Abstract
Aims
Proteins with a PDZ (for PSD-95, DLG, ZO-1) and one to three LIM (for Lin11, Isl-1, Mec-3) domains are scaffolding sarcomeric and cytoskeletal elements that form structured muscle fibres and provide for the link to intracellular signalling by selectively associating protein kinases, ion channels, and transcription factors with the mechanical stress–strain sensors. Enigma homolog (ENH) is a PDZ–LIM protein with four splice variants: ENH1 with an N-terminal PDZ domain and three C-terminal LIM domains and ENH2, ENH3, and ENH4 without LIM domains. We addressed the functional role of ENH alternative splicing.
Methods and results
We studied the expression of the four ENH isoforms in the heart during development and in a mouse model of heart hypertrophy. All four isoforms are expressed in the heart but the pattern of expression is clearly different between embryonic, neonatal, and adult stages. ENH1 appears as the embryonic isoform, whereas ENH2, ENH3, and ENH4 are predominant in adult heart. Moreover, alternative splicing of ENH was changed following induction of heart hypertrophy, producing an ENH isoform pattern similar to that of neonatal heart. Next, we tested a possible causal role of ENH1 and ENH4 in the development of cardiac hypertrophy. When overexpressed in rat neonatal cardiomyocytes, ENH1 promoted the expression of hypertrophy markers and increased cell volume, whereas, on the contrary, ENH4 overexpression prevented these changes.
Conclusion
Antagonistic splice variants of ENH may play a central role in the adaptive changes of the link between mechanical stress-sensing and signalling occurring during embryonic development and/or heart hypertrophy.
Keywords: Hypertrophy, PDZ–LIM protein, Alternative splicing
1. Introduction
PDZ–LIM proteins play important roles in muscle and heart development.1 These proteins combine two interaction modules, a PDZ domain (named for the proteins PSD-95, DLG, ZO-1)2 and one to three LIM domains (named for the proteins LIN-11, Isl-1, MEC-3).3
The PDZ–LIM protein family counts at least 10 members classified in three subfamilies.4 The first subfamily has five members: ALP (actinin-associated LIM proteins), Elfin, Mystique, LMO7, and RIL (reversion-induced LIM protein).5 Proteins in this subfamily carry a single PDZ domain and a single LIM domain. Two protein kinases form the second subfamily: LIM kinase 1 (LMK1) and LIM kinase 2 (LIMK2), characterized by a PDZ domain, two LIM domains and a tyrosine kinase domain.6 The members of the third subfamily are characterized by one N-terminal PDZ domain and three tandem C-terminal LIM domains. This family counts at least four members: Enigma (LMP-1), Enigma homolog (ENH), Cypher (ZASP, Oracle), and LMP-4 (LMP-2).7–9
In heart and skeletal muscle, PDZ and LIM domains are important elements of adaptors, which connect mechanical sensors, embedded in specialized cytoskeletal structures (e.g. Z-disc) to signalling proteins. In the heart, proteins with PDZ and LIM domains are necessary to allow embryonic development and adaptive remodelling in response to stress and strain.1 Indeed, Cypher and ALP have been shown to play two different but essential roles. ALP is essential in the embryonic development of the sarcomeric and cytoskeleton architecture in the right ventricular chamber.10 Cypher is not directly involved in heart development, but it is essential to maintain the Z-line structure of the sarcomere during muscle contraction.11
ENH is a PDZ–LIM protein expressed in the heart and other tissues. The PDZ domain of ENH1 interacts with α-actinin and localizes ENH at the Z-disc in neonatal rat cardiomyocytes.12 First discovered as a PKCβ interacting partner,8 its biological function was so far poorly understood. Recently, we identified two new binding partners for ENH1 in cardiomyocytes, namely protein kinase D1, which plays a critical role in the cardiovascular system13 and α1C, the pore subunit of the L-type voltage-gated calcium channel.14 Thus ENH1 connects a major signalling pathway with calcium entry required to sustain contractile activity. Although the precise functional importance of these interactions has yet to be demonstrated, these observations point towards a key role for ENH in heart muscle.
By alternative splicing, ENH is diversified into at least four isoforms: ENH18, ENH2, ENH3,12 and ENH415. ENH1 is the only isoform that contains the three LIM motifs at the C-terminal domain. ENH2, ENH3, and ENH4 only retain the N-terminal PDZ domain. The four ENH isoforms are expressed in heart and skeletal muscle and are conserved across rat, mouse, and human species, suggesting their physiological importance.8,12,15,16
What could be the functional relevance of the splice variants? To address this question, we have assessed quantitatively the expression of the ENH splice isoforms in heart at different developmental stages and in a mouse model for cardiac hypertrophy. We found that the four splice variants are differentially expressed in embryo, neonatal, and adult heart. ENH1 is predominant in embryonic heart, whereas the adult heart expressed mainly the truncated isoforms ENH2, ENH3, and ENH4. In heart, hypertrophy induced by transaortic constriction (TAC) in mice ENH isoform expression reverted to a pattern similar to neonatal/embryonic hearts. Furthermore, ENH1 overexpressed in neonatal cardiomyocytes was found to promote the development of hypertrophy, whereas silencing of ENH1 prevented hypertrophy. Surprisingly, overexpression of ENH4 also blocked the development of hypertrophy. Taken together, the data reported here show that alternative splicing of ENH transcripts produces antagonistic ENH isoforms during heart development: embryonic isoform ENH1 may cause, whereas the adult isoform ENH4 may prevent cardiac hypertrophy.
2. Methods
2.1. TAC surgery
Animals used in the present study were treated in conformity with the Guide for the Care and Use of Laboratory Animals published by the NIH (Publication No. 85-23, 1996). Protocols and use of animals were approved by Committee for Animal Experiments of both Nagaoka University of Technology and Tokyo Institute of Technology. Transverse thoracic aortic constriction was performed as described17 on 8-week-old adult mice (C57BL/6, the Jackson Laboratory, Bar Harbor, ME, USA). At hypertrophic phase (8 days after banding) after surgery, animals from the experimental and sham-operated groups were sacrificed and the hearts were removed. Left ventricles were weighed and quickly frozen in liquid nitrogen for total RNA extraction.
2.2. Cell culture
Neonatal rat ventricular myocytes were prepared as described.14 The cardiomyocytes were transferred to culture plates, and fibroblasts were removed by adhesion onto the plates for 1 h. The cardiomyocytes were then plated overnight and maintained in DMEM, containing 10% foetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin.
2.3. Real-time PCR
Total RNA (1 µg) was used for RT–PCR with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions. The synthesized cDNA (50 ng of RNA content), 250 nM primers pairs, 250 nM probes, and Absolute QPCR ROX Mix (AB gene) were mixed in a final volume of 25 µL and used for quantitative real-time PCR. The PCR reaction was carried out in triplicates in 96-well plates with ABIPrism 7000 Sequence Detection System (Applied Biosystems). The primers and TaqMan probes used in the experiments are shown in Table 1. As for standard curves, we amplified each ENH fragment with the primer pairs and inserted into pGEM T-easy (Promega). After confirmation of the sequence, the plasmids were used for standard curves.
Table 1.
Primers and probes sequences used for real-time PCR
| Forward 5′–3′ | Reverse 5′–3′ | Probe 5′–3′ | |
|---|---|---|---|
| Rat ENH1 | GGC TCC ACC TGG CAT GAC | AAA ATG TCT GCC CTT CCA AAC TT | FAM-TTG CTT TGT GTG CTC CGT GTG TTG TG-BHQ1/TAMRA |
| Rat ENH3 | ACC AGC GTG TGT TCC GAG T | CGT TTG GGT GGA ATT TTC ACA | FAM-CAG CAA AAT GGT AAC CCT GGC A-BHQ1/TAMRA |
| Rat ENH4 | GCT GTC CAG AAG AAA GCA CA | TGT GAT AAA ACT CCG TGT TGC | FAM-ACC CAA ACG CCC ACC AAG AA-BHQ1/TAMRA |
| Rat cyclophilin A | TCT GGG ATA CAC TTG GCA TGA T | TCA AAT TTC TCT CCG TAG ATG GAC TT | FAM-ACA CGC CAT AAT GGC ACT GGT GG-BHQ1/TAMRA |
| Mouse ENH1 | TCT GGG ATA CAC TTG GCA TGA T | GCC CTT CCA AAC TTT CAC AAC A | FAM-CTT GCT TTG TGT GCT CTG-BHQ1/TAMRA |
| Mouse ENH3 | AGA GGA TCC CAG GGT GAC ATT A | CGT TTA GGT GGA ATT TTC ACA | FAM-CAG CAA AAT GGT AAC CCT GGC A-BHQ1/TAMRA |
| Mouse ENH4 | CCG TCC AGA AGA AAA CAC AC | TGT GAT AAA ACT CCG TGT TGC | FAM-CCA CCT AAA CGC CCA CCA AGA-BHQ1/TAMRA |
| Mouse cyclophilin A | TGT GCC AGG GTG GTG ACT T | TCA AAT TTC TCT CCG TAG ATG GAC CT | FAM-ACA CGC CAT AAT GGC ACT GGC GG-BHQ1/TAMRA |
| Rat ANP | ATG GGC TCC TTC TCC ATC AC | CAT CTT CTC CTC CAG GTG GTC | |
| Rat BNP | GGG CTG TGA CGG GCT GAG GTT | AGT TTG TGC TGG AAG ATA AGA | |
| Rat betaMHC | GCA GCT TAT CAG GAA GGA ATA C | CTT GCG TAC TCT GTC ACT C |
2.4. RNA interference
The ‘hairpin strategy’ was used to design short-interfering RNA.18 The specific hairpin sequence of ENH1 corresponding to positions 1698–1718 (RNAi1) or 1452–1472 (RNAi2) of coding sequence were used to amplify the vector carrying the U6 promoter as described.14 We selected sequences unique for rat ENH1 in the NCBI cDNA database.
2.5. Adenovirus vector constructs
Recombinant human adenoviruses-5 encoding ENH1-FL, ENH4-FL, ENH1 RNAi, and control RNAi were constructed, amplified, and purified as described.14
2.6. RNase protection assay
RNase protection assay (RPA) was performed as described.14 Total RNA was extracted from neonatal rat cardiomyocytes with an acid phenol–guanidinium reagent (TRI-Reagent, Molecular Research Center, Inc.) according to the manufacturer's instructions. The riboprobe was prepared from the rat ENH cDNA and subcloned in pGEM-T-easy (Promega). Transcription with T7 RNA polymerase of the BamHI-linearized plasmid yielded a riboprobe of 366 nucleotides, and protected fragments of 207 nt for ENH1, 225 nt for ENH2, 258 nt for ENH3, and 276 nt for ENH4. A rat GAPDH probe was used as a loading control. The GAPDH plasmid digested with HinfI produced a riboprobe of 185 nt and a 154 nt protected band after RNase digestion. Autoradiographic signals were detected with a PhosphoImager (Molecular Dynamics, Inc.).
2.7. Patch-clamp
The capacitance of single neonatal rat cardiomyocyte was recorded by patch-clamp in the whole-cell configuration and voltage-clamp mode.14 The bath solution contained 125 mM N-methyl-glucamine, 5 mM 4-aminopyridine, 20 mM tetraethylammonium chloride, 2 mM CaCl2, 2 mM MgCl2, 10 mM d-glucose, and 10 mM HEPES, pH 7.4. The patch pipettes solution contained 130 mM CsCl, 10 mM EGTA, 3 mM MgATP, 0.4 mM LiGTP, and 25 mM HEPES, pH 7.2. The reference electrode was placed in a KCl solution linked to the bath with an agar bridge, reducing the liquid junction potential to negligible values. The cell was voltage-clamped using an Axopatch 200B amplifier (Axon CNS, Molecular Devices) at a holding potential of −90 mV. The currents were filtered at 1–2 kHz and sampled at 5 kHz using a Digidata 1322A (Axon CNS).
3. Results
3.1. Rat enh gene structure and splice variants expressed in the heart
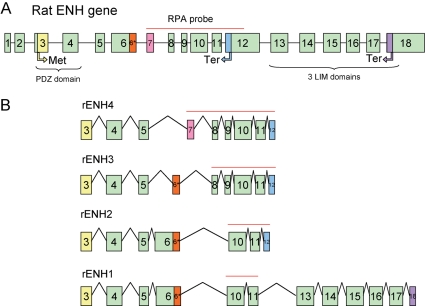
The enigma homolog (enh) gene is located on chromosome 2 (locus 2q44) of rat genome and the chromosome 3 (locus 3H3) of the mouse genome. The rat enh gene spans 11.96 kb and consists of 18 exons. Figure 1A shows a schematic representation of the genomic organization of the rat enh gene. The first two exons encode a 5′-untranslated region; the coding region extends from exon 3 to exon 18. Four splice variants encoded by the enh gene have been described.8,12,15,16 Figure 1B shows the schematic exonic composition of the four ENH splice variants. ENH1 is the only isofom containing the three LIM motifs in its C-terminal end. ENH2, ENH3, and ENH4 are truncated isoforms that only retain the N-terminal PDZ domain. Exon 12 contains a stop codon; hence skipping of exon 12 leads to the expression of the long isoform ENH1, whereas inclusion of exon 12 produces the three isoforms ENH2, ENH3, and ENH4 with truncated C-terminal. Differential expression between the truncated isoforms ENH2, ENH3, and ENH4 is the result of alternative skipping or rearrangement of exons 6–9.
Figure 1.
Scheme of the rodent ENH gene and the splice variants of its transcript. (A) Schematic representation of the genomic organization of the rat ENH gene with 18 exons. Translation start codon (Met) is located on the third exon. There are two stop codons (Ter): the first located in the 12th exon, the second in the 18th exon. The PDZ domain is encoded by the third and fourth exons; the three LIM domains are encoded by the 13th to 18th exons. The red line indicates the probe used RPA spanning coding sequences from exon 7 to 12. (B) Schematic representation of the exonic organization of the four known enh splice variants. The red lines indicate the length of the RPA probe protected from RNase digestion by hybridization to the various ENH isoforms.
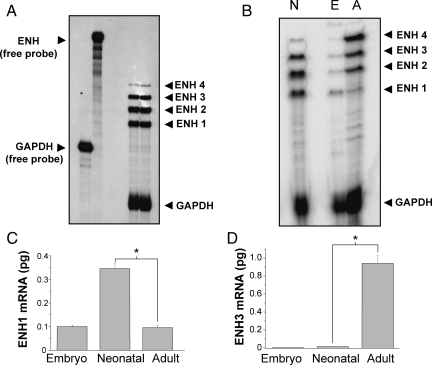
To detect which splice variants of ENH are expressed in heart, we designed a single anti-sense RNA probe spanning exons 6–12 (Figure 1A, red line). The probe and the protected RNA fragments after RNase digestion are indicated as a red line on each ENH splice variant in Figure 1A and B. Analysing total RNA extracted from isolated ventricular neonatal rat cardiomyocytes, this probe allowed us simultaneous detection of the four ENH splice variants by RPA (Figure 2A). ENH1 and ENH2 are strongly expressed, whereas the expression of ENH3 and ENH4 is much lower.
Figure 2.
Expression of the four ENH splice variants in heart at various stages of development. (A) Autoradiography of an RPA experiment using probes for ENH and for the housekeeping gene GAPDH as control. Twenty microgram of total RNA extracted from ventricular neonatal rat cardiomyocytes were hybridized with both the ENH and the GAPDH probes before RNase digestion. The autoradiography shown is representative of four independent experiments. (B) Autoradiography of an RPA experiment with 20 µg of total RNA extracted from adult, embryo or neonatal rat hearts hybridized to ENH and GAPDH probes. This particular autoradiography is representative of three independent experiments. (C and D) ENH1 (C) and ENH3 (D) mRNA measured by real-time PCR in 50 ng of total RNA extracted from embryonic, neonatal, and adult rat hearts. Shown are the means and SD of three independent experiments (*P< 0.002).
Using the same RPA probe, splicing of the ENH transcript was analysed in rat hearts at different developmental stages: embryonic, neonatal, and adult (Figure 2B). Striking changes in the expression pattern of ENH isoforms were observed between embryonic, neonatal, and adult heart. In the adult heart, expression of ENH3 and ENH4 was markedly increased compared with neonatal heart, whereas the expression of ENH1 and ENH2 was reduced. In contrast, in embryonic heart, ENH1 was the most abundant splice variant, different from the neonatal pattern in which the RPA signals from ENH1 and ENH2 are equivalent, and ENH3 is also strongly expressed. The comparison of the RPA signals suggests that during heart development splicing of ENH shifts from the embryonic isoform ENH1 towards the adult isoforms ENH2, ENH3, and ENH4.
For an alternative quantitative assessment of ENH splice variants by real-time PCR, we designed primers and probes to measure the expression of ENH1 and ENH3 variants. The results of real-time PCR (Figure 2C and D) corroborate the RPA data (Figure 2B). Relative to total RNA, the expression of ENH1 is the highest in neonatal heart, and similar in embryonic and adult heart. On the other hand, ENH3 is hardly detectable in embryonic heart tissue and low in neonatal heart, but highly expressed in adult heart tissue. This shift in the levels of ENH1 isoforms is best explained by a change of the alternative splicing of ENH in intron 11. In adult heart, exon 12 is included leading to the short isoforms, whereas in embryonic and neonatal heart, exon 12 with its stop codon is spliced out, leading to the expression of ENH1. Note that this latter type of splicing is maintained in adult heart tissue, possibly in immature cardiomyocytes.
3.2. Expression of ENH isoforms in diseased mouse heart
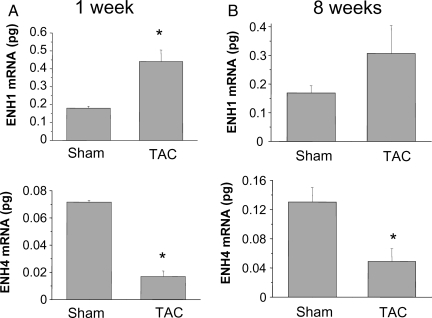
The expression of the embryonic isoform ENH1 in adult heart could be an indicator of heart de-differentiation linked to hypertrophic growth. To test this, we used hearts from mice operated by TAC leading to heart hypertrophy and heart failure. ENH1 expression was measured by real-time PCR in mice hearts following TAC or sham operation (Figure 3). In the hypertrophic phase (1 week after TAC, Figure 3A), the amount of ENH1 was significantly higher than in TAC-operated hearts compared with sham-operated hearts. On the contrary, the cardiac expression of ENH4 was significantly lower in the TAC-operated than in sham-operated animals. The cardiac expression of ENH3 was also lower in the TAC-operated than in sham-operated animals but not significantly (see Supplementary material online, Figure S1). Eight weeks after TAC when the mice have become prone to heart failure, ENH1 was no longer significantly elevated over sham-operated hearts (Figure 3B). However, expression of ENH3 (see Supplementary material online, Figure S1) and ENH4 were significantly lower in TAC vs. sham mice. These results suggest that in cardiac hypertrophy, the expression of ENH1 is favoured, whereas ENH4 is reduced. Hence, with regard to ENH isoform expression, hypertrophic resembles neonatal heart.
Figure 3.
Expression of the splice variants ENH1 and ENH4 in diseased heart of mice. ENH splice-variant expression was examined in mouse heart after transaortic constriction (TAC) leading to cardiac hypertrophy and heart failure. (A) ENH1 (upper) and ENH4 (lower) during the development of cardiac hypertrophy 1 week after TAC or sham operation. ENH splice variants were quantified by real-time PCR using 50 ng of total RNA. (B) ENH1 (upper) and ENH4 (lower) in mice prone to heart failure 8 weeks after TAC or sham operation. ENH splice variants were quantified by real-time PCR using 50 ng of total RNA. Shown are means and SD of three independent experiments from three mouse hearts for each experimental conditions; *significantly different from sham-operated control (P < 0.02).
3.3. Role of ENH1 in a neonatal rat cardiomyocyte model of hypertrophy
Adaptor PDZ–LIM proteins such as ENH are capable to translate mechanical stress signals sensed in the Z-disc to intracellular signalling cascades and gene expression.1 Indeed, in cardiomyocytes, ENH1 interacts with structural and signalling proteins, i.e. several isoforms of protein kinase C,8 protein kinase D1,14 and α-actinin.12 In the mouse disease model above, arterial constriction leading to mechanical stress caused heart hypertrophy. The associated changes in ENH splicing strongly suggested its implication.
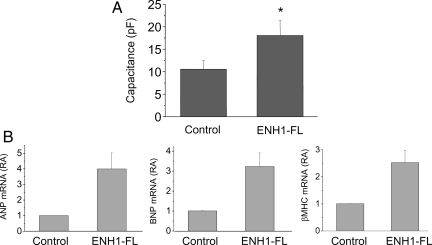
To test a possible causal role of ENH1 in the development of heart hypertrophy, we overexpressed a flag-tagged ENH1 in rat neonatal ventricular cardiomyocytes. Rat ENH1 cDNA was transferred using an adenoviral vector.14 To monitor quantitatively changes in cell volume, electrical capacitance of the whole-cell membrane was measured by the patch-clamp technique. As is shown in Figure 4A, overexpression of ENH1 significantly increased membrane capacitance and hence cell volume of neonatal rat cardiomyocytes in comparison to control cells.
Figure 4.
ENH1 overexpression induced hypertrophy of ventricular neonatal cardiomyocytes. (A) The volume of cardiomyocytes was determined by measuring the electrical capacitance of the plasma membrane using the whole-cell configuration of the patch-clamp technique. Cells were plated on glass coverslips and infected with ENH1-FL adenovirus. The capacitance was measured 48 h after infection. Shown are means and SD of four independent cell preparations (n= 25 cells per condition). *P < 0.01. (B) Hypertrophic marker gene expression. Cardiomyocytes were infected with ENH1-FL adenovirus for 48 h. The relative mRNA expression of three marker genes of cardiac hypertrophy, ANP (left), BNP (middle), and βMHC (right) were measured by real-time PCR using 100 ng of total RNA. Shown are the means and SD of three independent cell preparations.
Furthermore, the mRNA levels of three markers of cardiac hypertrophy, namely ANP, BNP, and βMHC, were also significantly increased upon ENH1 overexpression (Figure 4B). Taken together, these results suggest that ENH1 promotes the development of cardiomyocyte hypertrophy.
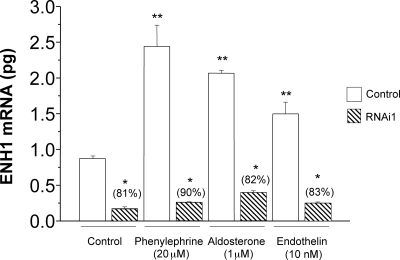
To verify that ENH1 indeed participates in the development of cardiomyocyte hypertrophy, we silenced the expression of ENH1 using small hairpin RNA (shRNA) targeting ENH1 and delivered by adenoviral vectors.14 We first validated the efficiency of this shRNA to knockdown the endogenous ENH1 expression as well as the stimulated increased expression of ENH1 in ventricular neonatal rat cardiomyocytes (Figure 5). Forty-eight hours after adenovirus treatment, the cardiomyocytes were exposed for 30 h to phenylephrine (20 µM), aldosterone (1 µM), or endothelin I (10 nM) used as hypertrophic stimuli. The expression of ENH1 was then measured by real-time PCR. All three stimuli induced a significant increase in the expression of ENH1 similar to the observation in TAC-operated mice (Figure 3). This effect was efficiently prevented by ENH1 shRNA.
Figure 5.
ENH1 silencing in neonatal rat cardiomyocytes. To silence the expression of ENH1, specific shRNA were delivered into ventricular neonatal rat cardiomyocytes by adenoviruses. Cardiomyocytes were first treated with ENH1-RNAi for 48 h and then stimulated with phenylephrine (20 µM), aldosterone (1 µM), or endothelin I (10 nM) for 30 h. The quantitative amount of ENH1 mRNA was determined by real-time PCR using 50 ng of total RNA. Data shown are means and SD of three independent measurements from three independent cell preparations (*P < 0.0002 and **P< 0.0001).
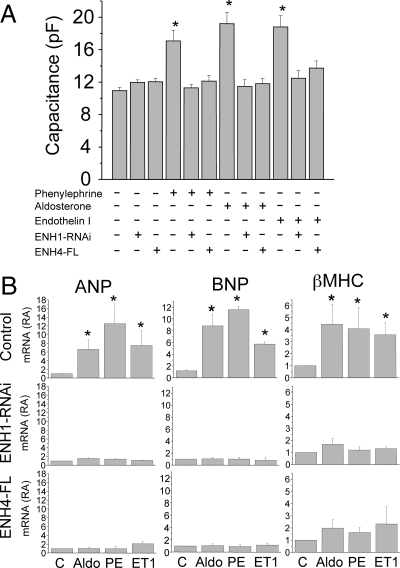
With this effective knockdown of ENH1, we investigated the role of ENH1 in the development of heart hypertrophy. As is shown in Figure 6A, cardiomyocyte cell size measured as whole-cell membrane capacitance was significantly increased by three hypertrophic stimuli: phenylephrine, aldosterone, or endothelin 1. These effects were not seen after ENH1 silencing. Without any treatment, ENH1 silencing did not result in any significant change in membrane capacitance (Figure 6A). These data support a central role of ENH1 in the increase of volume of cardiomyocytes upon stimulation with hypertrophic stimuli acting via various intracellular signalling pathways.
Figure 6.
ENH1 silencing or ENH4 overexpression prevent cardiomyocyte hypertrophy. (A) The volume of cardiomyocytes was determined by measuring the electrical capacitance of the plasma membrane using the whole-cell configuration of the patch-clamp technique. Cells were plated on glass coverslips then either exposed to adenovirus delivering either ENH1-specific shRNA for 48 h or adenovirus delivering cDNA coding for FLAG-tagged ENH4 (ENH4-FL) for 24 h. Cells were then stimulated with phenylephrine (20 µM), aldosterone (1 µM), or endothelin I (10 nM) for 30 h before proceeding to the whole-cell membrane capacitance measurements. Shown are means and SEM of four independent cell preparations (n = 20 to 30 cells per condition); *P < 0.02. (B) Hypertrophic marker gene expression was measured by real-time PCR. Cells were treated with phenylephrine (PE; 20 µM), aldosterone (Aldo; 1 µM), or endothelin I (ET1; 10 nM) for 30 h. The relative mRNA expression (RA) of cardiac genes was measured by real-time PCR using 100 ng of total RNA. Upper lane shows the expression of ANF (left), BNP (middle), and βMHC (right) in control conditions. Middle lane shows the expression of ANF (left), BNP (middle), and βMHC (right) in cells treated with ENH1 shRNA 48 h prior to stimulation. Lower lane shows the expression of ANF (left), BNP (middle), and βMHC (right) in cells overexpressing ENH4-FL for 24 h prior to stimulation. Shown are means and SD of three independent cell preparations; *P < 0.002.
As shown in Figure 6B, ENH1 silencing also affected the expression of three marker genes for cardiac hypertrophy: ANP, BNP, and βMHC. The mRNA levels of these genes are increased upon aldosterone, phenylephrine, and ET1 stimulation (Figure 6B, upper panels). ENH1-RNAi blocked completely the induction of the cardiac hypertrophic marker genes (Figure 6B, middle panels). These results consolidate the role of ENH1 in the regulation of ventricular cardiomyocyte expansion stimulated by hormones which signal vascular stress and hypertension. Interestingly, the silencing of ENH1 changed gene expression and expansion only of cardiomyocytes which were stimulated, whereas steady-state functions of non-stimulated cells appeared unaffected.
3.4. The alternatively spliced isoform ENH4 is an antagonist of ENH1
ENH expression in ventricular cardiomyocytes in vivo varies with development and vascular stress not only by changes in transcription but also by changes in alternative splicing. Vascular stress favours the ‘embryonic’ ENH1 isoform, which plays a causal role for the development of cardiac hypertrophy, whereas in normal adult heart, the short isoforms ENH2, ENH3, and ENH4, which are lacking the C-terminal LIM domains, are predominant. Furthermore, it has been suggested that the LIM domain-deficient ENH isoforms might act as antagonist of ENH1.12 To test this hypothesis, a FLAG-tagged ENH4 (ENH4-FL) was ectopically expressed in ventricular neonatal rat cardiomyocytes using adenoviral vector. Cardiomyocytes were then stimulated for 30 h with aldosterone (1 µM), phenylephrine (20 µM), or endothelin-1 (10 nM). All three stimuli increased cell size measured as whole-cell membrane capacitance. Stimulated cardiomyocyte hypertrophy was totally suppressed in ENH4-FL expressing cardiomyocytes after stimulation (Figure 6A). Importantly, in a non-stimulated state, no change in whole-cell membrane capacitance was observed upon ENH4-FL overexpression (Figure 6A, third column).
The stimulated increase in the expression of the hypertrophy marker genes ANP, BNP, and βMHC was also efficiently prevented by the overexpression of ENH4-FL in neonatal rat cardiomyocytes (Figure 6B, lower panels). Taken together, the results presented in Figure 6 strongly suggest that ENH4 can efficiently prevent cardiomyocyte hypertrophy most likely by acting as an antagonist of ENH1. This confirms the central role of ENH isoforms in the development of heart hypertrophy.
4. Discussion
We report here the changes in the expression of the ENH splice variants at three development stages of the heart (embryonic, neonatal, and adult) and in vascular stress-induced cardiac disease. We present evidence that the (embryonic) ENH1 isoform is not only up-regulated during cardiac hypertrophy, but plays a central role promoting the expression of marker genes and an increase in cardiomyocyte cell volume. Strikingly, overexpression of ENH4, a splice variant lacking the three LIM motifs, prevented ventricular cardiomyocyte hypertrophy induced by vascular stress hormones, suggesting that ENH4 acts as a natural antagonist for ENH1.
Recent genome-wide studies have shown that 70–80% of the human genes are alternatively spliced and that a substantial part of the alternative splicing events are conserved in evolution (10–20% between mouse and man).19 It is thus quite evident that the functions of many gene products can only be appreciated by considering the interplay between their various alternatively spliced isoforms. This is well illustrated here: the different isoforms of the PDZ–LIM domain scaffolding protein ENH have strikingly distinct adaptor functions: ENH2, ENH3, and ENH4 which lack the LIM domains will no longer associate with signalling proteins and transcription factors but will maintain the selective association with cytoskeletal elements such as actin and α-actinin.12–15,20 Given that we show here by gene silencing a causal relationship between ENH1 expression and ventricular cardiomyocyte hypertrophy (Figures 5 and 6), we could have anticipated that overexpression of a LIM-less ENH isoform such as ENH4 could prevent hypertrophy (Figure 6). Indeed, ENH4 most likely acts as a dominant-negative inhibitor of ENH1 by competing with its PDZ domain for the association with the cytoskeletal proteins such as actin and α-actinin. ENH4 may thus replace ENH1 in the Z-disc complex changing potentially composition, architecture and functioning of this important regulatory multi-protein assembly.1
Stimuli leading to cardiac hypertrophy favour the (embryonic) ENH1 isoform and repress ENH4. This suggests that hormones which signal vascular stress change the alternative splicing mechanism predominant in mature adult heart back to those which prevail in embryonic or neonatal heart. This is consistent with the transcriptional re-activation of several foetal genes during cardiac hypertrophy.21 A similar change in alternative splicing has been reported for the large embryonic isoform N2BA of the giant protein Titin whose expression is up-regulated in patients with dilated cardiomyopathy.22 Thus, ENH is a new example of genes whose expression during cardiac hypertrophy returns to foetal pattern.
PDZ–LIM protein family members other than ENH also generate various alternatively spliced isoforms.6,23 Alternative splicing allows a tissue specific expression of the different PDZ–LIM protein isoforms, e.g. the cardiac or skeletal muscle specific isoforms of Cypher gene products.23 Changes in the expression of the different splice variant isoforms during development occur not only for ENH (this study) but also for LIMK1 and LIMK2.6 Scaffolding by PDZ–LIM proteins is extremely versatile. Not only are there at least 10 genes which encode for PDZ–LIM proteins, but alternative splicing provides a large repertory of different isoforms. This makes it possible that PDZ–LIM proteins can selectively adapt elements of the cytoskeleton with mechanical stress sensors and signalling molecules relating vascular stress.1 Furthermore, the same family can also structure the specific and highly localized interaction between signalling protein kinases and ion channels.14 To characterize the precise function of each of these multiple PDZ–LIM protein isoforms in specific tissue and sub-cellular localization and to understand the functional importance of changes in splicing occurring during development and disease present an impressive challenge to future research.
ENH1 silencing prevented the stimulated hypertrophy of ventricular neonatal cardiomyocytes, whereas its overexpression promoted hypertrophy (Figures 5 and 6). ENH1 interacts with PKD114 a serine/threonine kinase required for pathological cardiac remodelling.24 In the heart, PKD1 phosphorylates transcription factors,25 histone deacetylase,26 sarcomere proteins,27 and a cardiac calcium channel.14 ENH1 might play a role in the scaffolding PKD1 at different cellular localization to reach its various targets. Moreover in the brain, ENH1 has been shown to interact with Id2,20 a transcriptional repressor, which also plays an essential role in cardiac differentiation.28 We are now investigating the precise role of these complex ENH1 functions in pathological cardiac remodelling. ENH illustrates well the challenge to describe in a comprehensive manner the molecular pathology of heart diseases which may involve a multitude of PDZ–LIM protein isoforms.
The mechanisms regulating the splicing of ENH are currently being investigated. Indeed, little is known about the splicing factors and mechanisms which control PDZ–LIM protein gene products. The splice factor ASF/SF2 has been shown to be required for the specific expression of the cardiac isoform of cypher.29 Given the decisive role for cardiomyocyte development and disease of the different ENH isoforms shown here, it would be of interest to identify the splicing factors and mechanism regulating the expression of the various ENH isoforms. Furthermore, it will be important to determine common elements which may govern splicing in the family of PDZ–LIM protein genes in various cells and tissues.
In summary, we have shown that the ENH splice variants are expressed differentially during the development of the heart and that in cardiac hypertrophy the pattern of expression is similar to that observed in embryonic or neonatal heart. ENH1 re-expressed during cardiac hypertrophy plays an active role in the remodelling of cardiomyocytes. Finally, we also show that ENH4, a short splice variant lacking the three LIM domains, acts as an antagonist of ENH1. This latter finding might provide a clue of how to target ENH1 for the prevention of cardiac hypertrophy.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by the Promotion of Environmental Improvement for Independence of Young Researchers, Special Coordination Funds for Promoting Science and Technology. Ministry of Education, Culture, Sports, Science, and Technology of Japan (to A.D.M.). The Takeda Science Foundation (to A.D.M.). The Fondation pour Recherches Médicales, Geneva and the Swiss National Science Foundation support T.Y. and W.S. S.W. was supported by a grant of Helse Sør-Øst. M.H. is supported by NIH grant (HL0811401).
Supplementary Material
Acknowledgements
We thank Drs Koichi Takimoto and Sigrid Skånland for critical reading of the manuscript.
Conflict of interest: none declared.
References
- 1.Hoshijima M. Mechanical stress–strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290:H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 3.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 4.te Velthuis AJ, Bagowski CP. PDZ and LIM domain-encoding genes: molecular interactions and their role in development. ScientificWorldJournal. 2007;7:1470–1492. doi: 10.1100/tsw.2007.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Q, Zhao Y. The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein–protein interaction. Biol Cell. 2007;99:489–502. doi: 10.1042/BC20060126. [DOI] [PubMed] [Google Scholar]
- 6.Scott RW, Olson MF. LIM kinases: function, regulation and association with human disease. J Mol Med. 2007;85:555–568. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, Ruiz-Lozano P, Martone ME, Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J Biol Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, et al. Protein–protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- 9.Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, et al. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ–LIM protein in limb and heart development. Dev Biol. 2004;273:106–120. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Pashmforoush M, Pomiès P, Peterson KL, Kubalak S, Ross J, Jr, Hefti A, et al. Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat Med. 2001;7:591–597. doi: 10.1038/87920. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q, Chu PH, Huang C, Cheng CF, Martone ME, Knoll G, et al. Ablation of Cypher, a PDZ–LIM domain Z-line protein, causes a severe form of congenital myopathy. J Cell Biol. 2001;155:605–612. doi: 10.1083/jcb.200107092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa N, Hoshijima M, Oyasu M, Saito N, Tanizawa K, Kuroda S. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem Biophys Res Commun. 2000;272:505–512. doi: 10.1006/bbrc.2000.2787. [DOI] [PubMed] [Google Scholar]
- 13.Avkiran M, Rowland AJ, Cuello F, Haworth RS. Protein kinase d in the cardiovascular system: emerging roles in health and disease. Circ Res. 2008;102:157–163. doi: 10.1161/CIRCRESAHA.107.168211. [DOI] [PubMed] [Google Scholar]
- 14.Maturana AD, Wälchli S, Iwata M, Ryser S, Van Lint J, Hoshijima M, et al. Enigma homolog 1 scaffolds protein kinase D1 to regulate the activity of the cardiac L-type voltage-gated calcium channel. Cardiovasc Res. 2008;78:458–465. doi: 10.1093/cvr/cvn052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niederlander N, Fayein NA, Auffray C, Pomiès P. Characterization of a new human isoform of the enigma homolog family specifically expressed in skeletal muscle. Biochem Biophys Res Commun. 2004;325:1304–1311. doi: 10.1016/j.bbrc.2004.10.178. [DOI] [PubMed] [Google Scholar]
- 16.Ueki N, Seki N, Yano K, Masuho Y, Saito T, Muramatsu M. Isolation, tissue expression, and chromosomal assignment of a human LIM protein gene, showing homology to rat enigma homologue (ENH) J Hum Genet. 1999;44:256–260. doi: 10.1007/s100380050155. [DOI] [PubMed] [Google Scholar]
- 17.Yasukawa H, Hoshijima M, Gu Y, Nakamura T, Pradervand S, Hanada T, et al. Suppressor of cytokine signaling-3 is a biomechanical stress-inducible gene that suppresses gp130-mediated cardiac myocyte hypertrophy and survival pathways. J Clin Invest. 2001;108:1459–1467. doi: 10.1172/JCI13939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuschl T. Expanding small RNA interference. Nat Biotechnol. 2002;20:446–448. doi: 10.1038/nbt0502-446. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- 20.Lasorellas A, Lavarone A. The protein ENH is a cytoplasmic sequestration factor for Id2 in normal and tumor cells from the nervous system. Proc Natl Acad Sci USA. 2006;103:4976–4981. doi: 10.1073/pnas.0600168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorn GW, 2nd, Robbins J, Sugden PH. Phenotyping hypertrophy: eschew obfuscation. Circ Res. 2003;92:1171–1175. doi: 10.1161/01.RES.0000077012.11088.BC. [DOI] [PubMed] [Google Scholar]
- 22.Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 23.Huang C, Zhou Q, Liang P, Hollander MS, Sheikh F, Li X, et al. Characterization and in vivo functional analysis of splice variants of cypher. J Biol Chem. 2003;278:7360–7365. doi: 10.1074/jbc.M211875200. [DOI] [PubMed] [Google Scholar]
- 24.Fielitz J, Kim MS, Shelton JM, Qi X, Hill JA, Richardson JA, et al. Requirement of protein kinase D1 for pathological cardiac remodeling. Proc Natl Acad Sci USA. 2008;105:3059–3063. doi: 10.1073/pnas.0712265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozgen N, Obreztchikova M, Guo J, Elouardighi H, Dorn GW, 2nd, Wilson BA, et al. Protein kinase D links Gq-coupled receptors to cAMP response element-binding protein (CREB)-Ser133 phosphorylation in the heart. J Biol Chem. 2008;283:17009–17019. doi: 10.1074/jbc.M709851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuello F, Bardswell SC, Haworth RS, Yin X, Lutz S, Wieland T, et al. Protein kinase D selectively targets cardiac troponin I and regulates myofilament Ca2+ sensitivity in ventricular myocytes. Circ Res. 2007;100:864–873. doi: 10.1161/01.RES.0000260809.15393.fa. [DOI] [PubMed] [Google Scholar]
- 28.Moskowitz IP, Banerjee SK, Lage ML, Huang XN, Smith SH, Saba S, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.