supported by the National Key Technology R & D Program (2006BAK02A03) and a Doctoral Grant from Henan Agricultural University (No. 30700321). The authors wish to thank Dr. David White from the FDA (USA) for his kind donation of the CVM1841 and CVM827 strains, which were used as positive controls for the amplification of the floR and cmlA genes.
References
- 1.Altekruse SF, Elvinger F, Lee KY, Tollefson LK, Pierson EW, Eifert J, Sriranganathan N. Antimicrobial susceptibilities of Escherichia coli strains from a turkey operation. J Am Vet Med Assoc. 2002;221:411–416. doi: 10.2460/javma.2002.221.411. [DOI] [PubMed] [Google Scholar]
- 2.Arcangioli MA, Leroy-Setrin S, Martel JL, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 3.Arcangioli MA, Leroy-Setrin S, Martel JL, Chaslus-Dancla E. Evolution of chloramphenicol resistance, with emergence of cross-resistance to florfenicol, in bovine Salmonella Typhimurium strains implicates definitive phage type (DT) 104. J Med Microbiol. 2000;49:103–110. doi: 10.1099/0022-1317-49-1-103. [DOI] [PubMed] [Google Scholar]
- 4.Bass L, Liebert CA, Lee MD, Summers AO, White DG, Thayer SG, Maurer JJ. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob Agents Chemother. 1999;43:2925–2929. doi: 10.1128/aac.43.12.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff KM, White DG, McDermott PF, Zhao S, Gaines S, Maurer JJ, Nisbet DJ. Characterization of chloramphenicol resistance in beta-hemolytic Escherichia coli associated with diarrhea in neonatal swine. J Clin Microbiol. 2002;40:389–394. doi: 10.1128/JCM.40.2.389-394.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco JE, Blanco M, Mora A, Blanco J. Prevalence of bacterial resistance to quinolones and other antimicrobials among avian Escherichia coli strains isolated from septicemic and healthy chickens in Spain. J Clin Microbiol. 1997;35:2184–2185. doi: 10.1128/jcm.35.8.2184-2185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blickwede M, Schwarz S. Molecular analysis of florfenicol-resistant Escherichia coli isolates from pigs. J Antimicrob Chemother. 2004;53:58–64. doi: 10.1093/jac/dkh007. [DOI] [PubMed] [Google Scholar]
- 8.Bolton LF, Kelley LC, Lee MD, Fedorka-Cray PJ, Maurer JJ. Detection of multidrug-resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348–1351. doi: 10.1128/jcm.37.5.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd D, Cloeckaert A, Chaslus-Dancla E, Mulvey MR. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob Agents Chemother. 2002;46:1714–1722. doi: 10.1128/AAC.46.6.1714-1722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs CE, Fratamico PM. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. 2nd ed. Wayne: Clinical and Laboratory Standards Institute (formly NCCLs); 2002. CLSI M31-A2. [Google Scholar]
- 12.Cloeckaert A, Baucheron S, Chaslus-Dancla E. Nonenzymatic chloramphenicol resistance mediated by IncC plasmid R55 is encoded by a floR gene variant. Antimicrob Agents Chemother. 2001;45:2381–2382. doi: 10.1128/AAC.45.8.2381-2382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloeckaert A, Baucheron S, Flaujac G, Schwarz S, Kehrenberg C, Martel JL, Chaslus-Dancla E. Plasmid-mediated florfenicol resistance encoded by floR gene in Escherichia coli isolated from cattle. Antimicrob Agents Chemother. 2000;44:2858–2860. doi: 10.1128/aac.44.10.2858-2860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloeckaert A, Sidi Boumedine K, Flaujac G, Imberechts H, D'Hooghe I, Chaslus-Dancla E. Occurrence of a Salmonella enterica serovar typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar agona. Antimicrob Agents Chemother. 2000;44:1359–1361. doi: 10.1128/aac.44.5.1359-1361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doublet B, Schwarz S, Nussbeck E, Baucheron S, Martel JL, Chaslus-Dancla E, Cloeckaert A. Molecular analysis of chromosomally florfenicol-resistant Escherichia coli isolates from France and Germany. J Antimicrob Chemother. 2002;49:49–54. doi: 10.1093/jac/49.1.49. [DOI] [PubMed] [Google Scholar]
- 16.Du X, Xia C, Shen J, Wu B, Shen Z. Characterization of florfenicol resistance among calf pathogenic Escherichia coli. FEMS Microbiol Lett. 2004;236:183–189. doi: 10.1016/j.femsle.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kehrenberg C, Schwarz S. Plasmid-borne florfenicol resistance in Pasteurella multocida. J Antimicrob Chemother. 2005;55:773–775. doi: 10.1093/jac/dki102. [DOI] [PubMed] [Google Scholar]
- 19.Keyes K, Hudson C, Maurer JJ, Thayer S, White DG, Lee MD. Detection of florfenicol resistance genes in Escherichia coli isolated from diseased chickens. Antimicrob Agents Chemother. 2000;44:421–424. doi: 10.1128/aac.44.2.421-424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan AA, Nawaz MS, Khan SA, Cerniglia CE. Detection of multidrug-resistant Salmonella typhimurium DT104 by multiplex polymerase chain reaction. FEMS Microbiol Lett. 2000;182:355–360. doi: 10.1111/j.1574-6968.2000.tb08921.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim E, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida. Microbiol Immunol. 1996;40:665–669. doi: 10.1111/j.1348-0421.1996.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 22.Shen J, Hu D, Wu X, Coats JR. Bioavailability and pharmacokinetics of florfenicol in broiler chickens. J Vet Pharmacol Ther. 2003;26:337–341. doi: 10.1046/j.1365-2885.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- 23.Singer RS, Patterson SK, Meier AE, Gibson JK, Lee HL, Maddox CW. Relationship between phenotypic and genotypic florfenicol resistance in Escherichia coli. Antimicrob Agents Chemother. 2004;48:4047–4049. doi: 10.1128/AAC.48.10.4047-4049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vassort-Bruneau C, Lesage-Descauses MC, Martel JL, Lafont JP, Chaslus-Dancla E. CAT III chloramphenicol resistance in Pasteurella haemolytica and Pasteurella multocida isolated from calves. J Antimicrob Chemother. 1996;38:205–213. doi: 10.1093/jac/38.2.205. [DOI] [PubMed] [Google Scholar]
- 25.White DG, Piddock LJ, Maurer JJ, Zhao S, Ricci V, Thayer SG. Characterization of fluoroquinolone resistance among veterinary isolates of avian Escherichia coli. Antimicrob Agents Chemother. 2000;44:2897–2899. doi: 10.1128/aac.44.10.2897-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White DG, Hudson C, Maurer JJ, Ayers S, Zhao S, Lee MD, Bolton L, Foley T, Sherwood J. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J Clin Microbiol. 2000;38:4593–4598. doi: 10.1128/jcm.38.12.4593-4598.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Chen S, White DG, Zhao S, McDermott P, Walker R, Meng J. Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J Clin Microbiol. 2004;42:3483–3489. doi: 10.1128/JCM.42.8.3483-3489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]