Abstract
In order to detect diseases such as cancer at an early stage while it still may be curable, it is necessary to develop a diagnostic technique which can rapidly and inexpensively detect protein and nucleic acid biomarkers, without making any sacrifice in the sensitivity. The authors have developed a technique, based on the use of bioactivated microfluidic channels integrated with electrodes for electrical sensing, which can be used to detect protein biomarkers, target cells, and DNA hybridization. In this article, they discuss the theoretical detection limits of this kind of sensor and also discuss various experimental considerations in the electrical characterization of our device. In particular, they discuss the temperature dependence, the impedance drift, the noise sources, and various methods for optimizing the signal to noise ratio.
INTRODUCTION
The lab on a chip implementation of rapid and inexpensive instrumentation for medical diagnostics and the study of genomics, proteomics, and cellomics can significantly reduce the costs and increase the efficiency of medical diagnostics in the clinical setting, and potentially alleviate the global health care crisis. In order for doctors and physicians to be able to diagnose diseases at an early stage, while the disease may still be curable, it is necessary that they have access to a versatile platform which can rapidly and inexpensively analyze a wide panel of biomarkers, biomolecular indicators which signal the state of a disease. Of particular importance are genomic biomarkers, protein biomarkers, and cell biomarkers. Genomic biomarkers are important in giving an indication of a patient’s predisposition to certain diseases based on the patient’s DNA and include nucleic acid biomarkers such as gene mutations, polymorphisms, and quantitative gene expression analysis. However, an understanding of the patient’s genome alone is insufficient for proper prediction of disease susceptibility due to the heterogeneous nature of diseases such as cancer between populations. Knowledge of the proteome,1 which includes both quantifying the various protein structures and also the functional interactions between the proteins, can give a more comprehensive understanding of the state of the disease in a patient. Finally, the ability to detect various target cells such as tumor cells or bacterial cells in blood can be of immense use for early disease diagnosis. The comprehensive knowledge of the genome, the proteome, and the cellome for individual patients will provide the information necessary to make personalized medicine feasible across heterogeneous populations.
Current detection techniques require expensive labeling, long incubation times,2 and bulky optical equipment for fluorescence scanning, only allowing for the analysis of a limited panel of biomarkers in the clinical setting.3 It has become more apparent that a small number of markers are insufficient in properly diagnosing diseases across populations. Thus, the need for developing rapid inexpensive platforms which can be multiplexed to analyze a wide panel of biomarkers has become ever pressing.
Various attempts have been made at detection of cells, proteins, and DNA using electrical impedance based sensors, which are advantageous given that they lower preparation time and reagent costs due to elimination of fluorescent labeling.4 Label-free detection of detection of changes in protein conformation has been reported recently using nanogap sensors.5 Many of the above mentioned techniques, and other electrical biosensors presented to date require numerous washing steps while lacking the ability for real time detection. The biggest problem with most electrical impedance biosensors is achieving consistency in results. The use of coulter counters6 makes possible the counting and sizing of particles in real time. The capture of target proteins on beads coated with probe molecules and the detection of their presence based on size changes has been demonstrated.7, 8 However, differentiation between two different proteins which may be similar in size is difficult.
We have developed a microfluidic platform capable of electrically detecting target cells,9 target proteins,10 and DNA (Ref. 11) in less than an hour with at least the same sensitivity as the current techniques, while maintaining the selectivity which current electrical detection techniques do not have. With this type of integrated platform capable of detecting these three different types of markers, biomarker detection, and discovery can be multiplexed to a level which the current techniques lack the capability thereof. In our previous work,9, 10, 11 we have demonstrated the proof of concept for detection of these various biomarkers. In this article, our intention is to discuss the performance limits of this device based on theoretical considerations and various experiments we have performed.
DEVICE OPERATION PRINCIPLES
The device (Fig. 1) consists of a microchannel integrated with electrodes (labeled A and C) for measuring the conductance along the channel. In one application, the channels are functionalized with a probe molecule, and a micron sized bead is functionalized with the target molecule. In the case of protein detection, the surface is functionalized with probe antibodies. The test sample is then injected into the channel, so that target antigens are captured by the probe antibodies. Finally, in order to amplify the current modulation, beads coated with secondary antibodies are injected into the channel to bind to the captured antigens. In the case of nucleic acid biomarkers, the surface is functionalized with probe DNA, and the target DNA can be immobilized onto micron sized beads in order to amplify the modulation of the current. A bead getting captured will result in a permanent change in the current across the channel, and a bead passing by without getting captured will result only in a transient change. By scaling the channel down to approximately the size of the bead, one can achieve single bead detection.
Figure 1.
Cross section schematic of gated microchannel with electrodes labeled A and B. The target particle specifically binds to the probe molecules which are immobilized on the base of the channel. (Bottom plot) Current between electrodes A and C during bead or cell capture.
THEORETICAL CONSIDERATIONS
The detection limit of the system is determined by two primary factors. The first is the rate at which target molecules are captured by the surface probe molecules. The second is the electronic detection limit of the system, that is, how many beads can be detected by the sensor. We will discuss both of them.
Reaction limited sensor
When applying a flow rate operating in the reaction limited regime, that is, where reagents are continuously being supplied to the active area surface, the target protein capture rate is limited by the speed of the antigen-antibody reaction, which can be modeled to the first order using Langmuir’s equation,12
| (1) |
where b is the number of probes which are bound by target molecules, is the total number of receptor molecules, and are the on and off rate constants, respectively, and is the target concentration at the sensor surface. Solving this equation results in the following:
| (2) |
where is the concentration of target analyte in the bulk solution and
| (3) |
Solving for the previous equation in equilibrium results in the following steady state relationship:
| (4) |
This relationship determines the concentration of target molecules in solution necessary for a given percentage of probe molecules to be occupied on the sensor surface. If one were to design a sensor which was electrically sensitive enough to detect a single target molecule in equilibrium, the molar concentration of the target molecule in bulk solution required would be
| (5) |
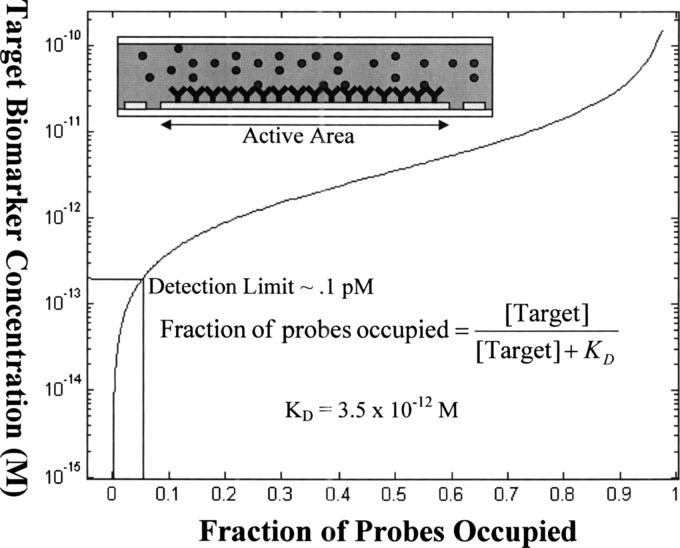
where A is the surface area of the active area of the sensor. This can be achieved if one were to completely eliminate the nonspecific binding of nontarget molecules and if the sensor was sensitive enough to detect a single molecule. However, in reality, our system is currently limited by the nonspecific binding of beads to the channel surface, which we previously reported to be 5%.10 This limits the detection limit of the sensor to (Fig. 2).
Figure 2.
Plot of Langmuir’s first order equation in equilibrium. The target biomarker concentration in solution is plotted against the fraction of probes which are occupied. Since the current setup is limited by 5% nonspecific binding, the detection limit is roughly .
Electronic noise sources
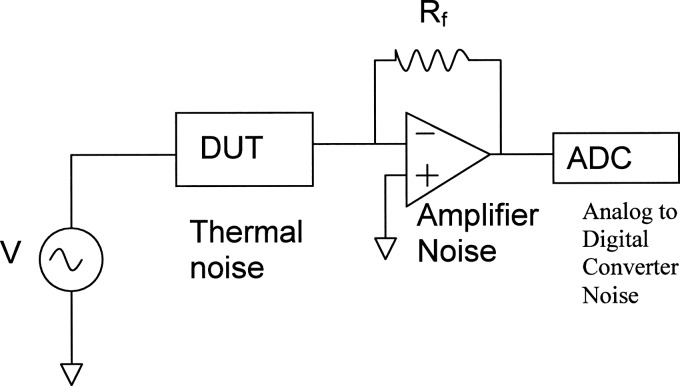
The sensor system consists of three noise sources (Fig. 3). The first is the noise from the sensor, the second is the amplifier noise, and the third is the noise due to the analog to digital converter, which can be expressed by the following equation:
| (6) |
The sensor has several sources of noise,13 however, since we operate the sensor at frequencies of tens of kilohertz, which is relatively high such that impedance contribution due to the double layer capacitance on the electrodes is negligible, and also since we are operating in a non-Faradaic regime, we can focus on the thermal noise due to the conductivity of the solution, which is equal to
| (7) |
Assuming a noise bandwidth of , and a solution resistance of , which have been obtained experimentally, comes out to be . As for the amplifier noise, in our system we use a SR570 low noise current amplifier, where for a gain of the noise current comes out to be . The analog to digital converter noise is negligible compared to the others, so from Eq. 6 the total output referred voltage noise is .
Figure 3.
Current sensor system has three sources of noise. The device under test (DUT) has thermal noise, then there is the noise from the current amplifier, and finally the A to D noise which is negligible compared to the other two.
The change in current due to the presence of a single particle of diameter d, and channel of size D, and electrode spacing of L can be estimated by the equation which was predicted by Bean and Deblois,14
| (8) |
A diameter particle in a channel with electrodes spaced apart as we have fabricated15 will cause a change of over a baseline current of . This change in current translates to a change of in the output voltage.
Experimental SNR
In order to determine which type of circuit design is best for optimizing the signal to noise ratio, we examine both a two electrode system and a four electrode system.
The two electrode measurement [Fig. 4a] consists of a function generator tied to the left electrode and a current preamplifier tied to the right electrode. The output of the amplifier is tied to a data acquisition card which is then processed using a custom written LABVIEW program. The four electrode setup is shown in Fig. 4b. This technique involves the use of four electrodes, the outer two are used for carrying the current, and the inner two are used for controlling the voltage. Separation of the current carrying and the voltage control ensures that current measured across the outer two electrodes will not be affected by the voltage drop due to the double layer at the surface of the electrodes, since the voltage drop occurs only between the inner two electrodes, and thus the sensing region of the sensor is limited to that region. The amplifier circuit design for this type of measurement configuration can be found in Carbonaro et al.7
Figure 4.
(a) Two point measurement with a voltage source tied to the left electrode and a current amplifier tied to the right electrode. (b) A four point probe measurement where the outer two electrodes are the current carrying electrodes and the inner two are for controlling voltage.
Bead capture
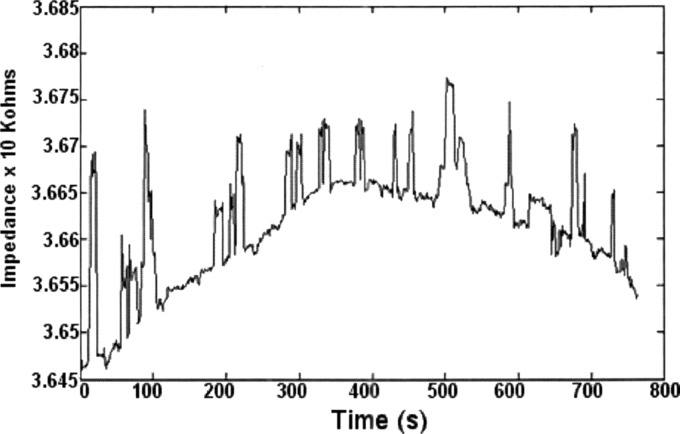
In Fig. 5 we show representative data of beads flowing through the channel without getting captured in a two electrode measurement. In this particular experiment, nonfunctionalized beads were flowing through the microchannel resulting in the peaks as shown. The variation in the peak size is a result of the fact that in some cases more than one bead is present in the active area of the sensor at a time.
Figure 5.
Representative data of impedance vs time for diameter beads passing through a channel.
A baseline drift in the impedance of is also apparent. This is likely due to several factors. One possibility is temperature dependence of solution conductance, as discussed in the next section. Another possibility is the fact that ions and other molecules in the electrolyte constantly adsorb and desorb from the bare gold surface. Possible solutions to this problem are the use of a differential measurement between a target and control channel, in order to reject the common mode effects. Another possibility is the use of self-assembled monolayer or an insulative layer of some sort that can prevent the adsorption and desorption of molecules on the gold surface. This issue is currently under study.
We repeated this type of experiment for four different cases. We did a two electrode and a four electrode measurement, using beads and beads, and we measured the SNRs in all four cases, as shown in Table 1. The performance of the two electrode system was only slightly better.
Table 1.
Signal to noise ratio for two point and four point measurements for 10 and diameter beads.
| Bead diameter | Signal to noise ratio | |
|---|---|---|
| Two point | Four point | |
| 10 | ||
| 20 | 6.2673 | 8.4314 |
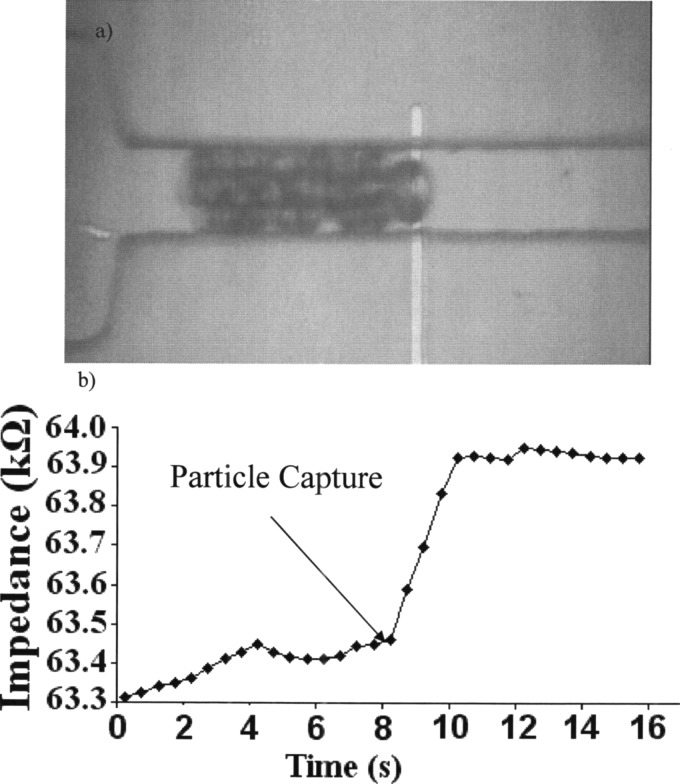
In Fig. 6b we show representative data of a bead flowing through the channel and then getting captured on the surface of the electrode. Once the bead gets captured, it blocks the rest of the beads from flowing through the channel behind it. The capture of the bead results in an instantaneous increase in the impedance across the electrodes at time [Fig. 6a].
Figure 6.
(a) Image of particles getting captured on the surface of the microchannel on top of the electrode. (b) The corresponding instantaneous steady state change in impedance as the particle has been captured.
Temperature dependence
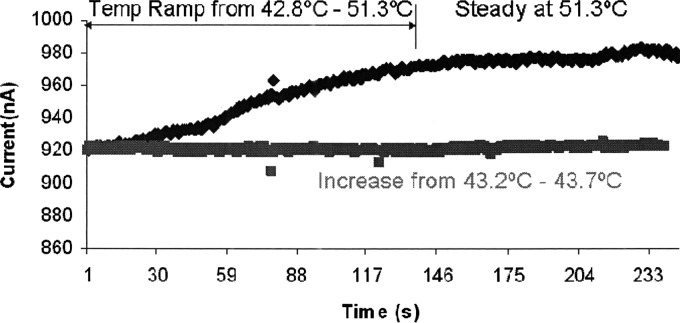
We also examined the performance of this device with respect to temperature fluctuations. In general, solution conductivity varies an average of . As seen in Fig. 7, we heated up our biochip to temperatures well above room temperature so that the temperature fluctuations in the room would not have any effects. In the case where we kept the temperatures changes to less than (red curve), a noticeable change was not observed in the current amplitude.
Figure 7.
Dependance of baseline current on temperature.
However, in the case where temperature was increased by more than (blue curve), a change greater than 2% was observed in the rms current level. Given that the greater change in temperature resulted in a greater change in current, it is reasonable to conclude that the swings in temperature of the environment are responsible for the impedance drift. Given that we measured the instantaneous impedance changes in this study, we were able to neglect this effect. However, in order to assure consistency in measurements, ultimately it would be necessary to alleviate this problem by either isolating the device such that external temperature changes were ineffective, or perhaps to incorporate some sort of a differential measurement system in a parallel control channel in order to subtract out the effects of temperature change on the conductivity of the solution.
CONCLUSION
In conclusion, we have developed a microfluidic platform capable of electrically detecting a wide variety of biomarkers. The sensor performance is limited by the electrical detection limit of the sensor and also the kinetics of the probe and target molecule interactions. Assuming that the electrical detection limit can be reduced to a single molecule, the performance limit will ultimately be set by the reaction kinetics. In this case, the electrical detection limit will determine the level of quantification noise.
With our current implementation, however, the nonspecific binding is the main factor limiting the performance. Given that 5% of the beads bind to the surface nonspecifically, the sensor is limited to a detection limit of . If we were to eliminate the nonspecific binding by optimizing the surface chemistry on the channel surface and the bead surface so that no beads were to bind nonspecifically, then the performance would be limited by the minimum number of detectable beads. If the electrical sensor performance were optimized to be able to detect a single bead, then the minimum detectable particle size would limit the performance. If that limit were to decrease to a single protein or DNA molecule, then as previously mentioned, the reaction kinetics would set the performance limit.
Acknowledgments
This research was supported by National Institutes of Health Grant P01 HG000205. The authors would like to thank Jessica Melin and the Stanford Microfluidics Foundry for their invaluable help in fabrication of the devices used in this study.
References
- MacBeath G., Nat. Genet. 32, 526 (2002). 10.1038/ng1037 [DOI] [PubMed] [Google Scholar]
- Burnette W. N., Anal. Biochem. 112, 195 (1981). 10.1016/0003-2697(81)90281-5 [DOI] [PubMed] [Google Scholar]
- Crowther J. R., The ELISA Guidebook (Humana, Totowa, NJ, 2001). [Google Scholar]
- Ayliffe H. E., Frazier A. B., and Rabbitt R. D., J. Microelectromech. Syst. 8, 50 (1999); 10.1109/84.749402 [DOI] [Google Scholar]; Gomez R. et al. , Biomed. Microdevices 3, 201 (2001); 10.1023/A:1011403112850 [DOI] [Google Scholar]; Gomez-Sjoberg R., Morisette D. T., and Bashir R., J. Microelectromech. Syst. 14, 829 (2005); 10.1109/JMEMS.2005.845444 [DOI] [Google Scholar]; Radke S. M. and Alocilja E. C., IEEE Sens. J. 4, 434 (2004); 10.1109/JSEN.2004.830300 [DOI] [Google Scholar]; Radke S. M. and Alocilja E. C., IEEE Sens. J. 5, 744 (2005); 10.1109/JSEN.2005.848138 [DOI] [Google Scholar]; Yang L. J., Li Y. B., Friffis C. L., and Johnson M. G., Biosens. Bioelectron. 19, 1139 (2004); 10.1016/j.bios.2003.10.009 [DOI] [PubMed] [Google Scholar]; Yang L. J., Ruan C. M., and Li Y. B., Biosens. Bioelectron. 19, 495 (2003). 10.1016/S0956-5663(03)00229-X [DOI] [PubMed] [Google Scholar]
- Ionescu-Zanetti C., Nevill J. T., Di Carlo D., Jeong K. H., and Lee L. P., J. Appl. Phys. 99, 024305 (2006); 10.1063/1.2161818 [DOI] [Google Scholar]; Lee W. C., Cho Y. H., and Pisano A. P., J. Microelectromech. Syst. 16, 802 (2007); 10.1109/JMEMS.2007.896706 [DOI] [Google Scholar]; Nevill J. T., DiCarlo D., Liu P., Jeong K. H., and Lee L. P., Digest of Technical Papers—International Conference on Solid State Sensors and Actuators and Microsystems, TRANSDUCERS’05 2, 1668 (2005); 10.1109/SENSOR.2005.1497410 [DOI] [Google Scholar]; Yi M. Q., Jeong K. H., and Lee L. P., Biosens. Bioelectron. 20, 1320 (2005). 10.1016/j.bios.2004.05.003 [DOI] [PubMed] [Google Scholar]
- Coulter W. H., Patent No. 2656508 (1953).
- Carbonaro A. and Sohn L. L., Lab Chip 5, 1155 (2005). 10.1039/b504827c [DOI] [PubMed] [Google Scholar]
- Gawad S., Schild L., and Renaud P., Lab Chip 1, 76 (2001); 10.1039/b103933b [DOI] [PubMed] [Google Scholar]; Saleh O. A. and Sohn L. L., Rev. Sci. Instrum. 72, 4449 (2001); 10.1063/1.1419224 [DOI] [Google Scholar]; Saleh O. A. and Sohn L. L., Proc. Natl. Acad. Sci. U.S.A. 100, 820 (2003). 10.1073/pnas.0337563100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanmard M., Talzsaz A. H., Nemat-Gorgani M., and Pease F., Biomicrofluidics 1, 044103 (2007). 10.1063/1.2815760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanmard M., Talasaz A. H., Nemat-Gorgani M., Pease F., Ronaghi M., and Davis R. W., Lab Chip 9, 1429 (2009). 10.1039/b818872f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanmard M.et al. , International Solid-State Sensors, Actuators and Microsystems Conference, 2007. TRANSDUCERS 2007 (unpublished).
- Squires T. M., Messinger R. J., and Manalis S. R., Nat. Biotechnol. 26, 417 (2008). 10.1038/nbt1388 [DOI] [PubMed] [Google Scholar]
- Hassibi A., Navid R., Dutton R. W., and Lee T. H., J. Appl. Phys. 96, 1074 (2004). 10.1063/1.1755429 [DOI] [Google Scholar]
- Deblois R. W. and Bean C. P., Rev. Sci. Instrum. 41, 909 (1970). 10.1063/1.1684724 [DOI] [Google Scholar]
- Javanmard M., Talasaz A. H., Nemat-Gorgani M., Huber D. E., Pease F., Ronaghi M., and Davis R. W., IEEE Sens. J. 9, 883 (2009). 10.1109/JSEN.2009.2022558 [DOI] [PMC free article] [PubMed] [Google Scholar]