Summary
Background
All AKR/J mice have a subtle defect that involves malformation of the central portion of hair fibres that is best visualized under white and polarized light microscopy.
Aims
This study sought to characterize the clinical and ultrastructural features of the hair interior defect (HID) phenotype and to determine the chromosomal localization of the hid mutant gene locus.
Methods
White and polarized light microscopy combined with scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were used to characterize the HID phenotype. Complementation testing and gene-linkage studies were performed to map the locus.
Results
Using SEM, the hair-fibre structure on the surface was found to be similar to hairs obtained from normal BALB/cByJ+/+and C57BL/6 J+/+mice. There were also no differences in sulphur content. TEM revealed degenerative changes in the medulla similar to that seen by light microscopy. This autosomal recessive mutation is called HID (locus symbol: hid). We mapped the hid locus to the distal end of mouse chromosome 1. No genes reported to cause skin or hair abnormalities are known to be within this interval except for the lamin B receptor (Lbr), which had been excluded previously as the cause of the hid phenotype in AKR/J mice.
Conclusion
A potentially novel gene or known gene with a novel phenotype resides within this interval, which may shed light on human diseases with defects in the inner structure of the hair fibre.
Introduction
The hair interior defect mutation (HID; locus symbol hid) arose spontaneously in the AKR/J mouse strain and over time became fixed, thus all AKR/J mice are homozygous for this mutation (hid/hid).1 Although hid is described as an autosomal recessive mutation, very little is known about its genetics. Clinically, hid/hid mice appear to be completely normal at all ages with no abnormal hair cycling, and the albino coat colour is not affected. The phenotype can only be recognized by microscopic examination of the pelage hair. Trigg1 observed that in most hair types the inner structure of the hair fibre, particularly the corticomedullary boundary, was affected, and in rare cases the diameter of the hair fibre was also reduced. The pathogenesis of HID was proposed to be due to a failure of cells within the fibre to change shape during early differentiation.1,2
Humans also have a variety of hair fibre disorders caused by defects of the inner structure of the hair fibre that can cause light-refraction changes. These include pili annulati, trichothiodystrophy, and other diseases.3–5 Finding mutant laboratory mice with similar phenotypes is a useful strategy for identifying the genes responsible for these diseases, and these mice may serve as preclinical tools to test hypotheses and ultimately therapeutic approaches.
We report the chromosomal localization of the hid locus, features of the hair fibres, and show by complementation analysis that this is not due to a mutation in Foxq1, another gene that when mutated results in abnormalities of the central region of the hair fibres.
Methods
All studies were performed using Institutional Animal Care and Use Committee approved protocols.
Mice
AKR/J-hid/hid (JR648), BALB/cByJ+/+(JR1026), C57BL/6 J+/+(JR664), and SB/LeJ-Foxq1sa/Foxq1sa (JR269) mice were obtained (Jackson Laboratory, Bar Harbor, ME, USA). Mice were maintained in an animal unit with controlled humidity, temperature and light cycle (12 h light/dark cycle) under specific pathogen-free conditions (http://jaxmice.jax.org/html/health/quality_control.shtmlAnimalhealth). Mice were housed in double-pen polycarbonate cages (0.033 m2 floor area) at a maximum capacity of four mice per pen. Mice were allowed free access to autoclaved food (NIH 31, 6% fat; LabDiet 5K52, Purina Mills, St. Louis, MO, USA) and acidified water (pH 2.8–3.2).
Microscopic examination of hair structure
To determine the earliest point at which the HID was detectable, hairs were manually plucked at 3-day intervals from the dorsal thoracic interscapular region from a minimum of two AKR/J-hid/hid mice aged 9–20 days. Additional specialized hairs were collected from the muzzle (vibrissae), eyelids (cilia), perianal and inguinal regions, around the feet (vibrissae), tail, and inner and outer surfaces of the ear pinna. The hair plucking did not traumatize the hair fibre (see below). Plucked hairs were collected in clean, labelled, screw-capped plastic tubes (Nunc; Nalge Nunc International, Denmark). For analysis, 20–30 hairs were mounted on labelled and precleaned glass slides (Fisherbrand; Fisher Scientific, Pittsburgh, PA, USA) moistened with xylene. In total, 20–30 plucked hairs were placed on each slide and separated, then 3–4 drops of mounting medium (Permount; Fisher Scientific) were placed on the bottom of the slide. A glass coverslip was placed on the edge, and the mounting medium pulled across the slide by capillary action to eliminate air bubbles. After drying, slides were examined with a photomicroscope using standard white and polarized light microscopy. Polarized light reveals banding patterns that may be characteristic of some diseases, such as the so-called ‘tiger stripes’ in trichothiodystrophy.3
Scanning electron microscopy
To evaluate structural integrity of the hair surfaces, scanning electron microscopy (SEM) and element analysis were performed on guard hairs from AKR/J-hid/hid, BALB/cByJ+/+, and C57BL/6 J+/+mice (2-month-old females, two mice from each strain). Hair was removed manually from the dorsal thoracic interscapular region of each mouse. The mouse hair cycles in waves, and in adults, if the easily removed, as in this case, the follicles are in telogen or exogen, which allows them to be removed without damage. This was confirmed by removing a 1-cm2 piece of dorsal skin and evaluating the nonmanipulated hairs in situ by SEM.
The hair fibres were mounted on aluminium stub using double-sided sticky tape, sputter-coated with a 4-nm layer of gold, and examined at 20 kV at a working distance of approximately 15 mm on an SEM (Hitachi S3000N VP; Hitachi Science Systems, Japan).6
Hair fibres were assessed for sulphur content7 by weight using an X-ray microanalysis system (EDAX; Mahwah, NJ, USA). Areas included the club, mid-shaft, and tip regions from at least two different hairs per mouse. Samples were examined for an average of at least 300 live seconds to ensure that a comprehensive reading was obtained.8
Transmission electron microscopy
In order to evaluate the cellular structure within the pelage hair fibres, additional hair samples from the groups evaluated by SEM as describe above were processed for transmission electron microscopy (TEM). Owing to the hard consistency of the hair fibres in plastic blocks, they were easily removed during sectioning, making evaluation impossible. In order to section the hair fibres they first needed to be extracted.
Typically ≥ 10 hair fibres per sample were incubated in 5–10 mL of 50 mmol/L sodium phosphate buffer pH 7.9, 2% sodium dodecyl sulphate, and 20 mmol/L dithiothreitol for 4 h at 65 °C. After recovery by low-speed centrifugation, the extracted hairs were fixed in 2.5% glutaraldehyde in 0.1 mol/L cacodylate buffer (pH 7.2), postfixed with buffered 1% osmium tetroxide overnight at 4 °C, dehydrated in a graded series of ethanol and infiltrated with Spurr resin. Blocks were orientated visually to produce longitudinal and cross-sections of the hair fibre. Sections were then examined by TEM (JEM-1230; Jeol USA Inc., Peabody, MA, USA) as previously described.9
Complementation (allelism) testing
Both the satin (Foxq1sa/Foxq1sa) and HID (hid/hid) mutant mice have abnormalities within their respective hair fibres. As the gene responsible for the satin mutation was determined to be Foxq1,10 intercrossing these two mutant mice was performed to determine if they were allelic mutations. Three AKR/J-hid/hid females were crossed with three male SB/LeJ-Foxq1sa/Foxq1sa mice. The progeny from these crosses had hairs plucked at 20, 30, and 45�days of age. The hairs were mounted and examined as described above. If the F1 progeny from the double homozygous cross had medulla defects, then the hid gene would be noncomplementary and hid would be an allelic mutation of Foxq1. However, if the progeny were clinically normal, then hid would be complementary and therefore due to a mutation in a different gene.
Gene mapping
Four AKR/J hid/hid female mice were mated with four BALB/cByJ+/+male mice, yielding 24 F1 progeny, which were used to create 12 breeding pairs. The phenotype of the F1 and F2 mice was determined by light microscopic examination of the hair fibres from the dorsal thoracic interscapular region.
The tail tips of the 12 mutant mice were amputated, DNA extracted using a modified hot sodium hydroxide protocol,11 and the DNA pooled. DNA was also collected from 10 clinically normal F2 progeny and 2 parental mice (AKR/J-hid/hid females and BALB/cByJ+/+males. DNA concentrations (optical density 260 nm) were determined on all DNA samples, and equal amounts of DNA from the 12 mutant mice were mixed to make a pooled sample. A similar pooled sample was assembled from the 12 normal siblings. These two pools were tested for simple sequence length polymorphism (SSLPs) allele ratios compared with parental and F1 controls using 87 markers spaced at 7–35 cM (mean spacing 14.4 cM) apart throughout the genome.12
Samples were amplified in a thermal cycler (PTC225 or PTC200; PerkinElmer, Waltham, MA, USA) with initial incubation at 94 °C for 3�min, followed by 38 cycles of melting at 94 °C for 30 s, annealing at 55 °C for 35 s at 72 °C for 30 s, and a final extension at 72 °C for 7min, after which the samples were held at 4 °C. Each reaction had a volume of 12 µL containing 3 ng DNA template and 0.025 units/µL Taq DNA Polymerase (Eppendorf, New York, NY, USA). The final concentration of the other reagents were 0.12 µmol/L for each primer (Invitrogen Corp., Carlsbad, CA, USA or Integrated DNA Technologies, Coralville, IA, USA), 1.5 mmol/L MgCl2, 200 µmol/L each dNTP (Amersham Biosciences, Piscatawy, NJ, USA), 50 mmol/L KCl, 10 mmol/L Tris-HCl pH 8.3, and 0.001% gelatin. Primer sequences and location are available upon request. PCR products were stored at −20 °C prior to electrophoresis. Fluorescent dye (0.5 × SYBR Green I Nucleic Acid Stain; Cambrex Corporation, East Rutherford, NJ, USA) was added to each 3.5% agarose gel (MetaPhor; Cambrex Corp., East Rutherford, NJ USA) and separated by electrophoresis in Tris/borate/EDTA buffer. Gels were run at 300 V for 1–3 h, and photographed using medium-wave ultraviolet light, using a high-resolution digital camera system (EDAS290; Kodak, Rochester, New York, NY, USA).
Markers that showed bias to the AKR/J allele in the mutant pool PCR, when the normal pool PCR showed either no bias or a bias toward the BALB/cByJ allele, were re-run on all individual DNAs to initiate the map-building. More markers from the resulting candidate interval were typed to refine the map.
Results
Clinical features/macroscopic examination
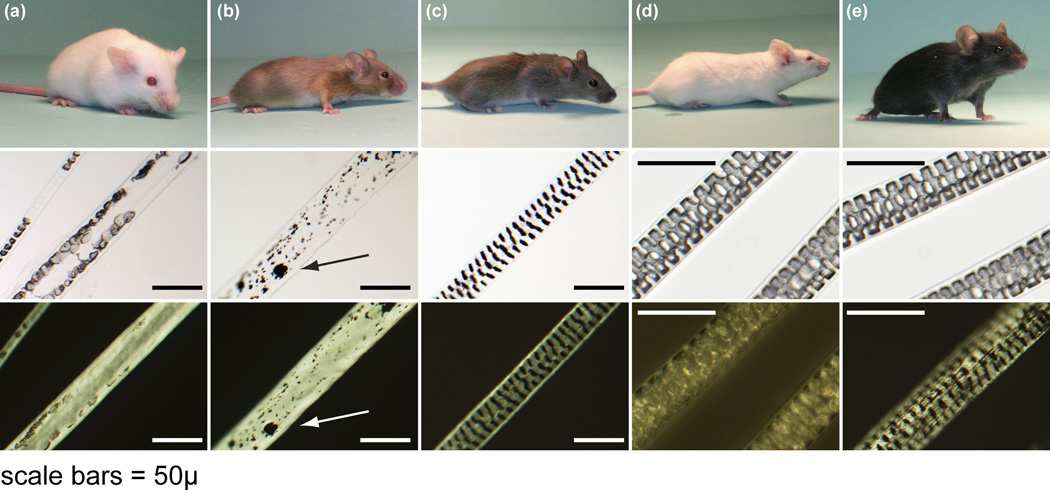
Mice of the AKR/J inbred strain were all homozygous for the HID (hid/hid) phenotype. No abnormal features were evident by gross examination at the time of birth. The first hair coat (completion of embryogenesis) emerged at 5 days postpartum as is commonly found in most inbred strains including AKR/J-hid/hid, BALB/cByJ+/+ (an unrelated inbred albino mouse strain), and C57BL/6 J+/+ (an unrelated inbred pigmented mouse strain).13 No abnormalities in coat texture, colour, or length of the hair fibres were discernable macroscopically when examined at various ages (from 5 days until 6 months of age) (Fig. 1). However, when the two albino strains were paired, the AKR/J mice appeared to have a softer appearance to their coats, which was similar to the satiny sheen observed in the satin (Foxq1sa/Foxq1sa) mutant mice (Fig. 1).14
Figure 1.
Hair colour, texture, and fibre quality in a complementation test. (a) AKR/J-hid/hid mice were crossed with (b) SB/LeJ-Foxq1sa/Foxq1sa, Lystbg/Lystbg mutant mice. (c) Their F2 progeny were brown and had normal guard hair fibres with septulate patterns similar to that of (d) normal BALB/cByJ+/+albino and (e) C57BL/6 J+/+black mice. Representative guard hairs are shown below each mouse under white and polarized light to emphasize the defects in the centre of the hair fibres in (a) the hair intereior defect and (b) satin mutant mice. Clumping of melanin pigment (arrow) in (b) the satin mice is because they are also homozygous for the tightly linked beige allelic mutation of the lysosomal trafficking regulator (Lystbg/Lystbg) gene responsible for the beige phenotype (bar = 50 µm).
Light microscopy
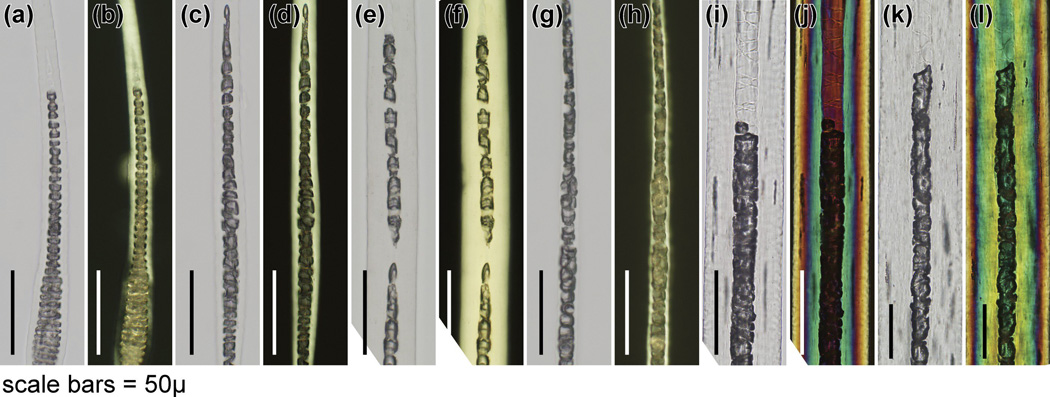
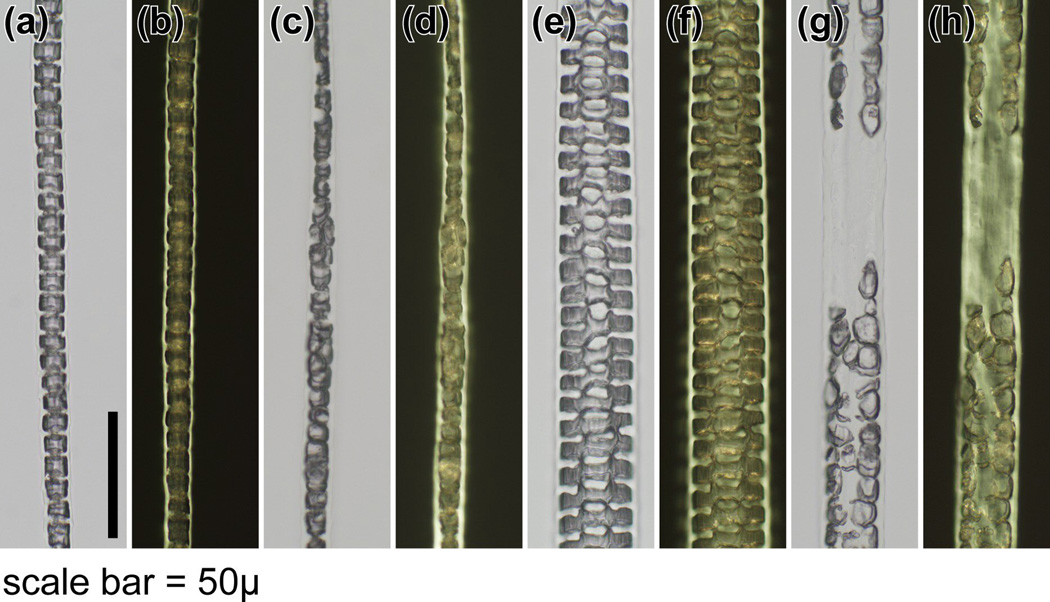
The HID was evident in mounted hairs obtained from mice at the earliest time they could be manually removed (9 days of age). Hair types from various anatomical sites (pelage, vibrissae, cilia, tail, ear, hair around feet, genital/perianal) were examined. The central region of the hair fibre in hid/hid mutant mice was most obvious in the wider pelage hairs, primarily the awl and guard hairs, which normally have a septate pattern. The central region of these fibres appeared to lack a cellular structure or cells that appeared to form the cortex were large, elongated or sometimes round, and extended into the central region. These abnormal cells had an irregular arrangement and shape and no compaction, with various degrees of severity between individual hairs from any one mouse compared with the unaffected BALB/cByJ+/+and C57BL/6 J+/+mouse hairs used as controls (Fig 1–3). In the thinner hair fibres that were septate, the abnormality was present, the septa were disorganized, and changes were very subtle compared with the unaffected control mice (Fig 2, Fig 3). Hairs tapered in diameter at their proximal and distal ends. When comparable regions were compared in photomicrographs, the AKR-hid/hid hairs were usually thinner (Fig 2, Fig 3).
Figure 3.
Histology of specialized hairs. (a,c,e,g,i,k) White and (b,d,f,h,j,l) polarized microscopy revealed very subtle differences in septation patterns in the centre of(a–d) cilia (eyelashes), (e–h) tail hairs, and (i–l) vibrissae. Left pair, BALB/cByJ+/+; rght pair AKR/J-hid/hid mice (bar = 50 µm).
Figure 2.
Histology of pelage hairs. (a,c,e,g) White and (b,d,f,h) polarized microscopy showed a normal (a,b) septate pattern in the medulla of zigzag hairs and (e,f) septulate pattern in guard hairs from a BALB/cByJ+/+mouse. IN contrast, the septations in zigzag hairs from AKR/J-hid/hid mice were disorganized, with (c, d) some of the cells being rounded rather than rectangular. These changes were prominent in (g,h) guard hairs, where the septulate pattern was completely missing. The medulla also seemed to be missing, and cells at the corticomedullary junction were rounded and intermittent (bar = 50 µm).
Ultrastructural analysis
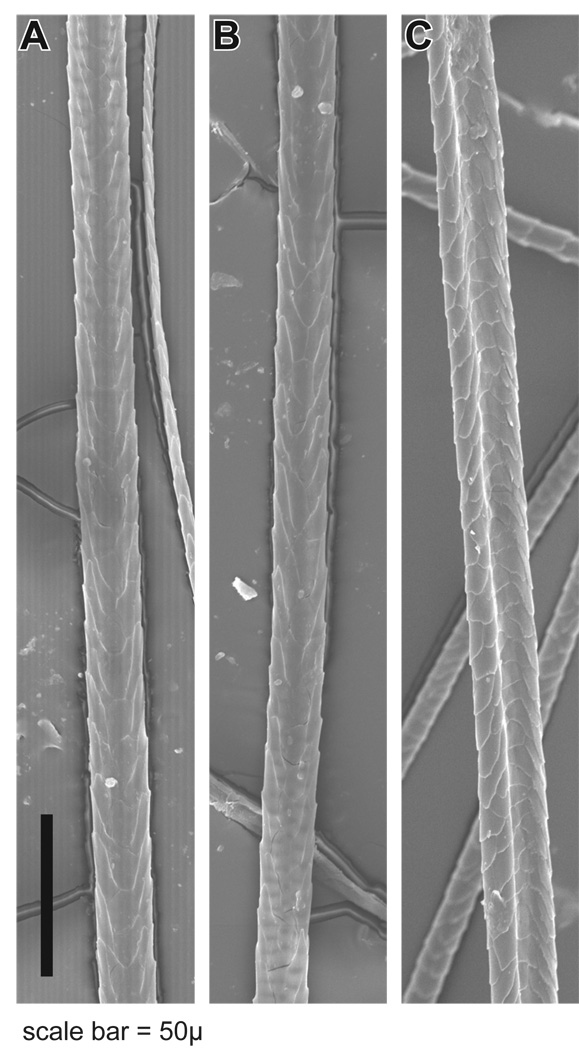
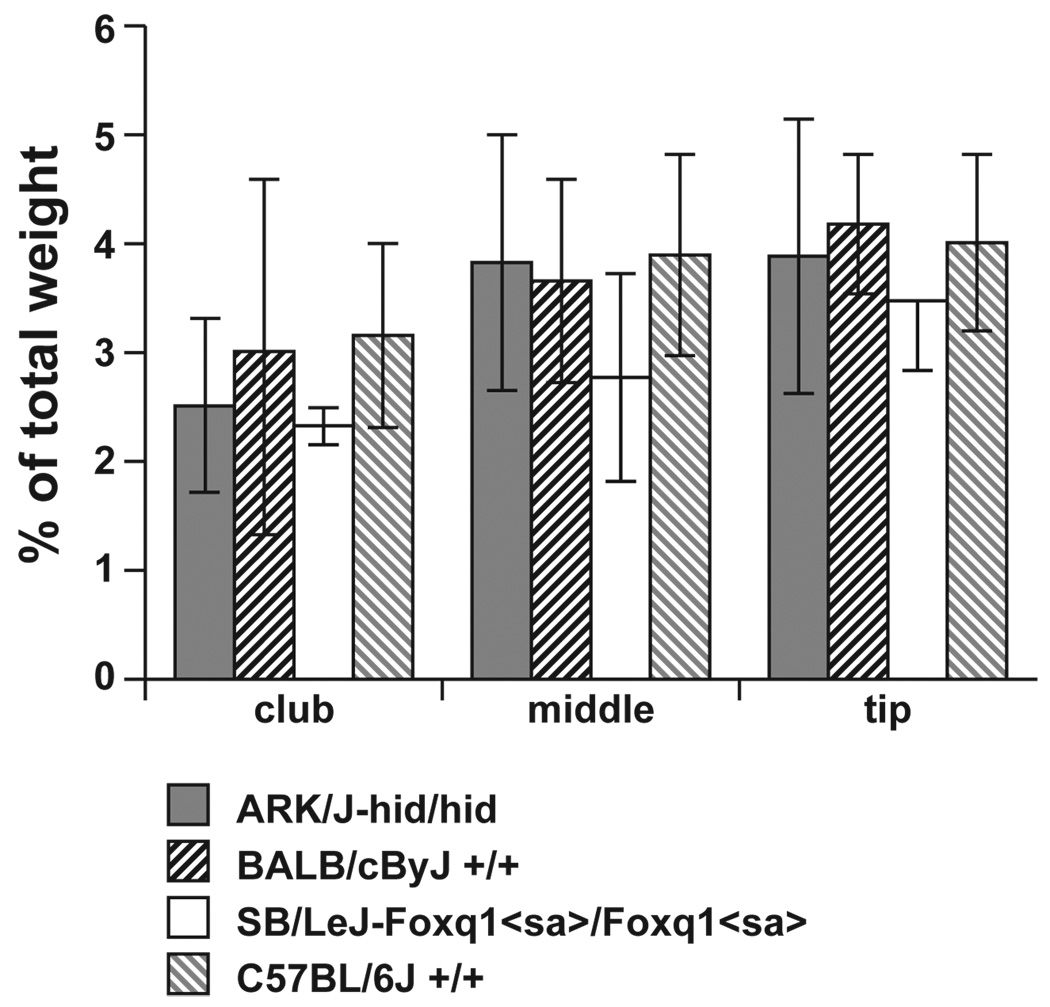
SEM is a useful approach to screen hair fibres for structural defects. In all four strains, AKR/J-hid/hid, BALB/cByJ+/+, C57BL/6 J+/+, and SB/LeJ-Foxq1sa/Foxq1sa, the hair-fibre surface structure was normal, with the cuticle formed in a regular pattern (Fig. 4). Element analysis was performed to determine if sulphur levels were abnormal, a sign of trichothiodystrophy, a common phenotype associated with structural defects in mouse hair fibres. Although there was variation in the sulphur concentration along the length of the fibres, the club region consistently had lower levels than the midshaft and distal tip. However, these differences were not significant between hairs from the four strains using the Kruskal–Wallis test (Fig. 5; club: P-value 0.671; middle: P-value 0.946; tip: P-value 0.946). Additionally there was no significant difference at various locations in each strain using the Friedman test (AKR/J P = 0.944; BALB/cByJ P = 0.944; SB/LeJ P = 0.500; C57BLP = 0.833).
Figure 4.
The surface structures of guard hairs from (a) BALB/cByJ, (b) AKR/J-hid/hid, and (c) SB/LeJ-Foxq1sa/Foxq1sa are essentially identical by scanning electron microscopy (bar = 50 µm).
Figure 5.
Element analysis of hair fibres. There were no significant differences in the sulphur levels, an indicator of structural integrity of hair fibres, between guard hairs in AKR/J-hid/hid, BALB/cByJ+/+, SB/LeJ-Foxq1sa/Foxq1sa, and(c) C57BL/6 J+/+ mice. Levels were reduced in the club region of the hair fibres in all four strains compared with the middle and distal tips but these were not significantly different. Kruskal–Wallis test; Club P = 0.671; middle P = 0.946; tip P = 0.946. Error bars indicate standard deviation.
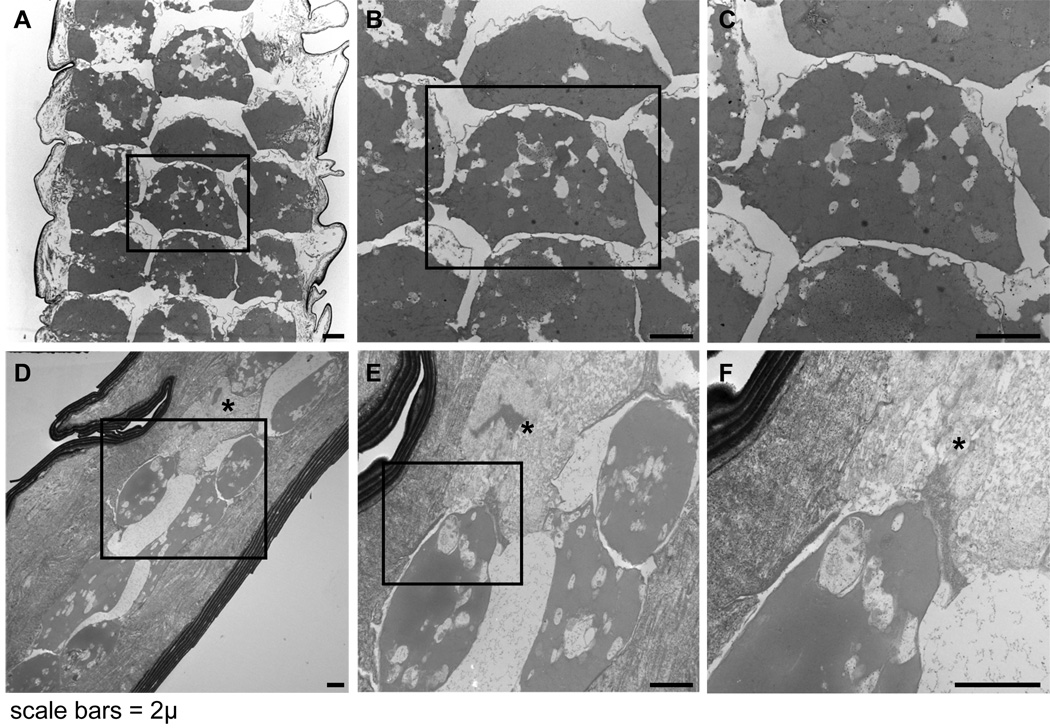
Detergent-extracted hairs examined by TEM revealed that in the control mice the widest hair fibres had a central region (medulla) of multiple, regularly spaced, parallel columns of roughly square to slightly rounded, desiccated cells. The regular separations between the cells created the septate and septulate patterns characteristic of normal hair fibres in mice. The outline of a central nucleus and bundles of electron opaque material with poor cytological detail were evident within the desiccated medullary cells (Fig. 6a–c). In contrast, the medulla of AKR/J-hid/hid mutant mice contained rounded desiccated cells with spaces containing fine granular material, presumably protein. These spaces appeared to be remnant spaces occupied by the cells before they dehydrated and decreased in size. Some of the cells were disintegrating. Most of those present contained an amorphous electron opaque (grey) cytoplasm with clear vacuoles of various sizes. The round to oval shape of these cells closely resembled the clear, rounded cells seen within the hairs by light microscopy (Fig. 6d–f). The cuticle in both the mutant and control mouse hairs consisted of a series of overlapping plate-like structures. The cortex was amorphous although ghost-like remnants of cell membranes were evident in AKR/J-hid/hid hairs but were not evident in the control hairs.
Figure 6.
Ultrastructural features of detergent extracted hair fibres. (a–c) Normal albino BALB/cByJ+/+ hairs have a well defined septulation pattern with roughly square medullary cells. These desiccated ‘ghost cells’ retain their basic structural features. (d–f) In contrast, AKR/J-hid/hid hairs have no defined layering pattern. The medullary cells are rounded, amorphous, or disintegrating (asterisk). Contraction causes large empty spaces to form in the outline of the cells (bars = 2 µm).
Complementation (allelism) testing
Three AKR/J-hid/hid females were crossed with three male SB/LeJ-Foxq1sb/Foxq1sb mice. In total, 21 progeny were produced. Hair fibres were examined and phenotyped by an investigator blinded to the background of the mice. None of these F1 mice had abnormalities of their hairs, indicating that these mutations were complementary and therefore not allelic mutations (Fig. 1). As Foxq1 was mapped to mouse chromosome 13,10 we determined this was not the location for the hid locus.
Mapping the HID locus
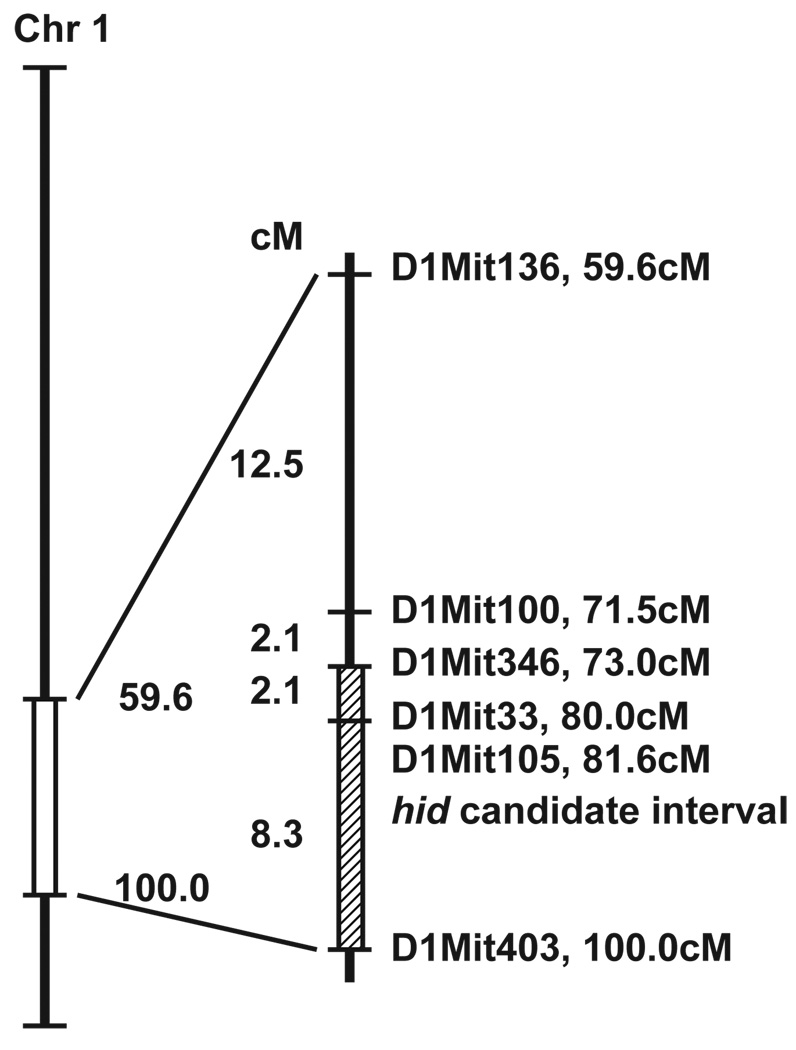
Four homozygous AKR/J-hid/hid female mice were mated with four BALB/cByJ+/+males, which yielded 24 F1 progeny. The hair fibres of these progeny were normal when examined macroscopically and by light microscopy, confirming that hid is a recessive mutation. Twelve F1 breeding pairs were then intercrossed to produce eight litters, comprising 69 F2 pups. The phenotype of the F2 progeny was determined by light microscopic examination of hair fibres from the dorsal interscapular region. Of the 69 F2 mice, 12 (17.3%) had the HID phenotype, which was lower than the anticipated 25% for a simple autosomal recessive mutation. Genome-wide screening using 87 SSLP markers, spaced at an average of 14.4-cM intervals, revealed linkage on the distal end of mouse chromosome 1. Individual typing of the 12 affected and 12 unaffected F2 progeny with a higher density of SSLP markers through the region further localized the mutation to the interval bounded by D1Mit346 on the proximal side and D1Mit403 on the distal side (Fig. 7). One F2 animal that was believed to be wild-type based on its hair morphology was found by genotyping to have homozygous AKR/J alleles throughout the candidate gene region. We tentatively concluded that this mouse was one of the expected mutants whose phenotype was not penetrant.
Figure 7.
Map location of the HID (hid) locus on the distal end of mouse chromosome 1.
Discussion
Mutant laboratory mice with skin defects have long been collected by mouse fanciers and studied by geneticists. They are usually very easy to identify, which is why so many exist. As with many diseases, abnormalities in one organ system can partially reflect other abnormal organ systems.15 Subtle lesions of the skin can easily be overlooked, a major concern when phenotyping genetically engineered mice. The satin mutation in mice gives hair a satiny appearance, which is evident when the mice are compared with other inbred strains (Fig. 1) and why these are highly sought after by the mouse fancier community (http://www.miceandrats.com/standmic.htm; http://www.afrma.org/micevar.htm; http://www.rmca.org/Articles/mousefancy.htm). The AKR-hid/hid mice have a subtle defect within their hair fibres, which has both similarities to and differences from defects in the hair of SB/LeJ-Foxq1sa/Foxq1sa mutant satin mice. When we intercrossed these mice, none of their offspring developed any type of hair-medulla defect, indicating they are not allelic mutations of Foxq1, thus confirming earlier work by Trigg.1
To study in more detail the defect in hair fibres, we performed ultrastructural and element analyses. This approach is useful to define major structural defects.8 Although the HID is obvious with even low magnification of mounted hairs examined with white and polarized light, the hairs were structurally normal on their surface when examined by SEM. Element analysis was also within normal limits. It was interesting to note that when the hair fibres were analysed along their length, mild but consistent differences were observed in both mutant and control hair samples. Such findings indicate that these types of measurements need to be taken at consistent sites on hair fibres for accurate comparisons to be made. The structural details of medullary cells were poorly preserved in both mutant and control mice, but nevertheless, the shape, organization, and survival of the cells were different in HID mutant mice. In both pigmented and albino wild-type hair fibres, the desiccated dead cells maintained their square shape and regular pattern. In contrast, the hid/hid medullary cells were more contracted, round to oval, and disintegrating to various degrees. These features together produce large and irregular empty spaces. These changes suggest that the cells were either not properly formed in the matrix or premedulla, or were fragile and were more susceptible to destruction in the normal maturation process of the hair fibre. Proteomic analyses in future studies may reveal the molecular defect in these hairs.
We mapped the hid locus to the distal end of mouse chromosome 1. Within this interval there are no published mutations that result in skin or hair abnormalities in mice with the exception of the ichthyosis mutation.16 This was shown to be due to a mutation in the lamin B receptor (Lbr) and excluded as the cause of the hid phenotype, because the spontaneous Lbric allelic mutation arose in the AKR/J strain.17 Another gene that maps within this interval is sterol O-acyltransferase 1 (Soat1). A spontaneous allelic mutation, adrenocortical lipid depletion (ald), is fixed in the AKR/J strain18 and was shown to be allelic with Soat1, so that the allele is now listed as Soat1a1d.19 Skin abnormalities in the form of xanthomatosis do occur in two different targeted allelic mutations (Soat1tm1Far and Soat1tm1Ishi), but only when compound homozygous mutants are created with mutations targeting either apolipoprotein E (Apoetm1Unc) or the low-density lipoprotein receptor (Ldlrtm1Her).20,21 No changes in hair fibres or hair follicles were described in these null mutations.
The low penetrance of affected mice in the F2 hybrids suggests that there is at least one modifier gene in addition to that responsible for the hid phenotype. Larger numbers of F2 mice are needed to generate a high-resolution map to positively identify the gene responsible for the HID phenotype, but it seems that a novel gene or a known gene not currently associated with hair biology is located at the distal end of mouse chromosome 1. Identification of this gene and comparison with various congenital human hair diseases will determine the value of this mutation as a biomedical tool.
Acknowledgements
We thank M. Barter for expert technical assistance in the gene mapping, and L. S. Bechtold for technical assistance with electron microscopy. This work was supported in part by mentorship and research grants from the North American Hair Research Society, the National Institutes of Health (RR000173 and AR053639), and National Natural Scientific Foundation of China (30400266). Shared scientific services were supported in part by a Basic Cancer Center Core Grant from the National Cancer Institute (CA34196).
Footnotes
Conflict of interest: none declared.
References
- 1.Trigg MJ. Hair growth in mouse mutants affecting coat texture. J Zool, Lond. 1972;168:165–198. [Google Scholar]
- 2.Sundberg JP. The hair interior defect (hid) mutation, chromosome? In: Sundberg JP, editor. Handbook of Mouse Mutations with Skin and Hair Abnormalities: Animal Models and Biomedical Tools. Boca Raton: CRC Press; 1994. pp. 289–290. [Google Scholar]
- 3.Whiting DA, Hosden FL. Color Atlas of Differential Diagnosis of Hair Loss. Cedar Grove, NJ: Canfield Publishing; 1996. [Google Scholar]
- 4.Giehl KA, Eckstein GN, Benet-Pages A, et al. A gene locus responsible for the familial hair shaft abnormality pili annulati maps to chromosome 12q24.32–24.33. J Invest Dermatol. 2004;123:1073–1077. doi: 10.1111/j.0022-202X.2004.23423.x. [DOI] [PubMed] [Google Scholar]
- 5.Giehl KA, Ferguson DJ, Dawber RP, et al. Update on detection, morphology and fragility in pili annulati in three kindreds. J Eur Acad Dermatol Venereol. 2004;18:654–658. doi: 10.1111/j.1468-3083.2004.01036.x. [DOI] [PubMed] [Google Scholar]
- 6.Bechtold LS. Ultrastructural evaluation of mouse mutations. In: Sundberg JP, Boggess D, editors. Systematic Characterization of Mouse Mutations. Boca Raton: CRC Press; 2000. pp. 121–129. [Google Scholar]
- 7.Bringans SD, Plowman JE, Dyer JM, et al. Characterization of the exocuticle a-layer proteins of wool. Exp Dermatol. 2007;16:951–960. doi: 10.1111/j.1600-0625.2007.00610.x. [DOI] [PubMed] [Google Scholar]
- 8.Mecklenburg L, Paus R, Halata Z, et al. FOXN1 is critical for onychocyte terminal differentiation in nude (Foxn1<nu>) mice. J Invest Dermatol. 2004;123:1001–1011. doi: 10.1111/j.0022-202X.2004.23442.x. [DOI] [PubMed] [Google Scholar]
- 9.Rice RH, Wong VJ, Pinkerton KE, et al. Cross-linked features of mouse pelage hair resistant to detergent extraction. Anat Rec. 1999;254:231–237. doi: 10.1002/(SICI)1097-0185(19990201)254:2<231::AID-AR9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Hong HK, Noveroske JK, Headon DJ, et al. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis. 2001;29:163–171. doi: 10.1002/gene.1020. [DOI] [PubMed] [Google Scholar]
- 11.Truett GE, Walker JA, Truett AA, et al. Preparation of PCR-quality DNA with hot sodium hydroxide and Tris (HOTSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 12.Taylor BA, Navin A, Phillips SJ. PCR-amplification of simple sequence repeat variants from pooled DNA samples for rapidly mapping new mutations of the mouse. Genomics. 1994;21:626–632. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- 13.Petkov PM, Ding Y, Cassell MA, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundberg JP. The satin (sa) mutation, chromosome 13. In: Sundberg JP, editor. Handbook of Mouse Mutations with Skin and Hair Abnormalities. Animal Models and Biomedical Tools. CRC Press: Boca Raton; 1994. pp. 413–416. [Google Scholar]
- 15.Sundberg JP. Handbook of Mouse Mutations with Skin and Hair Abnormalities. Animal Models and Biomedical Tools. CRC Press, Inc: Boca Raton, Florida; 1994. [Google Scholar]
- 16.Sundberg JP, Pittelkow MR. The ichthyosis (ic) mutation, Chromosome 1. In: Sundberg JP, editor. Handbook of Mouse Mutations with Skin and Hair Abnormalities. Animal Models and Biomedical Tools. CRC Press: Boca Raton; 1994. pp. 327–335. [Google Scholar]
- 17.Shultz LD, Lyons BL, Burzenski LM, et al. Mutations at the mouse ichthysosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huet anomaly. Hum Mol Genet. 2003;12:61–69. doi: 10.1093/hmg/ddg003. [DOI] [PubMed] [Google Scholar]
- 18.Arnesen K. The cytology of the adrenal cortex in mice with spontaneous adrenocortical lipid depletion. Acta Pathol Microbiol Scand. 1963;58:212–218. doi: 10.1111/apm.1964.60.4.487. [DOI] [PubMed] [Google Scholar]
- 19.Meiner VL, Welch CL, Cases S, et al. Adrenocorical lipid depletion gene (ald) in AKR mice is associated with an acyl-coA: cholesterol acyltransferase (ACAT) mutation. J Biol Chem. 1998;273:1064–1069. doi: 10.1074/jbc.273.2.1064. [DOI] [PubMed] [Google Scholar]
- 20.Accad M, Smith SJ, Newland DL, et al. Massive xanthomatosis and altered composition of atherosclerotic lesions in hyperlipidemic mice lacking acyl CoA: cholesterol acyltransferase 1. J Clin Invest. 2000;105:711–719. doi: 10.1172/JCI9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagyu H, kitamine T, Osuga J-I, et al. Absence of ACAT-1 attenuates atherosclerosis but causes dry eye and cutaneous xanthomatosis in mice with congenital hyperlipidemia. J Biol Chem. 2000;275:21324–21330. doi: 10.1074/jbc.M002541200. [DOI] [PubMed] [Google Scholar]