Abstract
Traditionally, expensive and time consuming techniques such as mass spectrometry and Western Blotting have been used for characterization of protein–protein interactions. In this paper, we describe the design, fabrication, and testing of a rapid and inexpensive sensor, involving the use of microelectrodes in a microchannel, which can be used for real-time electrical detection of specific interactions between proteins. We have successfully demonstrated detection of target glycoprotein–glycoprotein interactions, antigen-antibody interactions, and glycoprotein-antigen interactions. We have also demonstrated the ability of this technique to distinguish between strong and weak interactions. Using this approach, it may be possible to multiplex an array of these sensors onto a chip and probe a complex mixture for various types of interactions involving protein molecules.
Keywords: Electrical biosensor, electrical impedance detection, microfluidics, protein–protein interactions
I. Introduction
MANY biological functions such as signal transduction and gene regulation require highly specific interactions involving protein molecules. By studying many different proteins simultaneously, significant information can be gained as to how proteins interact with each other and with other types of molecules, both small and large, in order to control complex processes in cells. Current methods used for analyzing protein structures such as Western Blotting and ELISA, tend to be expensive and time consuming. Western Blots are based on gel electrophoresis and require labeled proteins. Labeling and reagent preparation incur significant costs, and the full process generally takes several hours [1]. Sandwich ELISAs are common for studying protein interactions involving antibodies. These too are expensive and time consuming techniques which involve lengthy procedures (typically several hours) [2].
The main challenge for rapid characterization of protein interactions rests in establishing an inexpensive and simple procedure requiring small reagent volumes capable of detecting real-time protein binding. Also of necessity is a technique which can be easily multiplexed allowing the simultaneous study of different proteins. Recently, protein microarrays [3] have become strong candidates for determining the presence and/or quantification of proteins in biological samples. Protein microarrays are advantageous since they open the possibility for multiplexed analysis of different proteins simultaneously. The disadvantage of using protein microarrays, as with all other fluorescence-based detection techniques, lies in the high reagent costs involved and the long incubation times. Recently, many efforts have been made toward the use of impedance-based sensors for biodetection and analysis [4]–[12]. Such sensors lower the reagent costs and preparation time since they eliminate the need for fluorescence labeling. Protein detection has been reported recently using nanogap sensors [13]–[16]. Many of the electrical impedance sensors presented to date require numerous washing steps and lack the ability for real-time detection. As for detection time, flow-based methods such as the use of coulter counters [17], can be used for counting and sizing particles in real-time. With the on-chip integration of microchannels and microelectrodes, the ability to capture target proteins on functionalized particles and detect their presence based on size changes has been demonstrated [18]–[21]. However, this type of device has difficulty in differentiating between two different types of proteins which may be similar in size.
Here, we describe a chip-based microfluidic device capable of electrical real-time detection of protein–protein interactions. In a previous study, we have demonstrated the ability of this technique to detect target cells without the need for any labeling [22]. We have demonstrated the detection selectivity of this technique for glycoprotein–glycoprotein interactions, antibody-antigen interactions, and also antigen-glycoprotein interactions by choosing specific examples. For studying antigen-antibody interactions, we used human chorionic gonadotropin (hCG) against anti-hCG antibody. For glycoprotein–glycoprotein interactions, we studied the binding between concanavalin A and lactoperoxidase. We also examined the affinity between hCG and ConA to study antigen-glycoprotein interactions. Future studies will be directed toward improving the detection limit and also exploring the potential use of this technique for practical applications such as the study of proteins on cell surfaces, and detection of protein and DNA biomarkers.
II. Theoretical Considerations
A. Device Operation
The basic device (Fig. 1) contains two electrodes to measure the impedance across the channel. The region in between electrodes A and C is the active area of the sensor. Protein receptors specific and complementary to the protein under study are immobilized on the base of the channel between electrodes A and C. The beads, which have been functionalized with proteins, are injected into the microchannel. If the desired interactions occur between the proteins, the beads will be captured in the active area of the sensor. This results in a partial occluding of the channel causing a decrease in the current across electrodes A and C. A possible optimization would be to pattern a gold region in between electrodes A and C to make the surface optimal for immobilization of protein receptors to the desired orientation. This is because gold is suitable for surface chemistry modifications, such as deposition of self assembled monolayers.
Fig. 1.
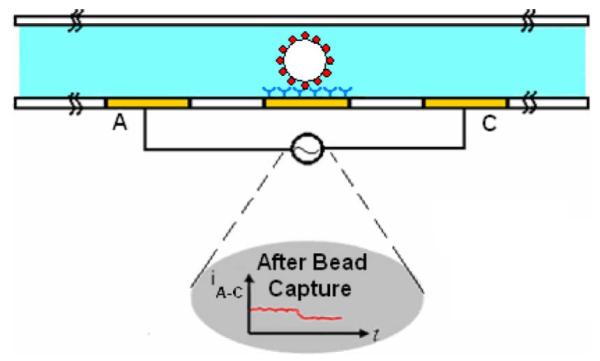
Cross section schematic of gated microchannel with electrodes labeled A and C. The functionalized beads specifically bind to the protein receptors which are immobilized on the gold electrode. (Bottom plot) Current between electrodes A and C during bead capture.
B. Performance Limits
We have developed a theoretical model for determining the detection limit of this biosensor. This type of model is useful in gaining design insight as to which parameters are the most significant in affecting the detection limit.
1) Electrical Detection Limit
By tailoring the geometry of the micro-channel to the bead size, the electrical detection limit can be improved to single-bead detection. The resistance change resulting from a particle passing through a channel is given by the following equation [23]:
| (1) |
where h is the diameter of the microsphere, ρsol is the resistivity of the solution, and Ac is the product of the height and the length of the channel. This equation works well for a bead positioned in the center of the active area of the sensor. The current change will be larger, however, for a bead which is positioned nearer to an electrode. To quantify this effect, we performed 2-D electrostatic simulations using Ansoft software (Ansys Inc., Pittsburg, PA), where we assumed a conductive media and a nonconducting sphere. As shown in Fig. 2, there is an increase in the magnitude of the current drop caused by the bead as the microsphere moves away from the center toward one of the electrodes.
Fig. 2.
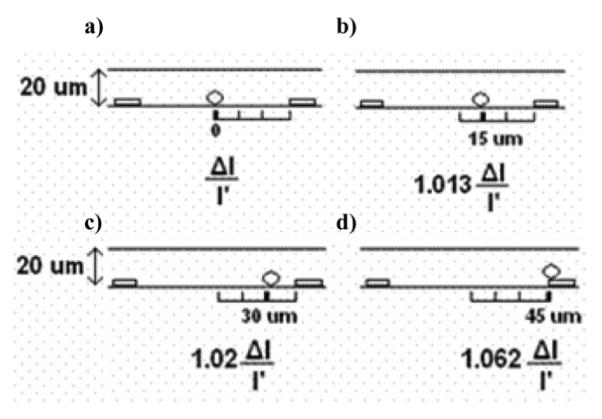
Influence of bead position on current change. (a) A 20 μm particle in the center of the channel causes a percentage current change of ΔI/I’. As the particles move closer and closer to the electrode, the resulting current change increases. (b) A sphere which is shifted 15 μm to the right results in 1.3% increase in current change with respect to a sphere in the center of the channel. (c) A sphere shifted 30 μm to the right results in 2% increase in the current change with respect to a sphere in the center. (d) A sphere sitting on top of the electrode results in 6% larger change in current in comparison to a sphere in the center of the channel.
There are three primary sources of noise in this system. They are the thermal noise resulting from the solution resistance of the electrolyte, the amplifier noise from the amplifying and read out circuitry, and the noise from the analog-to-digital converter. From our results below (Fig. 8), for a 50 μm × 50 μm sized channel and electrodes spaced 250 μm from each other, we measured the noise in the system to 1% of the baseline signal. Using (1), and assuming that the beads captured in the active region of the channel need to produce a resistance change of at least 1% in order to be detected, we approximate the minimum number of beads necessary to be captured in order produce a change in resistance greater than the noise level (Table I).
Fig. 8.

Representative data measured in the microchannel gated sensor. The instantaneous increase in impedance at corresponds to a lactoperoxidase coated CPG bead binding onto the active region of the device, as shown in Fig. 7. Noise level is 0.23% of the baseline impedance.
TABLE I.
Number of Beads Required for Detection. An Estimate of the Number of Beads Required to be Captured in the Active Region of the Channel in Order to Change the Baseline Channel Resistance by at Least the Noise Level (0.23%). Single Bead Detection can be Achieved With Bead Diameters Greater Than 24 μm.
| Bead Diameter (μm) |
1 | 5 | 10 | 15 | 20 | 25 | 30 |
| No. of Beads |
13,300 | 106 | 14 | 4 | 2 | 1 | 1 |
2) Bead Capture Rate
The rate at which particles are captured in the active area of the sensor is limited by two factors. The first is the hit rate of the beads passing through the channel in the active area. The second factor is the rate at which functionalized beads making contact with active area surface successfully bind to the immobilized receptor proteins, which is limited by the binding kinetics of the two interacting molecules.
The hit rate of the functionalized beads with the active area of the channel was calculated using a Matlab code we developed performing Monte Carlo Simulations of individual microspheres undergoing convection due to the laminar flow in the channel, and diffusion due to the Brownian motion of the particle in the channel.
At each time step in the simulation, the diffusion length and the drift length were calculated. The hit rate for a 1.2 μm sized particle with a diffusivity of 10−6 cm2/s is characteristic of an E. Coli cell, which would be a useful case study for characterization of cell surface proteins. The diffusion is modeled by applying a random displacement to each microsphere in the amount of
| (2) |
The convection of the bead in the channel is modeled by calculating the velocity profile in the channel. Our system closely resembles a horizontal pressure-driven flow between stationary parallel plates with no-slip boundary conditions. The velocity profile of the flow is dictated by
| (3) |
where Q is the volumetric flow rate which was assumed to be 100 nl/min, w the width of the channel, h the channel height, and y the vertical distance from the base of the channel. Thus, due to the convection, at each time step the microsphere is displaced by the amount of
| (4) |
We also assume that any microspheres colliding with the walls of the channel get reflected back into the channel. In Fig. 3, we plotted the relation between microsphere hit rate and channel height for various active area sizes, assuming a constant volumetric flow rate.
Fig. 3.
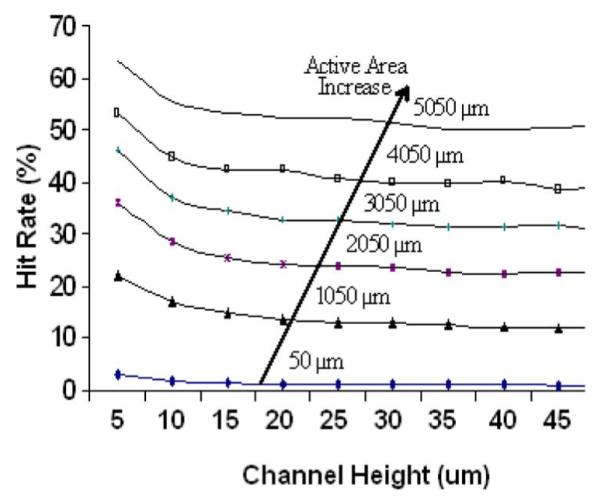
The hit rate of the 1.2 μm beads are plotted against the channel height for various active area lengths. The main parameter significantly affecting the hit rate is the active area size. An active area of 5050 μm can result in a hit rate greater than 50%.
As the channel height is increased beyond 10 μm, there is little decrease in the hit rate of the channel. Since the flow rate is constant, the velocity of the particle decreases as the channel height increases, which explains the little decrease in the hit rate. This information gives important insight into designing the geometry of the channel, in that it makes it clear that minimizing the channel size is not the best way to optimize the bead hit rate. At smaller channel geometries, nonspecific binding of beads and channel clogging becomes significant. The simulation results, however, indicate that the hit rate of particles is significantly increased with an increasing active area.
The active area of the sensor can be increased by increasing the spacing between the electrodes. However, this decreases the electrical sensitivity. Another method for increasing the active area size without compromising the electrical sensitivity involves integrating multiple sets of electrodes across the channel. A large active area length (> 5 mm) allows for the contact of over 50% of the beads passing through the channel.
The detection limit of this biosensor is determined by the number of beads required to pass through the channel before the minimum number of beads are captured in the active area of the sensor causing a change in impedance greater than the detection threshold of the sensor.
The minimum number of beads in the bulk solution required in the sample in order to be detected by the system, MNB, is given by the following equation:
| (5) |
where EDL is defined as the number of beads which can be electrically detected in the sensor active area and BCR is the rate at which beads are captured. Essentially, the detection limit is determined by the minimum number of beads required to be attached to the active area surface, and also capture rate of the beads passing through the channel onto the active area.
III. Fabrication
The microfluidic biochip used in this study is shown in Fig. 4. For our experiments, we fabricated 50 μm deep and 50 μm wide channels (Fig. 6). Au/Cr electrodes (2000Å/150Å) were micropatterned on a glass wafer using traditional photolithography, sputtering, and then liftoff processing. The glass was then cut into separate chips using a wafer saw. The microchannels were fabricated in PDMS (Stanford Microfluidics Foundry). The master mold for the microchannels was patterned onto a silicon substrate using SU-8 photoresist. PDMS (10:1 prepolymer:curing agent) was poured onto the master mold and allowed to cure. The glass chips and the PDMS slabs were aligned and then bonded together after oxygen plasma treatment [24].
Fig. 4.
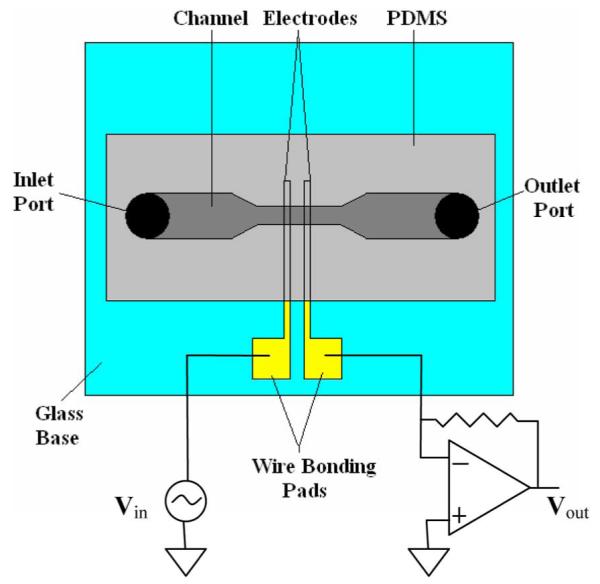
Schematic of microfluidic chip used in this study.
Fig. 6.
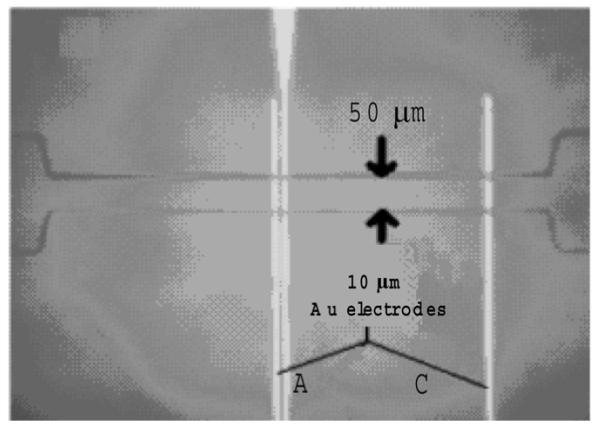
Image of 50 μm deep channel device integrated with outer electrodes labeled A and C. Electrical measurements were made across the channel using electrodes A and C, resulting in an active area of roughly 360 μm.
IV. Device Measurement and Characterization
Electrical impedance measurements were collected across the channel in the region between electrodes A and C. We applied a voltage signal to electrode A. We measured the current at electrode C using a current preamplifier (EI-400 Potentiostat Ensman Instruments, Bloomington, IN) and then the data was collected with a National Instruments data acquisition card and read by a custom-built Labview program. The channels were also monitored using optical microscopy in order to confirm that the signal changes were due to beads binding in between the electrodes.
The physical processes occurring at the interface between the electrode and the electrolyte and also the bulk solution directly dictate the impedance behavior [25]. The small separation of the layer of accumulated ions results in the double layer capacitance dominating the impedance at low frequencies [26]. Effects such as the Warburg impedance and the electron transfer resistance may also significantly affect the impedance at low frequencies [27].
In order to achieve our goal for detection of the presence of the microspheres due to the changes in channel resistance, the effect on the impedance resulting from all impedances except for the bulk solution resistance must be minimized. This can be achieved by working at sufficiently high frequencies. In a previous work [22], we measured the onset of bulk solution resistance dominance of the impedance at frequencies above 20 kHz. We found the optimum frequency to be approximately 30 KHz for our device.
A drift in the impedance also was observed, which we suspected was due to temperature fluctuations in the environment which are as large as 5 °C. In general, solution conductivity varies with temperature. In order to confirm this possibility, we studied the effect of temperature on the current levels. We heated up our biochip to temperatures well above room temperature so that the temperature fluctuations in the room would not have any effects. In the case where we kept the temperature changes to less than 1 °C, a noticeable change was not observed in the current amplitude. However, in the case where temperature was increased by more than 8 °C, a change greater than 2% was observed in the rms current level. Since the greater change in temperature resulted in a greater change in current, it is reasonable to conclude that the swings in temperature of the environment are responsible for the impedance drift.
Given that the impedance changes measured in this study were instantaneous and very rapid compared to the baseline drift, we were able to neglect this effect. However, in order to assure consistency in measurements it would be necessary to alleviate this problem by either isolating the device such that external temperature changes were ineffective, or perhaps to incorporate some sort of a differential measurement system in a parallel control channel in order to subtract out the effects of temperature change on the conductivity of the solution.
V. Real-Time Capture of Functionalized Beads
In this study, we used our biosensors to examine two types of protein–protein interactions. We examined antibody-antigen interactions and glycoprotein–glycoprotein interactions. We covalently coated microscopic beads with the various target proteins of interest. We activated our channel surface by physically adsorbing the receptor protein of interest onto the glass base of our channel.
A. Latex Bead Preparation
We prepared beads with three different target proteins. For studying antigen-antibody interactions, we attached hCG to the microspheres and measured its interactions with polyclonal anti-hCG antibodies which were physically adsorbed onto the glass base of the channel. Glycoprotein–glycoprotein interactions were tested by examining the interactions of the glycoprotein lactoperoxidase, which was immobilized onto the microspheres, and conA, a glycoprotein with specific affinity to sugar molecules, which was immobilized on the surface of the micro-channel.
A volume of 1.5 ml of latex particles (COOH-functionalized, 10.36 μm, 10% solid, Bangs Laboratories) were added to 5 ml of 30 mM MES buffer, pH 5.5, and the suspension was washed several times by centrifugation/resuspension in this buffer. The washed particles were suspended in a final volume of 18 ml of the MES buffer in a 50 ml BD plastic tube, containing 108 mg of EDC and 55 mg of sulfo-NHS, and shaken on a horizontal shaker at room temperature, fixed at a medium speed, for 55 min, while making sure that the particles remained suspended without any precipitation throughout this activation step. The NHS-activated latex particles were then precipitated by centrifugation and washed twice with 80 mM MOPS, pH 8.6, and finally suspended in 9 ml of this buffer. To 3 ml of this suspension, 0.5 ml of a 1 mg/ml hCG or lactoperoxidase, separately made in the MOPS buffer, were added and the suspensions were left on the horizontal shaker at room temperature, fixed at a medium speed, for 5.5 h, again making sure that no precipitation of the particles took place during this period. Finally, the bead suspensions consisting of latex particles (now with the proteins covalently attached to them) were washed several times with PBS and each finally suspended in 0.5 ml of the buffer and stored in the refrigerator for future use.
B. CPG Bead Preparation
Covalent coupling of the proteins to NH2-activated controlled pore glass (CPG) beads was carried out in a one-step reaction in PBS buffer. For lactoperoxidase, the reaction mixture contained 1 mg of the beads, 2.5 mg of the protein, 7 mg of EDC and 7 mg of sulpho-NHS, in a final volume of 1.5 ml PBS. The suspensions were left on a horizontal shaker for 6 h at room temperature, making sure that no precipitation took place during this period. The beads were then washed extensively with PBS by centrifugation followed by resuspension. They were finally suspended in 1 ml of PBS and stored in the refrigerator for future use.
C. Monitoring Protein–Protein Interactions
We performed protein–protein interaction experiments with both 10 latex beads and 10 μm CPG beads. Although we calculated 25 μm sized beads to be adequate in size for sensing a single bead, we chose 10 μm beads since they are easier to work with as they result in less channel clogging. Both the CPG and the latex beads were covalently coated with lactoperoxidase. Lactoperoxidase has an affinity for binding to Con A which we used (at 10 mg/ml) as the probe molecule for immobilization onto the glass base in the microchannel. Nonspecific binding to bare glass surface was very problematic for the latex beads coated with lactoperoxidase, thus we used CPG beads. The functionalized beads were suspended in Hepes buffer and then injected into the channel at a flow rate of less than 50 nl/min. A salt concentration of 200 mM KCl was used in this case to demonstrate the ability of this technique to work at high salt concentrations without degradation in performance. The electrical impedance was measured between electrodes A and C. As functionalized CPG beads became attached onto the electrode (Fig. 7), the electrical impedance measured between electrodes A and C instantaneously increased (Fig. 8).
Fig. 7.

Optical micrograph of electrodes in microchannel at t < 5 s after lactoperoxidase coated CPG bead binds to electrode A. Electrode C not shown.
D. Monitoring Antigen-Antibody Interactions
The antigen-antibody interaction studies were performed using 9 μm diameter latex beads coated with hCG. HCG is a biomarker for pregnancy, and its determination is used for detection of early pregnancy.
The microspheres functionalized with hCG were tested against the hCG antibody (diluted to 10 mg/ml), which was immobilized onto the base of the channel using physical adsorption.
The hCG coated beads attached very well to the antibodies immobilized at the base of the channel. The electrical impedance was measured between electrodes A and C and similar results were obtained as the protein–protein interaction experiments (Fig. 9). Microspheres passing between the electrodes without binding to the surface cause a transient increase in the current (at t = 16 s), and then a return to the original value after they leave the active area of the sensor. At t = 27 s, the peak corresponds to many beads passing across the sensor with only a fraction of them getting captured. The beads which are captured in the active area cause a permanent change in the measured resistance, as seen after t = 27 s.
Fig. 9.

Representative data measured for hCG and anti-hCG interactions. The instantaneous increase in impedance at t = 27 s corresponds to hCG coated latex beads binding onto the active region of the device. The peak at t = 16 s correspond to several beads passing across the sensor without getting capture. The sharp spike at t = 27 s corresponds to many beads passing across the sensor with only some of them getting captured, and then leveling off at approximately 76 kΩ.
VI. Evaluation of Binding Strength
An added advantage of this technique is that the relative binding strength between the proteins can be determined. In general, it is possible to distinguish between specific protein–protein interactions and nonspecific interactions based on the binding strengths. It is also possible to distinguish between various types of protein interactions. Typically, the binding strength resulting from specific antigen-antibody interactions is stronger than that of nonspecific interactions. The fluid flow rate in the channel is also directly proportional to the drag force being applied to the microsphere attached to the base of the channel. The drag force required to pull off the beads from the base of the channel is proportional to the binding strength of the proteins interacting with each other. This means that a larger binding force requires a higher flow rate to unbind the attached microspheres. Thus, by measuring the flow rate required to detach the beads from the base of the channel for various interactions, it is possible to determine the binding strength relative to each other.
In order to examine the binding strength for antigen-antibody interactions and also glycoprotein-antigen interactions, we measured the binding strengths holding the beads for various channel and bead surfaces. For each protein assay, we incubated the functionalized microspheres in the active region of the sensor, until they came to rest at the glass base of the channel. The flow rate of the channel was incrementally increased until the microspheres became detached from the base of the channel. The mean flow rates required for dislodging all of the beads for the various assays and the corresponding standard error bars are shown in Fig. 10.
Fig. 10.
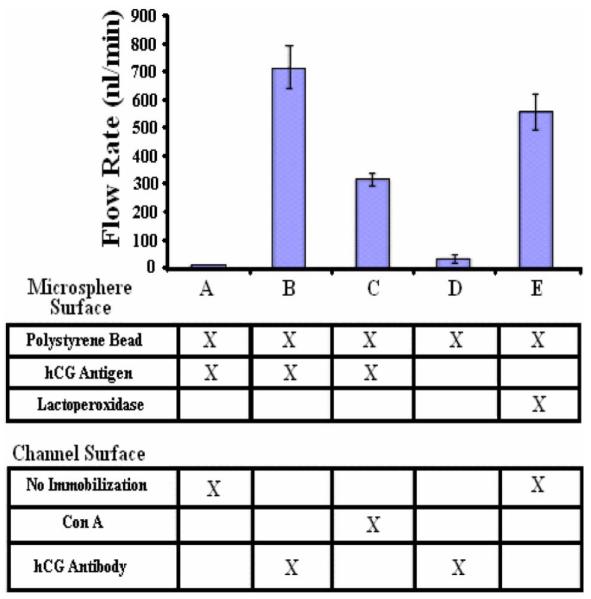
The relative binding strength measured for a variety of protein–protein interactions. In column A, the result of the control experiment is shown, where a hCG coated bead is tested against an untreated channel. In column B, hCG coated beads are tested against a channel with anti-hCG antibody immobilized on the active area. The high flow rate demonstrates the high affinity resulting from specific antibody-antigen interactions. In column C, antigen-glycoprotein interactions are examined. hCG coated beads are tested against a channel with ConA immobilized on the surface. In column D, another control experiment is performed where a plain latex bead is tested against a surface which has anti-hCG antibody immobilized on it. In column E, another control experiment is performed where beads covered with lactoperoxidase are tested against an untreated channel surface, showing high nonspecific binding.
Column A corresponds to the control experiment, where polystyrene beads were functionalized with hCG and they were incubated in a channel which was not bioactivated with any probe molecules. As a result, the beads were removed with a flow rate of 10 nl/min, demonstrating that the binding force between the beads and the surface is negligible.
Column B corresponds to the study of specific interactions between hCG and anti-hCG. Latex beads were functionalized with hCG and tested against a channel bioactivated with anti-hCG antibodies. The microspheres became detached as an average flow rate of 714 nl/min was applied. Binding strengths this large were expected due to the high affinity of specific antigen-antibody interactions.
Column C corresponds to the study of antigen-glycoprotein interactions. HCG functionalized latex beads were incubated in a channel bioactivated with Con A. Given that hCG is a glycoprotein in nature, it was interesting to measure its affinity with ConA compared to its specific interaction with anti-hCG antibody. An average flow rate of 300 nl/min was required to unbind the microspheres. While the affinity is significant, it was not as significant as that of column B, which confirms that specific antigen-antibody interactions are greater in strength than glycoprotein–glycoprotein interactions.
Column D corresponds to another control experiment, where plain latex beads were tested against a channel functionalized with anti-hCG antibody. A low flow rate of 33 nl/min was sufficient to dislodge the beads, confirming that the binding force between the beads and the surface is nonspecific and can therefore be neglected.
Column E corresponds to a third control experiment where latex beads functionalized with lactoperoxidase were tested against a bare channel surface. The binding strength holding down the beads was unexpectedly high, requiring an average flow rate of 560 nl/min to dislodge the beads. It is possible that this large affinity results from charge interactions between the glass and the lactoperoxidase. This nonspecific affinity is quite significant, and in a practical device it would be quite difficult to distinguish between this and specific interactions, which is problematic. It is interesting that the CPG beads which were coated with lactoperoxidase did not have the same nonspecific binding issues which the polystyrene beads faced. This phenomenon can be understood if we analyze the surface charge of the beads. The glass surface of the microchannel and the CPG beads have an isoelectric point (PI) of 3.5 [28], meaning that the surface charge is negative at the pH the system operates. The surface of the polystyrene beads, however, has a PI of 6.5 [29], meaning that it is less negative compared to the CPG beads, almost neutral at the operating pH. Lactoperoxidase has a theoretical PI of 8.3 [30], meaning that the surface charge is positive at the operating pH. Thus, the lactoperoxidase will result in the surface of the latex beads having an overall larger positive charge than the CPG, giving the polystyrene beads a greater affinity to the surface of the glass bead. By optimizing the surface chemistry taking into account the pI information in order to minimize the charge difference between the beads surface and the channel surface, nonspecific binding can be minimized. Also, we have demonstrated elsewhere that nonspecific binding can be minimized using an appropriate blocking buffer [31].
As predicted, the results presented in this section indicate that the binding strength resulting from specific antigen-antibody interactions are much greater than that of nonspecific interactions due to beads coming to rest on the surface of the channel. For the most part, selective binding can be achieved primarily by optimizing the flow rate in the channel. One exception is strong electrostatic interactions which occur due to PI difference of the channel surface and the bead surface, as shown in column E, which have a relatively large nonspecific binding affinity. However, by using the appropriate blocking buffer, these interactions can be minimized, and this problem can be resolved.
VII. Conclusion
We have demonstrated the proof of concept of an impedance-based electrical method for the real-time analysis of protein–protein interactions. By probing solution resistance changes caused by blockage of ionic current due to functionalized beads binding on the surface of a bioactivated microchannel, we successfully detected antigen-antibody interactions, glycoprotein–glycoprotein interactions, and antigen-glycoprotein interactions. We would like to emphasize, however, that this method is easily generalized for the study of a wide array of protein–protein interactions. A key advantage of this technique is its selectivity in bead capture, allowing for the possibility of multiplexed detection of a wide panel of protein–protein interactions using an array of sensors on a single chip.
The selectivity of this technique can be enhanced by immobilizing a primary antibody onto the microsphere and the secondary antibodies on the channel surface. This will allow for an extra level of specificity given that the bead is functionalized only with the protein of interest, making it suitable for analyzing a complex mixture of proteins. The binding of functionalized beads to the antibodies immobilized on the glass base in regions outside of the active area of the sensor also limits sensitivity. Detection limit can be enhanced by effectively increasing the active area of the device by integrating multiple sets of electrodes across the channel. Further enhancements of sensitivity may be achieved by adopting an immobilization procedure which results in antibodies being immobilized predominantly on the gold electrodes, as opposed to the entire channel length. Further studies will be dedicated to multiplexing an array of sensors onto a single chip to be used for separation of proteins in a complex mixture.
Fig. 5.
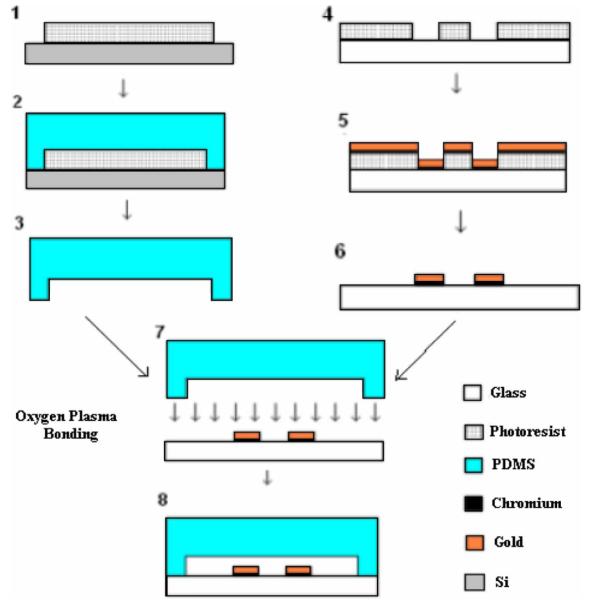
Fabrication process for microchannels with integrated electrodes. In order to fabricate the microchannel. (1) The master mold of the channel is micro patterned onto the Silicon wafer using SU-8 photoresist. (2) PDMS is poured onto the master mold in gel form and then cured. (3) PDMS layer is then peeled off. The gold electrodes are fabricated onto a glass wafer. (4) The electrodes are micro patterned onto the glass wafer using photoresist. (5) The wafer is then sputtered with a layer of chromium and then gold. (6) Liftoff processing is used to remove the unwanted gold and photoresist. (7) The glass chip with electrodes and the PDMS chip are then cleaned in an oxygen plasma oven and aligned, (8) and then bonded. Figure not drawn to scale.
ACKNOWLEDGMENT
The authors would like to thank J. Melin and the Stanford Microfluidics Foundry for their invaluable help in fabrication of the devices used in this study.
This research was supported in part by the National Institutes of Health under Grant P01 HG000205. The associate editor coordinating the review of this paper and approving it for publication was Prof. Anita Lloyd Spetz.
Biography
 Mehdi Javanmard received the B.S. degree with highest honors from the Georgia Institute of Technology, Atlanta, in 2002, and the M.S. and Ph.D. degrees from Stanford University, Stanford, CA, in 2004 and 2008, respectively, all in electrical engineering.
Mehdi Javanmard received the B.S. degree with highest honors from the Georgia Institute of Technology, Atlanta, in 2002, and the M.S. and Ph.D. degrees from Stanford University, Stanford, CA, in 2004 and 2008, respectively, all in electrical engineering.
He has held research positions in Georgia Tech Research Institute, Lawrence Livermore National Laboratory, Stanford Linear Accelerator Center, and Stanford Genome Technology Center. His main research areas are biosensors, biolectronics, and microfluidics with an emphasis on detection of pathogenic bacteria, genetic biomarkers, and protein biomarkers.
 Amirali H. Talasaz received the B.S. degree from Sharif University of Technology, Tehran, Iran, in 2001, and the M.S. and Ph.D. degrees from Stanford University, Stanford, CA, in 2003 and 2007, respectively, all in electrical engineering.
Amirali H. Talasaz received the B.S. degree from Sharif University of Technology, Tehran, Iran, in 2001, and the M.S. and Ph.D. degrees from Stanford University, Stanford, CA, in 2003 and 2007, respectively, all in electrical engineering.
He currently serves as a Research Associate at the Stanford Genome Tecnhology Center, Stanford University.
 Mohsen Nemat-Gorgani received the B.S. degree in biochemistry from London University, London, U.K., in 1970, the M.S. degree in molecular enzymology and the Ph.D. degree in biochemistry from Warwick University, Warwick, U.K., in 1972 and 1974, respectively.
Mohsen Nemat-Gorgani received the B.S. degree in biochemistry from London University, London, U.K., in 1970, the M.S. degree in molecular enzymology and the Ph.D. degree in biochemistry from Warwick University, Warwick, U.K., in 1972 and 1974, respectively.
He served as a Postdoctoral Scholar at Vanderbilt University from 1974 to 1975. He has held various academic and industrial positions since then. He is currently a Professor at the Institute of Biochemistry and Biophysics, University of Tehran and also a Visiting Professor/Research Associate at the Stanford Genome Technology Center, Stanford University.
 David E. Huber received the Ph.D. degree from Stanford University, Stanford, CA, in 2006 after completing his dissertation, which was an investigation of transport and dispersion phenomena in an electrokinetic concentration technique called microfluidic temperature gradient focusing (TGF).
David E. Huber received the Ph.D. degree from Stanford University, Stanford, CA, in 2006 after completing his dissertation, which was an investigation of transport and dispersion phenomena in an electrokinetic concentration technique called microfluidic temperature gradient focusing (TGF).
He is an Engineering Research Associate at the Stanford Genome Technology Center. Prior to joining the center, he was a Postdoctoral Appointee at Sandia National Laboratories, where he worked on the development of a nanochannel-based DNA hybridization assay, nanochannel fabrication, and numerical modeling of fluid flow and heat transfer within microfluidic devices. He was a member of the Stanford University Microfluidics Laboratory. Previously, he held a series of engineering and management positions at Echelon Corporation, Genemachines, and Affymax Research Institute.
 Fabian Pease received the B.A., M.A., and Ph.D. degrees from Cambridge University, Cambridge, U.K., in 1960, 1962, and 1964, respectively. His Ph.D. thesis was on High-Resolution Scanning Electron Microscopy.
Fabian Pease received the B.A., M.A., and Ph.D. degrees from Cambridge University, Cambridge, U.K., in 1960, 1962, and 1964, respectively. His Ph.D. thesis was on High-Resolution Scanning Electron Microscopy.
After graduating, he was an Assistant Professor of Electrical Engineering at University of California, Berkeley, for three years, where he continued his microscopy research. In 1967, he joined the Technical Staff of Bell Laboratories, where he first worked on digital television and later led a group that developed the processes for electron beam lithographic mask manufacture, and demonstrated a pioneering LSI circuit built with electron beam lithography. Since 1978, he has been a Professor of Electrical Engineering at Stanford University. His group’s areas of research include micro and nanofabrication and their application to electronic and magnetic devices and structures. This work has included the original demonstration of lithography with the scanning tunneling microscope, exploring the limits of resolution of deep ultraviolet lithography, the invention of the microchannel heat sink and nonconventional electron beam technology for semiconductor manufacturing. He was appointed the William E. Ayer Professor of Electrical Engineering in March 2001. He has published over 200 articles and authored several book chapters.
Dr. Pease has served the IEEE in a variety of capacities. He is also a member of the National Academy of Engineering. With his student, D. Tuckerman, he received the first IEEE Paul Rappaport Award. He was also the recipient of the IEEE Cledo Brunetti Award in 2001, for advancing high-resolution patterning technologies, high-performance thermal management, and scanning electron microscopy for microelectronics. Other honors include the Richard P. Feynman Prize for Microfabrication, which he shared with a student, T. Newman, for writing a page of text in a 6 micron square with 25 nm linewidths, and a Title A Fellowship from Trinity College, Cambridge.
 Mostafa Ronaghi received the Ph.D. degree from the Royal Institute of Technology, Stockholm, Sweden.
Mostafa Ronaghi received the Ph.D. degree from the Royal Institute of Technology, Stockholm, Sweden.
He is a molecular biologist, specializing in DNA sequencing methodology. He is currently the Chief Technology Officer and Senior Vice President at Illumina. Prior to this position, he was a Principal Investigator and Senior Research Associate at the Stanford Genome Technology Center, Stanford University, focusing on developing analytical techniques for molecular diagnostics. He is an inventor of Pyrosequencing Technology and co-founded Pyrosequencing AB in 1997 (renamed to Biotage in 2003), and co-invented Molecular Inversion Probe assay and co-founded ParAllele BioScience to develop this multiplexed technology for genetic testing. ParAllele was acquired by Affymetrix in May 2005. In 2005, he co-founded NextBio, a search engine for life science data. He also serves on the Board of Directors of Microchip Biotechnologies and Aurora Biofuels.
 Ronald W. Davis received the B.S. degree in mathematics, physics, chemistry, and botany from East Illinois University, Charleston, in 1964, and the Ph.D. degree in chemistry from California Institute of Technology, Pasadena, in 1970.
Ronald W. Davis received the B.S. degree in mathematics, physics, chemistry, and botany from East Illinois University, Charleston, in 1964, and the Ph.D. degree in chemistry from California Institute of Technology, Pasadena, in 1970.
He is considered to be a world leader in biotechnology, and the development and application of recombinant DNA and genomic methodology to biological systems. His laboratory has developed many of the techniques currently used in academic and industrial biotechnology laboratories. His laboratory was also instrumental in the development of lambda vectors, which were commonly used for the primary cloning of DNA molecules in E. coli. In 1983 he became a member of The National Academy of Sciences.
Dr. Davis is also considered to be a world expert in the electron microscopy of nucleic acids and has developed many of the mapping methods for which he received the Eli Lilly Award in Microbiology in l976. His laboratory also developed many of the yeast vectors and helped to develop yeast as a host for recombinant DNA for which he received the United States Steel Award in l98l, presented by the National Academy of Sciences. He was a coauthor on a publication that first described a new approach for conducting human genetics and for the construction of a human genetic linkage map for which he received the Rosentiel Award for Work in Basic Medical Research. His laboratory is now conducting genomic analysis of Saccharomyces cerevisiae for which he received the 2004 Lifetime Achievement Award from the Yeast Genetics Society. His laboratory is developing many new technologies for the genetic, genomic, and molecular analysis of model organisms and human with a focus on clinical medicine for which he received the 2004 Sober Award from the American Society for Biochemistry and Molecular Biology (ASBMB/IUBMB).
REFERENCES
- [1].Burnette WN. Western blotting electropheretic blotting electrophoretic transfer of proteins from sodium dodecyl sulfate poly acrylamide gels to unmodified cellulose and radiographic detection with antibody and radio iodinated protein a. Anal. Biochem. 1981;vol. 112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- [2].Crowther JR. The ELISA Guidebook. Humana Press; Totowa, NJ: 2001. [Google Scholar]
- [3].MacBeath G. Protein microarrays and proteomics. Nature Genetics. 2002;vol. 32:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- [4].Ayliffe HE, Frazier AB, Rabbitt RD. Electric impedance spectroscopy using microchannels with integrated metal electrodes. J. Microelectromech. Syst. 1999;vol. 8:50–7. [Google Scholar]
- [5].Cheng IF, Hsien-Chang C, Hou D, Hsueh-Chia C. An integrated dielectrophoretic chip for continuous bioparticle filtering, focusing, sorting, trapping, and detecting. Biomicrofluidics. 2007;vol. 1:021503–15. doi: 10.1063/1.2723669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gomez R, Bashir R, Sarikaya A, Ladisch MR, Sturgis J, Robinson JP, Geng T, Bhunia AK, Apple HL, Wereley S. Microfluidic biochip for impedance spectroscopy of biological species. Biomed. Microdevices. 2001;vol. 3:201–9. [Google Scholar]
- [7].Gomez-Sjoberg R, Morisette DT, Bashir R. Impedance microbiology-on-a-chip: Microfluidic bioprocessor for rapid detection of bacterial metabolism. J. Microelectromech. Syst. 2005;vol. 14:829–38. [Google Scholar]
- [8].Liu YS, Walter TM, Chang WJ, Lim KS, Yang LJ, Lee SW, Aronson A, Bashir R. Electrical detection of germination of viable model Bacillus anthracis spores in microfluidic biochips. Lab on a Chip. 2007;vol. 7:603–610. doi: 10.1039/b702408h. [DOI] [PubMed] [Google Scholar]
- [9].Radke SM, Alocilja EC. Design and fabrication of a microimpedance biosensor for bacterial detection. IEEE Sensors J. 2004;vol. 4:434–440. [Google Scholar]
- [10].Radke SM, Alocilja EC. A microfabricated biosensor for detecting foodborne bioterrorism agents. IEEE Sensors J. 2005;vol. 5:744–750. [Google Scholar]
- [11].Yang LJ, Li YB, Johnson CLGG. Interdigitated microelectrode (IME) impedance sensor for the detection of viable Salmonella typhimurium. Biosensors Bioelectronics. 2004;vol. 19:1139–47. doi: 10.1016/j.bios.2003.10.009. [DOI] [PubMed] [Google Scholar]
- [12].Yang LJ, Ruan CM, Li YB. Detection of viable Salmonella typhimurium by impedance measurement of electrode capacitance and medium resistance. Biosensors Bioelectronics. 2003;vol. 19:495–502. doi: 10.1016/s0956-5663(03)00229-x. [DOI] [PubMed] [Google Scholar]
- [13].Ionescu-Zanetti C, Nevill JT, Carlo DD, Jeong KH, Lee LP. Nanogap capacitors: Sensitivity to sample permittivity changes. J. Appl. Phys. 2006;vol. 99:1–5. [Google Scholar]
- [14].Lee WC, Cho YH, Pisano AP. Nanomechanical protein concentration detector using a nanogap squeezing actuator with compensated displacement monitoring electrodes. J. Microelectromech. Syst. 2007;vol. 16:802–808. [Google Scholar]
- [15].Nevill JT, Carlo DD, Liu P, Jeong KH, Lee LP. Proc. Digest of Tech. Papers—Int. Conf. Solid State Sensors, Actuators, Microsyst., TRANSDUCERS ’05. vol. 2. 2005. Detection of protein conformational changes with a nanogap biosensor; pp. 1668–1671. [Google Scholar]
- [16].Yi MQ, Jeong KH, Lee LP. Theoretical and experimental study towards a nanogap dielectric biosensor. Biosensors Bioelectronics. 2005;vol. 20:1320–1326. doi: 10.1016/j.bios.2004.05.003. [DOI] [PubMed] [Google Scholar]
- [17].Coulter W. U. S. Pat. 1953;Vol. 2656508
- [18].Carbonaro A, Sohn LL. A resistive-pulse sensor chip for multianalyte immunoassays. Lab on a Chip. 2005;vol. 5:1155–1160. doi: 10.1039/b504827c. [DOI] [PubMed] [Google Scholar]
- [19].Gawad S, Schild L, Renaud P. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab on a Chip. 2001;vol. 1:76–82. doi: 10.1039/b103933b. [DOI] [PubMed] [Google Scholar]
- [20].Saleh OA, Sohn LL. Quantitative sensing of nanoscale colloids using a microchip coulter counter. Rev. Scientific Instruments. 2001;vol. 72:4449–51. [Google Scholar]
- [21].Saleh OA, Sohn LL. Direct detection of antibody-antigen binding using an on-chip artificial pore. Proc. Nat. Acad. Sci. United States of America. 2003;vol. 100:820–824. doi: 10.1073/pnas.0337563100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Javanmard M, Talasaz AAH, Nemat-Gorgani M, Pease F, Ronaghi M, Davis RW. Targeted cell detection based on microchannel gating. Biomicrofluidics. 2007;vol. 1:044103–1. doi: 10.1063/1.2815760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Koch M, Evans AGR, Brunnschweiler A. Microfluidic Technology and Applications. Baldock Research Studies Press Ltd.; Baldock, U.K.: 2000. [Google Scholar]
- [24].McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA, Whitesides GM. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;vol. 21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- [25].Wu J, Ben YX, Chang HC. Particle detection by electrical impedance spectroscopy with asymmetric-polarization AC electroosmotic trapping. Microfluidics and Nanofluidics. 2005;vol. 1:161–167. [Google Scholar]
- [26].Wu J, Ben YX, Battigelli D, Chang HC. Long-range AC electroosmotic trapping and detection of bioparticles. Ind. Eng. Chem. Res. 2005;vol. 44:2815–2822. [Google Scholar]
- [27].Katz E, Willner I. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: Routes to impedimetric immunosensors, DNA-sensors, and enzyme biosensors. Electroanalysis. 2003;vol. 15:913–947. [Google Scholar]
- [28].Maheshwari R, Bhavani R, Dhathathreyan A. Solid-liquid interfacial energy as a tool to estimate shifts in isoelectric points of adsorbed proteins on solid surfaces. J. Colloid and Interface Sci. 2006;vol. 293:500–504. doi: 10.1016/j.jcis.2005.06.076. [DOI] [PubMed] [Google Scholar]
- [29].Somasundaran T, Deo N, Somasundaran P. Zwitterionic latex particles as an effective carrier for DNA. J. Colloid and Interface Sci. 2002;vol. 246:223–226. doi: 10.1006/jcis.2001.7947. [DOI] [PubMed] [Google Scholar]
- [30].Lindh L, Svendsen IE, Svensson O, Cárdenas M, Arnebrant T. The salivary mucin MUC5B and lactoperoxidase can be used for layer-by-layer film formation. J. Colloid and Interface Sci. 2007;vol. 310:74–82. doi: 10.1016/j.jcis.2007.01.086. [DOI] [PubMed] [Google Scholar]
- [31].Javanmard M, Talasaz AAH, Nemat-Gorgani M, Pease F, Ronaghi M, Davis RW. Electrical immunoassay using protein functionalized microchannels. Lab on a Chip J. 2009 doi: 10.1039/b818872f. [DOI] [PMC free article] [PubMed] [Google Scholar]