Abstract
We present a statistically innovative as well as scientifically and practically relevant method for automatically segmenting magnetic resonance images using hierarchical mixture models. Our method is a general tool for automated cortical analysis which promises to contribute substantially to the science of neuropsychiatry. We demonstrate that our method has advantages over competing approaches on a magnetic resonance brain imagery segmentation task.
Keywords: Automated cortical analysis, Magnetic resonance imagery, Mixture model, Classification, Segmentation
1. Introduction
Gaussian mixture modelling is ubiquitous throughout laboratories in the world for image analysis and segmentation, and has played a core role in medical image analysis, and in particular Human Brain Mapping for the past several decades. A fundamental aspect of Human Brain Mapping research today is cortical reconstruction of brain volumes into their macroscopic components, including gray matter (G), white matter (W), or cerebrospinal fluid (C). Throughout the field of brain mapping, almost without exception the construction of cortical manifolds requires some form of segmentation via probabilistic model fitting. Reconstruction of volumetric submanifolds such as the thalamus, hippocampus, deep nuclei, and other structures are being performed via automatic segmentation methodologies by many investigators (Fischl et al., 2001; Grabowski et al., 2000; Holmes et al., 1998; Kapur et al., 1996; Joshi et al., 1999; Miller et al., 2000; Ratnanather et al., 2001; Shattuck et al., 2001; Robb, 1999; Teo et al., 1997; Wells-III et al., 1996; Xu et al., 1999). Such approaches boil down to some form of classical Bayesian segmentation and Neyman–Pearson likelihood-ratio testing on Gaussian models and Gaussian mixture models. Even segmentation methods which apply segmentation implicitly through mapping without direct Bayesian hypothesis testing, such as those approaches based on deformable template matching (Christensen et al., 1997) or active surface methods (Yezzi et al., 2002; Westin et al., 2000; Tagare, 1997; Shen et al., 2001; Sclaroff and Liu, 2001; Schultz and Conradsen, 1998; Montagnat et al., 1999, 2001; Montagnat and Delingette, 1997; Mignotte and Meunier, 2001; Kervrann and Heitz, 1999; Jain et al., 1996; Garrido and de la Blanca, 1998, 2000; Chen and Metaxas, 2000; Caunce and Taylor, 2001; Xu et al., 2000; Xu and Prince, 2000; Pham et al., 2000)are usually based on some form of Gaussian mixture modelling. Usually, the data models are implicitly conditionally Gaussian, with the resulting matching functions of the quadratic-norm type.
At the heart of such brain mapping efforts is mixture modelling in which each tissue class is modelled as a single Gaussian component. Sometimes additional “partial volume” components are included to account for the inadequacy of the single Gaussian model. Of course, selecting the dimension of the model, i.e. the number of partial volume fits, reduces to the model selection problem; as well there is no reason to believe that a single model order fits all tissue and or imaging modality types. The focus of this paper is to propose a method which allows for the flexibility of a Gaussian mixture model—with model complexity selected adaptively from the data—for each tissue class. Our procedure involves modelling each class as a semiparametric mixture of Gaussians. Thus, for each subject j, the subject-specific marginal fj is taken to be a hierarchical mixture model—a mixture of Gaussian mixtures. For each of the three classes c ∈ {C, G, W,} the density fj has one mixture component fjc, and each of these components is itself a mixture of Gaussians. The major difficulty solved in this paper associated with employing such semi-parametric methods is to solve dynamically the model selection problem. The crucial step of determining class-conditional mixture complexities for (unlabeled) test data is hopeless in the unsupervised case—without labeled voxel observations known to belong to the three classes, the hierarchical mixture model is hopelessly unidentifiable. We accomplish this by matching models to a predefined data base of hand labelled experimental tissue samples. Thus, the use of a registry of expertly generated data becomes a fundamental part of our solution. This approach of course builds on the entire philosophy now emerging through the Biomedical Informatics Research Network (http://www.nbirn.net) initiatives in which data bases are federated and available for laboratory comparison and analysis. As we show, the performance improvement obtained is significant, implying that the emergence of hand labelled information from laboratories across the country which are combined with automated methods for analysis promise tremendous improvements in future automated brain analysis methods.
Of course, the importance of such improvements is in that a fundamental aspect of human brain mapping today is the subtle examination of small changes in brain morphometry. For instance, to investigate whether schizophrenia or Alzheimer's disease alters the structure of gray matter in the cingulate gyrus of affected subjects, accurate segmentations into tissue types of the cingulate gyrus for both affected and control subjects are required. Such segmentations are currently obtained via labor- and cost-intensive hand-segmentation of all images in a sample. For the purpose of investigating subtle differences between the affected and control populations, larger sample sizes afford greater statistical power and are, of course, desirable. This leads to a conflict between the desire for a large sample size and the desire to keep the number of hand-segmentations small. This manuscript presents a method for taking a hand-segmented training subset of the images to be segmented and producing accurate automated segmentations for the remaining images in the sample. This will allow for a larger sample of segmented images, thereby increasing the power of subsequent statistical analyses.
Our method proceeds according to the following outline.
(1) For each subject/class pair in the available training data set, estimate the marginal subject-specific class-conditional probability densities. Notice that for this supervised step (since these are training images, individual voxel class labels are available) semi-parametric mixture complexity estimation is appropriate.
(2) Find the “closest” training model to the (unlabeled) test data.
(3) Fit a mixture to the test data, using the training model obtained in step (2) to determine the class-conditional mixture complexities and starting locations.
(4) Classify voxels from the test data using the plug-in Bayes rule, where the mixture component class labels are inherited from the selected training model (but the mixture itself is estimated from the test data in step (3)).
This manuscript is structured as follows. In Section 2 we describe a data set of MR images to be investigated. Section 3 gives the mixture modelling methodology in detail. Section 4 describes the experimental protocol employed, and Section 5 reports the results of our investigations. Finally, in Section 6, we conclude with a discussion of the implications of our results and our methodology.
2. Data
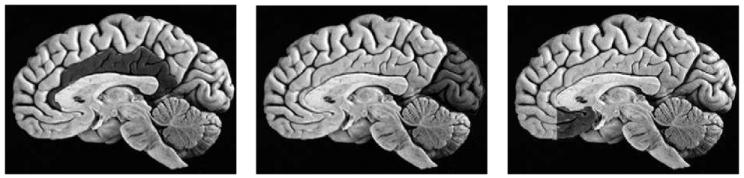
Dysfunction of specific subregions of the cerebral cortex has been implicated in the pathophysiology of several neuropsychiatric disorders, including schizophrenia. Although widespread thinning of the gray matter cortical mantle has been observed in subjects with schizophrenia (Zipursky et al., 1992), specific abnormalities of neuronal architecture may exist within portions of the cortical mantle and form the neuroanatomical basis for specific cognitive deficits or symptom groupings. Shown in Fig. 1 are examples of cingulate, occipital and medial prefrontal in the human brain, which are studied here. Such brain areas consist of different mixtures of gray matter, white matter, and cerebrospinal fluid.
Fig. 1.
Panel 1 shows the cingulate gyrus, panel 2 is the occipital gyrus, and panel 3 is the medial prefrontal gyrus. (Adapted from Duvernoy, 1999).
The cingulate gyrus, a large C-shaped gyrus that courses around the corpus callosum and cerebral ventricles, has attracted considerable interest because of its role in attention and other cognitive functions that appear to be critical for understanding schizophrenia and other neuropsychiatric disorders (Csernansky and Bardgett, 1998). The medial prefrontal cortex has been implicated in a variety of recent functional and structural neuroimaging studies of both normal affect regulation and differences associated with affective disorders such as major depressive disorder or bipolar affective disorder (Swerdlow and Koob, 1987; Drevets et al., 1992; Drevets and Todd, 1997; Botteron and Figiel, 1997; Drevets, 1999). The occipital cortex has been the focus of several major functional neuroimaging studies of visual perception; see, e.g. (Yantis, 2001; Tootell et al., 1998).
High-resolution MR imaging now affords an unprecedented opportunity to acquire detailed images of the neuroanatomical configurations and tissue characteristics of the living human brain. Substantial advances in our understanding of neuropsychiatric disorders are anticipated as this technology is used more widely in populations of individuals with such illnesses. However, most neuropsychiatric researchers still apply manual methods to assess the boundaries and internal tissue segmentations of cortical subregions in MR scans. These manual methods are limited by their dependency on the constant availability of human experts and by between rater error (e.g., Bartzokis et al., 1993; Haller et al., 1997).
We investigate first a set of cingulate gyrus images provided by John Csernansky of the Washington University School of Medicine, St. Louis. High-resolution MR scans were acquired using a turbo-FLASH sequence (TR = 20 ms, TE = 5.4 ms, flip angle = 30°, ACQ-1, Matrix 256 × 256, Scanning Time = 13.5 min) with 1 mm × 1 mm × 1 mm isotropic resolution across the entire cranium. (See, for instance, Cho et al. (1993) for MR imaging background.) MR image data were reformatted via intensity scale adjustment to maximize tissue contrast, using the voxel intensities in the corpus callosum and the lateral ventricle as limiting values. MR scans were then interpolated into 0.5 mm × 0.5 mm × 0.5 mm isotropic resolution using trilinear interpolation.
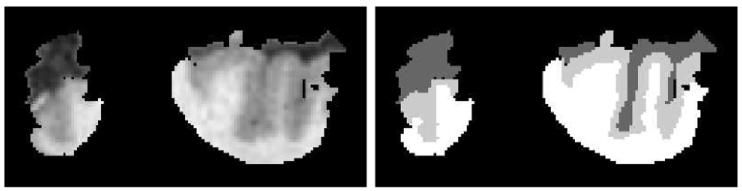
We consider J = 10 subjects; five with schizophrenia (labeled “CG 1” through “CG 5”) and five control subjects (labeled “CG 6” through “CG 10”). For each subject, the voxels in the cingulate gyrus of the right hemisphere, have been hand-segmented by expert neuroanatomists into one of three tissue classes; 𝓒 ={C, G, W}. (See Figs. 2 and 3.) Hand segmented files were created using Analyze (Robb, 1999) and a region of interest (ROI) defined by expert raters to isolate the cingulate gyrus. The ROI included white matter, gray matter and cerebrospinal fluid of one hemisphere that encompassed the cingulate gyrus and immediate surrounding region. A three-dimensional ROI encompassing the entire cingulate gyrus in the right hemisphere was outlined. The ROI was constructed in coronal sections. In each MR section, an enclosure which consisted of the gray matter of the right cingulate gyrus and its neighboring gray matter, white matter, and cerebrospinal fluid was drawn by hand. This was a fast procedure as it did not require manual segmentation of exact tissue types or precise anatomical structures. An example of such an ROI section is shown in Fig. 2; note that some voxels were not labeled and thus not included in the ROI. The right cingulate gyrus was manually delineated, according to anatomical rules. For each scan, a set of cingulate manual segmentations by an expert rater was obtained in the ROI. The manual segmentations consisted of three tissue classes. Voxel classifications by the rater were based on visual inspection of morphological cues such as gyri curvature and connectivity, as well as visual intensity cues between three tissue classes.
Fig. 2.
Panel 1 shows the original MRI, and Panel 2 is the hand-segmentation of (one slice of) CG 9 into C (dark gray), G (light gray), W (white); black is unsegmented region.
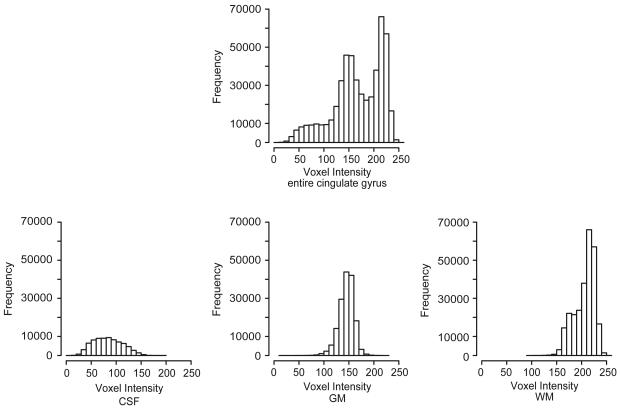
Fig. 3.
Frequency histograms for CG 9: entire cingulate gyrus, right hemisphere (top), and C (bottom left), G (bottom center), and W (bottom right).
There are between 390,000 and 700,000 class-labeled voxels per subject. The smallest subject-specific class-conditional sample size is 45,709 (C for subject CG 6). See Table 1 for details.
Table 1.
Sample sizes for 10 cingulate gyri
| Number of voxels |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CG 1 | CG 2 | CG 3 | CG 4 | CG 5 | CG 6 | CG 7 | CG 8 | CG 9 | CG 10 | |
| C | 150,128 | 90,035 | 91,452 | 85,063 | 121,138 | 45,709 | 108,617 | 84,402 | 76,886 | 125,409 |
| G | 210,636 | 186,529 | 136,856 | 124,227 | 130,717 | 154,993 | 181,538 | 169,583 | 161,138 | 131,230 |
| W | 333,341 | 359,746 | 263,401 | 243,651 | 274,943 | 193,633 | 320,293 | 276,141 | 264,607 | 339,410 |
| Total | 694,105 | 636,310 | 491,709 | 452,941 | 526,798 | 394,335 | 610,448 | 530,126 | 502,631 | 596,049 |
3. Semiparametric mixture modelling
Model selection in Gaussian mixtures—the estimation of mixture complexity—is of fundamental importance in many applications of finite mixture models. An enormous body of literature exists regarding the application, computational issues, and theoretical aspects of mixture models when the number of components is known (see, e.g., McLachlan and Peel, 2000; Everitt and Hand, 1981; McLachlan and Basford, 1988; Titterington et al., 1990), but estimating the unknown number of components remains an area of intense research effort. See, for instance, Chen and Kalbfleisch (1996); Dacunha-Castelle and Gassiat (1997, 1999); Escobar and West (1995); Henna (1985); James et al. (2001); Keribin (2000); McLachlan (1987); Priebe and Marchette (2000); Roeder and Wasserman (1997) for recent progress in this area.
We model the class-conditional probability density functions via the “alternating kernel and mixture” (AKM) method (Priebe and Marchette, 2000; James et al., 2001). This methodology results in an automatic estimate of mixture complexity. We do not claim AKM is necessarily the best approach; but it is reasonable and illustrative, and can have advantages over other techniques. In Priebe and Marchette (2000) AKM is compared to the Bayesian methodology of Roeder and Wasserman (1997), while James et al. (2001) presents a comparison with numerous competing approaches.
3.1. Modelling
For each subject j ∈ 𝒥: ={1, …, J }, consider magnetic resonance voxel observations 𝒳j : ={Xj1, …, Xjnj}; the subject-specific sample sizes are denoted by nj. Let the marginal probability density function for subject j voxel observations be given by
| (1) |
That is, fj=πjCfjC+πjGfjG+πjWfjW. The subject-specific class-conditional marginals are denoted fjc, and the subject-specific class-conditional mixing coefficients πjc are nonnegative and sum to unity. We assume that the Xji are identically distributed according to fj (but not independent).
Each subject-specific class-conditional marginal fjc is itself modelled as a mixture of normals
| (2) |
where the subject-specific class-specific mixing coefficients satisfy πjct ≥0 for each j, c, t, and for each j, c, and denotes the Gaussian probability density function with mean μjct variance . Thus the subject-specific marginals fj are modelled as hierarchical mixtures—mixtures of Gaussian mixtures;
| (3) |
This model is precisely analogous to the model used in “mixture discriminant analysis” (Hastie and Tibshirani, 1996). The subject-specific class-conditional mixture complexities kjc are to be estimated from the data via AKM.
3.2. Estimation
Given class-labelled training data, we have available subject-specific class-conditional sample sizes njc such that nj = Σcnjc. For the subject-specific mixing coefficients πjc we use the empirical estimate
| (4) |
the ratio of the subject-specific class-conditional sample size to the total subject-specific sample size. Let 𝒳jc denote that subset of ᵋj for which the class label is c.
We estimate the subject-specific class-conditional mixture complexities kjc, mixing coefficients πjct, and mixture components φjct, via AKM. This estimation is semiparametric; the mixture complexities k̂jc are estimated from the data. AKM employs an iterative estimation scheme, comparing the k-component mixture estimate against the k + 1-component mixture estimate. When the improvement obtained by adding a k + 1st component is negligible (less than some penalty term) the iteration is terminated and the resulting k-component mixture is used as the estimate. This general version of model selection—looking for the “elbow” or “knee” in a complexity vs. penalized estimation accuracy curve—is quite common. A distinguishing feature of AKM is the process of using successive kernel estimates to guide the successive mixture estimates.
The filtered kernel estimator (Marchette et al., 1996) extends the basic kernel estimator by allowing multiple bandwidths driven by a pilot mixture model. Given a Gaussian mixture
| (5) |
and a bandwidth h, define the filtered kernel estimator f̃(·; 𝒳, f, h) based on the mixture f and using the data 𝒳 = {X1, …, Xn} to be
| (6) |
where is the variance of the tth component of the mixture f and φ0 is the standard zero mean, unit variance normal. This allows for different bandwidths, guided by the mixture model f.
Let f̂1 be the single normal component with mean and variance determined by the sample moments of 𝒳jc—that is, f̂1 is a trivial one-component mixture. Let f̃1 be the filtered kernel estimate based on f̂1—that is, f̃1 is the standard kernel estimate using the normal reference rule to determine the bandwidth. For k = 2, 3, …, define in turn f̂k first to be the k-component mixture best matched to the nonparametric estimate f̃k−1
| (7) |
where is the integrated squared error and 𝓕k is the class of k-component Gaussian mixtures. Subsequently define f̃k to be the filtered kernel estimate based on the mixture f̂k.
Let
| (8) |
(This would be the log-likelihood if the observations were independent; alas, this is not the case for the brain voxels. We address the issue of spatial dependence in the next subsection.) The estimate of mixture complexity used here (analogous to the estimates proposed in Priebe and Marchette (2000) and James et al. (2001))is given by
| (9) |
The penalty term a(njc, k + 1) in the above equation—a function of sample size and model complexity—is the key practical issue in this version of model selection. A simple choice for a(njc, k + 1) is 3 log(njc). Asymptotic considerations require a(njc, k + 1)/njc → 0 as njc → ∞ for fixed k, but not too fast. In practice, the choice of a(njc, k + 1) drives the parsimoniousness of the resultant model. Akaike (1974), Rissanen (1978), and many others since (see, notably, George and Foster, 2000) have weighed in on the choice of this penalty term; nevertheless, for finite sample sizes the choice quite resembles an artistic balancing of parsimony and fit—the conventional bias-variance trade-off.
3.3. Spatial dependence
Because there is correlation amongst the voxel observations, and thus ℓ(·, ·) defined above is not the log-likelihood, simple interpretation of the penalty term a(njc, k+1) as a function of sample size calls for conditional covariance modelling of the three-dimensional spatial process. Covariograms (Cressie, 1993) for the subject-specific class-conditional random fields indicate that significant spatial correlation is evident at a distance of up to 10 voxels. (Stationarity and isotropy are indicated as well.) This suggests that the effective sample sizes are significantly smaller than the number of voxels njc.
One way to look at the effect of dependent observations on our estimate of mixture complexity is to note that, if the observations in the subject-specific class-conditional sample 𝒳jc are positively correlated, then the terms ℓ(f̂k+1, 𝒳jc) and ℓ(f̂k, 𝒳jc) in Eq. (9) should be scaled accordingly. That is, if the “effective number of independent observations” is n′jc:=njc/δ for 1≤δ≤njc, then substituting δ−1 ℓ(·, ·) into Eq. (9) will account for the correlation. The case δ = 1 represents independence and, as (positive) spatial correlation increases, δ increases as well. This can be presented as
| (10) |
The idea of accounting for spatial correlation via an “effective number of independent observations” (Cressie, 1993) is analogous to the “resolution elements” found in Worsley et al. (1992), e.g. Solid mathematical justification for adopting this approach is lacking except in the most restricted cases (Leadbetter et al., 1983). Direct modelling of the dependency structure, via methods such as Markov random field models, is the subject of intense ongoing research. Our ad hoc approach is meant as a simple and straight-forward method enabling semiparametric hierarchical mixture modelling.
Estimating δ is a sticky wicket. Nevertheless, as will be seen in the experimental results presented below, a significant performance improvement can be obtained by employing even a crude covariogram-based estimate δ̂ in the complexity selection methodology. Furthermore, Occam's Razor may suggest a preference to err on the side of parsimony. Our rule of thumb is to choose δ̂ to be the number of voxels in a ball whose radius is given by the distance at which the omnidirectional empirical covariogram drops below some threshold, rounded to the nearest integer.
Using a(n, k) = 3 log(n), we arrive at the estimate of mixture complexity
| (11) |
Given k̂jc selected in this manner, the mixture model estimate of the marginal probability density for subject j, tissue class c, is then given by
| (12) |
the k̂jc-component mixture identified in the AKM procedure. Results will be reported below for three choices of δ̂: 1, 25, and 99. The case δ̂ = 1 corresponds to ignoring the dependence; the choices δ̂ = 25 and δ̂ = 99 are obtained with threshold values 2/3 and 1/3,respectively.
4. Segmentation method
We consider segmenting the cingulate gyrus for subject γ ∈ 𝓙; in this case, we have J − 1 training subjects corresponding to indices j ∈ 𝓙γ := 𝓙\{γ}.
4.1. Training
For j ∈ 𝓙γ, we obtain
| (13) |
via AKM. For γ we have the test data set 𝒳γ.
Write
| (14) |
(This would be the likelihood if the subject-specific voxel observations were independent.) We proceed by choosing the index γ* specifying the training subject “closest” to the test data set. However, even though we proceed under the assumption that the subject-specific class-conditional tissue mixture models f̂jc for one training subject can be usefully transferred to the test sample, there is good reason to treat the subject-specific class-conditional mixing coefficients as nuisance parameters; while the tissue class probability distributions may be transferrable, we wish to assume that the percentage of gray matter, say, may vary from training subject to test subject. Toward this end we define
| (15) |
the collection of densities which agree with f̂j up to subject-specific class-conditional mixing coefficients. Then
| (16) |
The selection of training subject γ* provides the test subject γ with class-conditional complexity estimates k̂γ*c, as well as initial conditions obtained from the f̂γ*c and nγ*c/nγ*, for use in modelling 𝒳γ. (There are, of course, choices other than the likelihood-based option (Eq. (14)) for determining the “closest” training subject γ*.)
4.2. Testing
4.2.1. Mixture modelling
Given γ* selected as above, we estimate fγ by estimating the parameters
| (17) |
via
| (18) |
Notice that this involves estimation of parameters. Algorithmically, this M-estimate is obtained using the EM algorithm (see, for instance, McLachlan and Krishnan, 1997) with the training model f̂γ* as the starting point;
| (19) |
The estimate obtained in this manner can be written as
| (20) |
Class complexities and component labels are inherited from f̂γ*, so that (for c = G, for instance) the mixing coefficient π̂γG indicates the amount (proportion) of gray matter in the test image while provides a model for that gray matter.
This unsupervised modelling of the test subject mixture allows estimation of the proportion and character of the different tissue types as they appear in the test subject, with a goal toward improving the segmentation for this subject.
4.2.2. Classification
We next consider the Bayes plug-in classifier
| (21) |
The voxel x is to be labelled as belonging to the class which maximizes posterior probability of class membership. See Devroye et al. (1996) for a thorough discussion of the Bayes plug-in classifier.
This is (operationally) equivalent to the likelihood ratio test procedure (our estimates are indeed marginal density estimates) given by considering
| (22) |
and
| (23) |
Our automatic segmentation is then given by the following rules:
r1(x) > 1 implies voxel x is to be labelled as tissue class C.
r2(x) < 1 implies voxel x is to be labelled as tissue class W.
r1(x) < 1 & r2(x) > 1 implies voxel x is to be labelled as tissue class G.
- r1(x) > 1 & r2(x) < 1 should not occur.
- ○ (Fig. 3 suggests stochastic ordering, C<st G<stW; this event is labelled “unknown” in the event that it does occur.)
(Ties are to be broken arbitrarily.)
5. Results
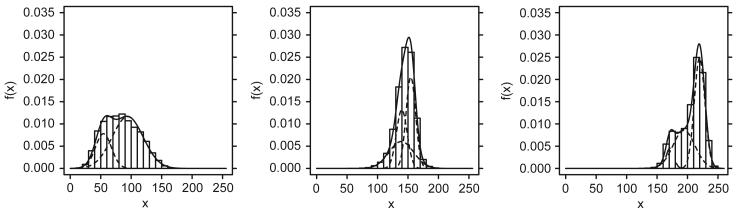
An example of the subject-specific, class-conditional marginal density estimates obtained via the AKM procedure presented above is presented in Fig. 4. Table 2 presents the complexity estimation results k̂jc for the 10 cingulate gyri under consideration for three choices of δ̂. From Fig. 4 we see that these (representative) fits are quite accurate. From Table 2 we see that δ̂ = 1 leads to a very high complexity, while δ̂ = 25 and δ̂ = 99 provide a reasonable framework to investigate the parsimony-fit trade-off.
Fig. 4.
Depicted are mixtures (δ̂ = 25) and histograms for CG 9. From left to right: C, G, W. Estimated model complexities are k̂CG9C = 2, k̂CG9G = 3, k̂CG9W = 3.
Table 2.
Mixture complexity estimation results for 10 cingulate gyri
| Estimated number of components |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CG 1 | CG 2 | CG 3 | CG 4 | CG 5 | CG 6 | CG 7 | CG 8 | CG 9 | CG 10 | |
| δ̂ = 1 | ||||||||||
| C | 3 | 1 | 4 | 3 | 3 | 2 | 26 | 3 | 2 | 2 |
| G | 2 | 16 | 3 | 1 | 3 | 1 | 1 | 1 | 3 | 3 |
| W | 3 | 13 | 5 | 16 | 17 | 13 | 6 | 16 | 12 | 11 |
| δ̂ = 25 | ||||||||||
| C | 3 | 1 | 2 | 3 | 3 | 1 | 2 | 2 | 2 | 2 |
| G | 1 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 3 | 3 |
| W | 3 | 2 | 3 | 4 | 3 | 2 | 3 | 4 | 3 | 5 |
| δ̂ = 99 | ||||||||||
| C | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| G | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 2 |
| W | 1 | 2 | 3 | 3 | 3 | 2 | 2 | 2 | 3 | 3 |
There is no “gold standard” available for this investigation; that is, we do not know the true voxel class labels. The expert's hand-segmentation is the “lead standard” against which we compare. Competing segmentation methodologies include:
An overly-simplistic model in which each tissue class is modelled with but a single normal.
Partial volume approaches (PV1 and PV2 in Table 3).
These two partial volume methodologies are sketched here.
Table 3.
Segmentation results for 10 cingulate gyri
| Probability of misclassification |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CG 1 | CG 2 | CG 3 | CG 4 | CG 5 | CG 6 | CG 7 | CG 8 | CG 9 | CG 10 | |
| PV1 | 0.15 | 0.09 | 0.23 | 0.20 | 0.28 | 0.15 | 0.15 | 0.16 | 0.20 | 0.24 |
| PV2 | 0.09 | 0.06 | 0.13 | 0.11 | 0.18 | 0.09 | 0.09 | 0.11 | 0.12 | 0.15 |
| AKM (δ̂ = 25) | 0.13 | 0.07 | 0.12 | 0.09 | 0.11 | 0.10 | 0.07 | 0.10 | 0.09 | 0.11 |
| AKM (δ̂ = 99) | 0.10 | 0.07 | 0.13 | 0.10 | 0.15 | 0.09 | 0.07 | 0.10 | 0.08 | 0.10 |
The partial volume approaches PV1 (Ratnanather et al., 2001) and PV2 (Ratnanather et al., 2004) involve a five-component Gaussian mixture fit via the EM algorithm; initial conditions are hand-selected based on the frequency histogram for the entire data set (e.g., the top panel in Fig. 3). One mixture component is initialized to account for each of the three tissue classes, and a “partial volume” mixture component is initialized between C & G and another between G & W. The difference between the two approaches is in the manner in which these partial volumes are allocated to the various tissue classes. These partial volume approaches are competitive with the most successful semi-automated MR brain image segmentation methodologies available in the literature, and are in use in current brain segmentation projects (Ratnanather et al., 2001, 2004).
In method PV1, τC/G = 1/2 of the C/G partial volume component is allocated to C and the other 1 − τC/G to G. That is, a voxel observation x for which the C/G partial volume component maximizes (over the five mixture components) posterior probability of class membership is assigned to tissue class C (resp. G) if it takes a value smaller (resp. larger) than the mean of the C/G partial volume component. The G/W partial volume component is allocated analogously.
Method PV2, on the other hand, optimizes probability of misclassification over τC/G ∈ [0, 1] and τG/W ∈ [0, 1]. For the cingulate gyrus data set under consideration herein, this optimization results in and .
Table 3 presents the probability of misclassification results for the 10 cingulate gyri under consideration for the AKM hierarchical mixture model with two choices of δ̂, as well as analogous results for the two competing partial volume approaches described above. Despite the small sample size of just 10 subjects, Table 3 suggests that the hierarchical mixture model methodology may provide superior automatic segmentation results to the competing approaches. For example, a paired one-sided t-test of AKM (δ̂ = 99) vs. PV2 yields a p-value of p = 0.03.
6. Discussion and conclusions
Voxel segmentation is one step of a sequence of procedures used in cortical analysis. Knowledge of volumes of gray matter, the coordinates of gray matter voxels and vertices of the gray/white surface is essential in a careful scientific analysis of cortical regions in normal and diseased subjects.
A real constraint on the ability to detect differences in two populations of MR brain imagery, and hence to investigate important neuropsychiatric hypotheses, is the cost of obtaining hand-segmentated brain volumes. This cost restricts the available sample sizes. Automated segmentation promises to allow for significantly larger sample sizes, and thus to advance the state of the science.
A hierarchical mixture modelling approach is demonstrated to provide automatic segmentation of magnetic resonance images superior to that of competing partial volume approaches. Our modelling methodology provides a key aspect missing in the methods currently in use: automatic subject-specific, class-conditional model complexity selection. The statistically innovative aspect of our work is the use of classified training data to obtain a model complexity for the unclassified test data, and hence allowing for richer test data models (and, subsequently, superior segmentation performance).
While specific details of the model fitting employed herein may be beneficially altered for particular applications, the general approach of (1) semiparametric estimation of subject-specific class-conditional marginal densities for a set of training volumes, (2) nearest neighbor matching of the test data to the training models providing for automated class-conditional mixture complexities, (3) parameter fitting of the selected training model to the test data, and (4) plug-in Bayes classification of unlabelled voxels, provides an advance in the state-of-the-art for automated MR brain image segmentation.
It is instructive to know which voxels are misclassified. In fact, for AKM the misclassified voxels are likely to be near class boundaries. This knowledge should be exploited to improve performance. However, such improvements may need to take into account that the “lead-standard” against which we test is based on expert hand-segmentation, and must not be considered a perfect “gold-standard.”
A useful generalization of our methodology involves “combining classifiers”; that is, rather than considering just the closest training model, one can perform the classification as above for each training model and then combine the results by weighting the classification obtained via the jth training model inversely proportional to max f ∈ 𝓕j 𝓛(γ,f). For our cingulate gyrus investigation this alteration improves the performance for seven of the 10 subjects. Note, however, that computational requirements increase proportional to the number of models used in the combination.
One aspect of our methodology for which further investigation is particularly warranted is our ad hoc accounting for spatial dependence (our δ̂). It will be beneficial to develop and use a more sophisticated spatially varying model to replace this crude covariogram estimate of the “effective number of independent observations” used herein.
The application of our methodology is not limited to just the cingulate gyrus. Experiments have been performed on medial prefrontal gyrus and occipital gyrus data sets; results indicate, again, the potential advantage of AKM. Indeed, MR brain image segmentation via AKM is a general tool for automated cortical analysis.
Footnotes
The work of CEP was partially supported by Office of Naval Research Grant N00014-01-1-0011. The work of MIM and JTR was partially supported by NIH grants P41 RR15241, R01 MH56584-04A1, P20 MH62130-A1, R01 MH62626-01, R01 MH60883-01, R01 EB00975, NSF NPACI and by ONR N00014-00-1-0327. The authors are grateful to their colleagues at the Washington University School of Medicine, St. Louis, and to anonymous reviewers.
The magnetic resonance brain image data set investigated in this manuscript is available at (http://www.cis.jhu.edu/data.sets/cortical_segmentation_validation/).
References
- Akaike H. A new look at the statistical model identification. IEEE Trans. Autom. Contr. 1974;19:716–723. [Google Scholar]
- Bartzokis G, Mintz J, Marx P, Osborn D, Gutkind D, Chiang F, Phelan CK, Marder SR. Reliability of in vivo volume measures of hippocampus and other brain structures using MRI. Magnet. Resonance Imaging. 1993;11:993–1006. doi: 10.1016/0730-725x(93)90218-3. [DOI] [PubMed] [Google Scholar]
- Botteron KN, Figiel GS. The neuromorphometry of affective disorders. In: Krishnan KRR, editor. Brain Imaging in Clinical Psychiatry. Dekker; New York, NY: 1997. pp. 145–184. [Google Scholar]
- Caunce A, Taylor CJ. Building 3d sulcal models using local geometry. Med. Image Anal. 2001;5:69–80. doi: 10.1016/s1361-8415(00)00033-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Kalbfleisch JD. Penalized minimum distance estimates in finite mixture models. Can. J. Statist. 1996;24:167–175. [Google Scholar]
- Chen T, Metaxas D. Medical Image Computing and Computer-Assisted Intervention—Miccai 2000, Vol. 1935. Lecture Notes in Computer Science. Springer; Berlin; 2000. Image segmentation based on the integration of markov random fields and deformable models; pp. 256–265. [Google Scholar]
- Cho ZH, Jones JP, Singh M. Foundations of Medical Imaging. Wiley; New York: 1993. [Google Scholar]
- Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE Trans. Med. Imaging. 1997;16:864–877. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- Cressie NAC. Statistics for Spatial Data. Wiley; New York: 1993. [Google Scholar]
- Csernansky JG, Bardgett ME. Limbic-cortical neuronal damage and the pathophysiology of schizophrenia. Schizophrenia Bull. 1998;24:231–248. doi: 10.1093/oxfordjournals.schbul.a033323. [DOI] [PubMed] [Google Scholar]
- Dacunha-Castelle D, Gassiat E. The estimation of the order of a mixture model. Bernoulli. 1997;3:279–299. [Google Scholar]
- Dacunha-Castelle D, Gassiat E. Testing the order of a model using locally conic parameterization: population mixtures and stationary ARMA processes. Ann. Statist. 1999;27:1178–1209. [Google Scholar]
- Devroye L, Gyorfi L, Lugosi G. A Probabilistic Theory of Pattern Recognition. Springer; Berlin: 1996. [Google Scholar]
- Drevets WC. Prefrontal cortical-amygdalar metabolism in major depression. Ann. NY Acad. Sci. 1999;877:614–637. doi: 10.1111/j.1749-6632.1999.tb09292.x. [DOI] [PubMed] [Google Scholar]
- Drevets WC, Todd RD. Depression, mania and related disorders. In: Guze SB, editor. Adult Psychiatry. Mosby; St Louis, MO: 1997. pp. 99–142. [Google Scholar]
- Drevets WC, Videon TO, Price JL, Preskorn SH, Carmichael ST, Raichle ME. A functional anatomical study of unipolar depression. J. Neurosci. 1992;12:3628–3641. doi: 10.1523/JNEUROSCI.12-09-03628.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy HM. The Human Brain: Surface, Three-Dimensional Sectional Anatomy With MRI, and Blood Supply. Springer; New York: 1999. [Google Scholar]
- Escobar MD, West M. Bayesian density estimation and inference using mixtures. J. Amer. Statist. Assoc. 1995;90:577–588. [Google Scholar]
- Everitt BS, Hand DJ. Finite Mixture Distributions. Chapman and Hall; London: 1981. [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated manifold surgery: Constructing Geometrically Accurate and Topologically Correct Models of the Human Cerebral Cortex. IEEE Trans. Med. Imaging. 2001;20:70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Garrido A, De la Blanca NP. Physically-based active shape models: initialization and optimization. Patt. Recogn. 1998;31:1003–1017. [Google Scholar]
- Garrido A, de la Blanca NP. Applying deformable templates for cell image segmentation. Patt. Recogn. 2000;33:821–832. [Google Scholar]
- George EI, Foster DP. Calibration and empirical bayes variable selection. Biometrika. 2000;87:731–747. [Google Scholar]
- Grabowski TJ, Frank RJ, Szumski NR, Brown CK, Damasio H. Validation of partial tissue segmentation of single-channel magnetic resonance images of the brain. Neuroimage. 2000;12:640–656. doi: 10.1006/nimg.2000.0649. [DOI] [PubMed] [Google Scholar]
- Haller JW, Banerjee A, Christensen GE, Gado M, Joshi S, Miller MI, Sheline Y, Vannier MW, Csernansky JG. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R. Discriminant analysis by Gaussian mixtures. IEEE Trans. Patt. Anal. Machine Intell. 1996;18:607–616. [Google Scholar]
- Henna J. On estimating of the number of constituents of a finite mixture of continuous distributions. Ann. Inst. Statist. Math. 1985;37:235–240. [Google Scholar]
- Holmes CJ, Hoge R, Woods RP, Evans AC, Toga AW. Enhancement of MR images using registration for signal averaging. J. Comput. Assist. Tomogr. 1998;22:324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Jain AK, Zhong Y, Lakshmanan S. Object matching using deformable templates. IEEE Trans. Patt. Anal. Mach. Intell. 1996;18:267–278. [Google Scholar]
- James LF, Priebe CE, Marchette DJ. Consistent estimation of mixture complexity. Ann. Statist. 2001;29:1281–1296. [Google Scholar]
- Joshi M, Cui J, Doolittle K, Joshi S, Van Essen D, Wang L, Miller MI. Brain segmentation and the generation of cortical surfaces. Neuroimage. 1999;9:461–476. doi: 10.1006/nimg.1999.0428. [DOI] [PubMed] [Google Scholar]
- Kapur T, Grimson WEL, Wells-III WM, Kikinis R. Segmentation of brain tissue from magnetic resonance images. Med. Image Anal. 1996;1:109–127. doi: 10.1016/S1361-8415(96)80008-9. [DOI] [PubMed] [Google Scholar]
- Keribin C. Consistent estimation of the order of mixture models. Sankhya. 2000;62:49–62. [Google Scholar]
- Kervrann C, Heitz F. Statistical deformable model-based segmentation of image motion. IEEE Trans. Image Process. 1999;8:583–588. doi: 10.1109/83.753745. [DOI] [PubMed] [Google Scholar]
- Leadbetter MR, Lindgren G, Rootzen H. Extremes and Related Properties of Random Sequences and Processes. Springer; New York: 1983. [Google Scholar]
- Marchette DJ, Priebe CE, Rogers GW, Solka JL. The Filtered Kernel Estimator. Comput. Statist. 1996;11:95–112. [Google Scholar]
- McLachlan GJ. On bootstrapping the likelihood ratio test statistic for the number of components in a normal mixture. Appl. Statist. 1987;36:318–324. [Google Scholar]
- McLachlan GJ, Basford KE. Mixture Models: Inference and Applications to Clustering. Marcel Dekker; New York: 1988. [Google Scholar]
- McLachlan GJ, Krishnan T. The EM Algorithm and Extensions. Wiley; New York: 1997. [Google Scholar]
- McLachlan GJ, Peel D. Finite Mixture Models. Wiley; New York: 2000. [Google Scholar]
- Mignotte M, Meunier J. A multiscale optimization approach for the dynamic contour-based boundary detection issue. Comput. Med. Imaging and Graph. 2001;25:265–275. doi: 10.1016/s0895-6111(00)00075-6. [DOI] [PubMed] [Google Scholar]
- Miller MI, Massie AB, Ratnanather JT, Botteron KN, Csernansky JG. Bayesian construction of geometrically based cortical thickness metrics. Neuroimage. 2000;12:676–687. doi: 10.1006/nimg.2000.0666. [DOI] [PubMed] [Google Scholar]
- Montagnat J, Delingette H. Cvrmed-Mrcas'97, Vol. 1205. Lecture Notes in Computer Science. Springer; Berlin: 1997. Volumetric medical images segmentation using shape constrained deformable models; pp. 13–22. [Google Scholar]
- Montagnat J, Delingette H, Malandain G. Medical Image Computing and Computer-Assisted Intervention, Miccai'99, Proceedings, Vol. 1679. Lecture Notes in Computer Science. Springer; Berlin: 1999. Cylindrical echocardiographic image segmentation based on 3d deformable models; pp. 168–175. [Google Scholar]
- Montagnat J, Delingette H, Ayache N. A review of deformable surfaces: topology geometry and deformation. Image and Vision Comput. 2001;19:1023–1040. [Google Scholar]
- Pham D, Xu C, Prince J. Current methods in medical image segmentation. Ann. Rev. Biomed. Eng. 2000;2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- Priebe CE, Marchette DJ. Alternating kernel and mixture density estimates. Comp. Statist. Data Anal. 2000;35:43–65. [Google Scholar]
- Ratnanather JT, Botteron KN, Nishino T, Massie AB, Lal RM, Patel SG, Peddi S, Todd RD, Miller MI. Validating cortical surface analysis of medial prefrontal cortex. Neuroimage. 2001;14:1058–1069. doi: 10.1006/nimg.2001.0906. [DOI] [PubMed] [Google Scholar]
- Ratnanather JT, Wang L, Nebel MB, Hosakere M, Han X, Csernansky JG, Miller MI. Validation of semiautomated methods for quantifying cingulate cortical metrics in schizophrenia. Psychiat. Res. Neuroimaging. 2004 doi: 10.1016/j.pscychresns.2004.07.003. in press. [DOI] [PubMed] [Google Scholar]
- Rissanen J. Modeling by shortest data description. Automatica. 1978;14:465–471. [Google Scholar]
- Robb RA. Biomedical Imaging, Visualization and Analysis. Wiley; New York: 1999. [Google Scholar]
- Roeder K, Wasserman L. Practical Bayesian density estimation using mixtures of normals. J. Amer. Statist. Assoc. 1997;92:894–902. [Google Scholar]
- Schultz N, Conradsen K. 2d vector-cycle deformable templates. Signal Process. 1998;71:141–153. [Google Scholar]
- Sclaroff S, Liu LF. Deformable shape detection and description via model-based region grouping. IEEE Trans. Patt. Anal. Mach. Intell. 2001;23:475–489. [Google Scholar]
- Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. NeuroImage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- Shen DG, Herskovits EH, Davatzikos C. An adaptive-focus statistical shape model for segmentation and shape modeling of 3-d brain structures. IEEE Trans. Med. Imaging. 2001;20:257–270. doi: 10.1109/42.921475. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Koob GF. Dopamine, schizophrenia, mania and depression: toward a unified hypothesis of cortico-striato-pallido-thalamic function. Behav. Brain Sci. 1987;10:197–245. [Google Scholar]
- Tagare HD. Deformable 2-d template matching using orthogonal curves. IEEE Trans. Med. Imaging. 1997;16:108–117. doi: 10.1109/42.552060. [DOI] [PubMed] [Google Scholar]
- Teo PC, Sapiro G, Wandell BA. Creating connected representations of cortical gray matter for functional MRI visualization. IEEE Med. Trans. 1997;16:852–863. doi: 10.1109/42.650881. [DOI] [PubMed] [Google Scholar]
- Titterington DM, Smith AFM, Makov UE. Statistical Analysis of Finite Mixture Distributions. Wiley; New York: 1990. [Google Scholar]
- Tootell R, Hadjikhani N, Mendola J, Marett S, Dale A. From retinotopy to recognition: fMRI in human visual cortex. Trends Cog. Sci. 1998;2:174–183. doi: 10.1016/s1364-6613(98)01171-1. [DOI] [PubMed] [Google Scholar]
- Wells-III WM, Grimson WEL, Kikinis RL, Jolesz FA. Adaptive segmentation of MRI Data. IEEE Trans. Med. Imaging. 1996;15:429–442. doi: 10.1109/42.511747. [DOI] [PubMed] [Google Scholar]
- Westin CF, Lorigo LM, Faugeras O, Grimson WEL, Dawson S, Norbash A, Kikinis R. Medical Image Computing and Computer-Assisted Intervention—Miccai 2000, Vol. 1935. Lecture Notes in Computer Science. Springer; Berlin: 2000. Segmentation by adaptive geodesic active contours; pp. 266–275. [Google Scholar]
- Worsley KJ, Evans AC, Marrett S, Neelin P. A three dimensional statistical analysis for CBF activation studies in human brain. J. Cereb. Blood Flow Metabol. 1992;12:900–918. doi: 10.1038/jcbfm.1992.127. [DOI] [PubMed] [Google Scholar]
- Xu C, Prince JL. Handbook of Medical Imaging. Academic Press; San Diego, CA: 2000. Gradient vector flow deformable models. [Google Scholar]
- Xu C, Pham DL, Rettmann ME, Yu DN, Prince JL. Reconstruction of the human cerebral cortex from magnetic resonance images. IEEE Trans. Med. Imaging. 1999;18:467–480. doi: 10.1109/42.781013. [DOI] [PubMed] [Google Scholar]
- Xu C, Pham DL, Prince JL. SPIE Handbook on Medical Imaging—Vol. III. Medical Image Analysis; SPIE: 2000. Medical image segmentation using deformable models; pp. 129–174. [Google Scholar]
- Yantis S. Visual Perception. Psychology Press; Philadelphia, PA: 2001. [Google Scholar]
- Yezzi A, Tsai A, Willsky A. A fully global approach to image segmentation via coupled curve evolution equations. J. Visual Commun. Image Representation. 2002;13:195–216. [Google Scholar]
- Zipursky RB, Lim KO, Sullivan EV, Brown BW, Pfefferbaum A. Widespread cerebral gray volume deficits in schizophrenia. Arch. Gen. Psychiatry. 1992;49:195–205. doi: 10.1001/archpsyc.1992.01820030027004. [DOI] [PubMed] [Google Scholar]