Abstract
The human CCCTC-binding factor, CTCF, organizes and regulates transcription of the genome by colocalizing distant DNA elements on the same and even different chromosomes. This protein consists of 11 zinc fingers flanked by polypeptide segments of unknown structure and function. We purified recombinant terminal fragments and observed that both are extended, monomeric, and predominantly consist of unordered content. We thus speculate that the role of the terminal extensions, and perhaps all of CTCF, is to act as a scaffold for the assembly of other proteins on a specific binding site.
Keywords: CTCF, chromatin, transcription, insulator
Introduction
The human CCCTC-binding factor, CTCF, regulates gene expression with different influences in a context-dependent manner. The same protein can act as a repressor,1 activator,2 or insulator3 when bound to different DNA sites. The genome-wide distribution of CTCF occupancy also suggests variation in function. ∼10,000 binding sites localize mostly to intergenic regions, but also to introns, promoters, exons, and untranslated regions.4–6 Furthermore, both context-dependent activity and ubiquitous global distribution are conserved throughout vertebrates and insects.7–13 Recent studies of CTCF suggest that the mechanism of this multifunctional transcriptional regulation may involve higher order chromatin structure.
CTCF organizes the human genome in the three-dimensional space of the nucleus by bringing together DNA elements from the same and even different chromosomes. The chromosome conformation capture methodology14 has identified cases in which the CTCF protein or CTCF-binding sites are required for gathering distant DNA within close proximity of each other.15,16 In the mouse genome, the imprinted H19/Igf2 locus in particular has yielded several insights into chromosomal organization mediated by CTCF. Not only has intrachromosomal association of elements within the locus been detected,17,18 but interchromosomal association between the locus and elements on different chromosomes has also been observed.19,20 Loss of a CTCF-mediated loop is sometimes also correlated with reduced transcription.15,16,19 One could imagine that the effect of a chromosome loop on gene expression may be defined by the DNA elements colocalized, perhaps explaining the context dependence of transcriptional regulation by CTCF.
How does CTCF perform such a myriad of regulatory functions in different genetic contexts? The protein contains 11 zinc fingers, 10 C2H2 and one C2HC, flanked by polypeptide segments of unknown structure. The full-length human protein contains 727 amino acids; the N- and C-terminal segments are ∼265 and ∼148 residues in length, respectively. We sought to elucidate molecular mechanisms of function by determining the structures of recombinant fragments. Our work took an unexpected turn when, much to our surprise, we discovered that the terminal fragments of CTCF are natively unstructured. Only the 11 zinc fingers fold into structural domains. We discuss the implications of this molecular architecture on possible functions of CTCF in transcriptional regulation and genome organization.
Results
Recombinant purification of CTCF terminal fragments
We obtained sufficient amounts of well-behaved CTCF protein fragments for biochemical experiments. Both terminal fragments can be purified to homogeneity as judged by SDS–PAGE [Fig. 1(A)]. The N-terminal fragment could be readily expressed and purified in a standard manner with metal affinity and size exclusion chromatography. The C-terminal fragment was proteolytically sensitive, predominantly yielding a ∼10 kDa truncation product after one affinity column. A second affinity step using a different purification tag placed at the other end of the fragment was used to isolate a full-length polypeptide. Both terminal fragments migrate anomalously slowly on a polyacrylamide gel. The N-terminal polypeptide is observed at ∼60 kDa instead of 31 kDa as expected; the C-terminal polypeptide is observed at ∼30 kDa instead of 19 kDa as expected. Similar aberrant migration has been observed before,21 but we nonetheless confirmed the identity of the proteins with Edman sequencing, which also revealed that the N-terminal methionine is processed in both cases. After purification, we can obtain milligram amounts of both fragments. Although a single band on a gel speaks to homogeneity of protein composition, we look for monodisperse behavior during size exclusion chromatography as our more rigorous standard of biochemical purity.
Figure 1.
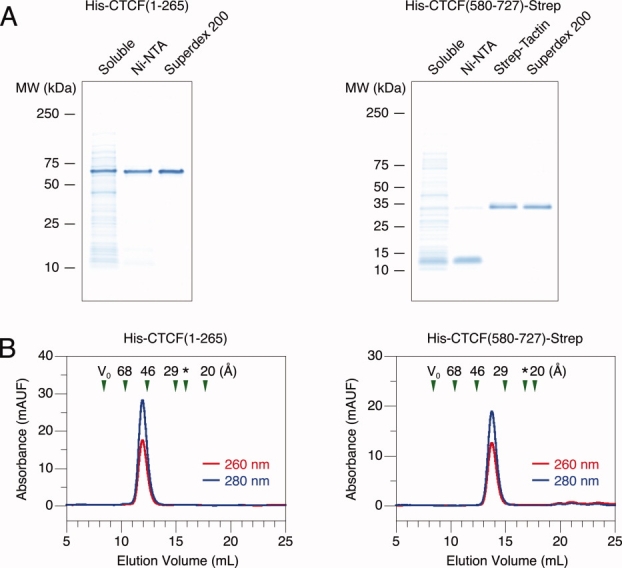
Purification of recombinant CTCF terminal fragments. Purification of the N- and C-terminal fragments from bacteria. (A) Protein fractions after different stages of purification resolved on a 4–20% SDS–PAGE gel. Positions of molecular mass standards are denoted on the left. Lane 1, soluble cell lysate. Subsequent lanes, protein fractions after affinity or size exclusion chromatography. (B) Size exclusion chromatography. Red and blue lines indicate UV absorbance at 260 and 280 nm, respectively. V0 marks the excluded void volume. Green triangles mark elution times of standards with known Stokes radii. An asterisk marks the predicted elution time of a globular protein with the same molecular mass as the CTCF fragment examined.
Measurements of hydrodynamic behavior reveal larger than expected radii for both terminal fragments. When examined with size exclusion chromatography, both fragments elute as single peaks [Fig. 1(B)]. Repeated experiments with different concentrations of protein over a 100-fold range did not alter the elution time (data not shown), suggesting the presence of a single monodisperse species as opposed to multiple species in rapid equilibrium. We also note that we do not observe any nucleic acid contaminants. Although we performed these experiments to ascertain protein purity, the same data also allows gleaning of a biophysical property, the protein hydrodynamic radius. The behavior of the N-terminal fragment revealed a Stokes radius of 50.7 ± 0.2 Å (N = 10); a 31 kDa protein is expected to migrate with a radius of 25 Å if globular.22 A similar result was observed with the C-terminal fragment, which revealed a Stokes radius of 37.3 ± 0.3 Å (N = 6); the expected measurement for a 19 kDa protein is 21 Å if globular.22 Larger than predicted radii may result from two potential molecular properties that give rise to increased hydrodynamic drag. Either the proteins are oligomeric or extended in conformation. We thus proceeded to measure the oligomerization state and secondary structure of the terminal fragments.
Biochemical properties of CTCF terminal fragments
Both terminal polypeptide fragments of CTCF are monomeric in solution. We directly measured the molar mass of each purified species with multi-angle light scattering coupled with size exclusion chromatography to determine the oligomeric state. Both terminal fragments yield a value close to that expected for a single subunit [Fig. 2(A)]. The N-terminal fragment measured 30 ± 1 kDa (N = 5); the calculated molar mass of a monomer is 31 kDa. The C-terminal fragment measured 20 ± 1 kDa (N = 8); the calculated molar mass of a monomer is 19 kDa. We also did observe, however, that ∼10% of the N-terminal fragments form disulfide-dependent dimers at high concentrations (data not shown). Because cysteine oxidation in a portion of a protein preparation is usually a spurious in vitro artifact, we doubt that these dimers are found in vivo. Given that the terminal fragments do not form multimers, oligomerization does not explain the large hydrodynamic radii observed.
Figure 2.
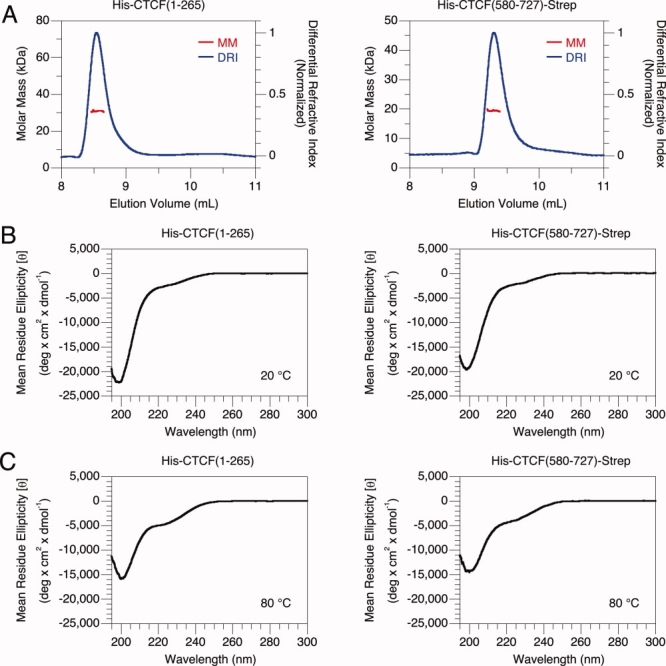
Biochemical properties of CTCF terminal fragments. Molar mass determination and secondary structure estimation of the N- and C-terminal fragments. (A) Multi-angle light scattering coupled with size exclusion chromatography. Red and blue lines indicate the molar mass and differential refractive index, respectively. (B) Far UV circular dichroism at 20°C. (C) Far UV circular dichroism at 80°C.
Both terminal segments of CTCF predominantly consist of unordered polypeptides. Instead of oligomerization, adoption of an extended conformation could yield a large hydrodynamic radius. We estimated the amount of secondary structure in the terminal fragments with circular dichroism spectroscopy. The observed spectra of both terminal polypeptides at 20°C [Fig. 2(B)] are consistent with a population predominantly comprised of unordered content. Random coils yield a distinct minimum at ∼197 nm not found with α helices or β strands.23 For both terminal polypeptides, a strong minimum is observed at ∼199 nm. Qualitative examination of spectral shape is further supported by quantitative analysis. Computational comparison to proteins of known fractional secondary structure content24 suggests the presence of mostly unordered polypeptide for both terminal fragments. The N-terminal polypeptide is estimated to contain 5 ± 0% helix, 10 ± 1% strand, 6 ± 1% turns, and 79 ± 1% unordered content (N = 8). The C-terminal polypeptide is estimated to contain 6 ± 1% helix, 15 ± 2% strand, 9 ± 1% turns, and 71 ± 2% unordered content (N = 8). Similar results were obtained with other algorithms (data not shown). The accuracy of this calculation should be regarded carefully because many caveats complicate the estimation of unordered content.25,26 The rough estimate, however, is clear and suggests that the bulk of both terminal fragments are unordered. Unfolded polypeptides are extended and likely account for the large hydrodynamic radii observed.
Circular dichroism measurements with both CTCF terminal fragments suggest an apparent increase of secondary structure content at higher temperature. Spectra of the terminal polypeptides at 80°C [Fig. 2(C)] differ from those at 20°C. Negative signal at 220 nm, generally indicative of ordered content, increases. The transitions are noncooperative and fully reversible (data not shown). We also note that negative signal at 200 nm, generally indicative of unordered content, decreases. Computational analysis, however, suggests that any gain of secondary structure may be quite small. The N-terminal polypeptide is estimated to contain 8 ± 1% helix, 12 ± 1% strand, 8 ± 1% turns, and 72 ± 2% unordered content (N = 5). The C-terminal polypeptide is estimated to contain 8 ± 0% helix, 16 ± 1% strand, 11 ± 0% turns, and 66 ± 1% unordered content (N = 4). While somewhat counterintuitive because one generally expects secondary structure to be lost with increasing temperature, similar behavior has been observed with intrinsically disordered proteins.27 Although we do not understand the biochemical basis of these peculiar observations, temperature-dependent changes in circular dichroism of the terminal fragments are reminiscent of unordered polypeptides.
The terminal segments of CTCF do not interact with each other. Given the possibility of intramolecular assembly, we attempted to detect an interaction between the terminal fragments that may assist in folding of secondary structures. After coexpression of the two polypeptides in bacteria, affinity purification of each did not yield the other (data not shown). Mixing the two fragments did not change the secondary structure as measured by circular dichroism (data not shown). Unfortunately, we could not purify monodisperse preparations of the zinc finger domains for similar experiments (data not shown). We interpret our results as excluding strong binding between the two terminal fragments.
Discussion
Functional implications of CTCF molecular architecture
No domains, defined in the classical biochemical sense as autonomously folding units of stable secondary structure, exist at the terminal segments of the protein CTCF. The terminal polypeptides comprise ∼413 residues, more than half of this 727 amino acid protein. We initially began protein expression efforts with the goal of attempting crystallographic studies. During the course of our investigation, however, biochemical experiments revealed that the terminal fragments of CTCF predominantly consisted of extended, monomeric, and unordered polypeptide. The large hydrodynamic radius for each fragment is closer to that expected for unfolded rather than native proteins of the same molecular weight.22,28 The circular dichroism spectra are similar to those observed for coil-like natively unfolded polypeptides;28 changes in circular dichroism as a function of temperature also resemble the response of intrinsically disordered proteins.27 Analysis of the CTCF sequence with disorder prediction algorithms29,30 even identified regions in the terminal domains as likely to be unstructured (data not shown). We cannot rule out the possible existence of isolated helices or strands, but these elements are neither abundant nor assemble into an ordered fold. Our biochemical data reaches a simple yet definitive conclusion. No domains exist in the terminal segments of CTCF. We may now deduce that no enzymatic functions are present. No known folds that bind small molecules ligands can be found. The possible functions of these segments are thus limited to those achievable by unordered structures. What then, could the functions of the terminal segments be?
Unstructured polypeptides often serve as molecular recognition elements for other proteins. Many natively unfolded polypeptides interact with other macromolecules.31 Indeed, several potential interacting proteins have been identified for CTCF.32,33 We have attempted to reconstitute recombinant complexes with some of the reported cofactors, but have yet to identify binding events that produce disorder to order transitions in the terminal fragments (data not shown). We remain encouraged, however, because circumstantial evidence suggests that the terminal segments do act as molecular recognition elements. Interactions are sometimes regulated by post-translational modifications that alter the binding interface. Suggestively, CTCF function can be disrupted by mutation of residues in the terminal segments that are modified by phosphorylation34,35 and sumoylation.36 The N-terminal segment serves as a substrate for poly(ADP-ribosyl)ation in vitro.37 Moreover, the classical zinc fingers of CTCF bind DNA,1,38 but these domains in other transcription factors also bind proteins.39,40 We now know that CTCF predominantly consists of zinc fingers and unstructured polypeptides, so we surmise that perhaps the entire protein functions as a scaffold or adaptor. Extreme speculation could even envisage recruitment regulated by post-translational modifications similar to that observed with histone tails or the RNA polymerase II C-terminal extension. Recruitment of different cofactors to different DNA sites may yield context-dependent genome organization and transcriptional regulation. We hope that our examination of CTCF architecture helps motivate future studies of regulated molecular recognition.
Materials and Methods
Molecular cloning
The terminal segments of CTCF were defined by excluding the 11 predicted zinc fingers using a minimum consensus motif.41 Fragments of the CTCF gene were obtained by PCR amplification from human cDNA clone TC115994 (Origene). All initial isolates differed from the reference human sequence1 with cytosine instead of thymine at position 1919; this conflict was resolved with Quikchange Site-directed Mutagenesis (Stratagene) prior to the subcloning that generated expression plasmids. Additional sequences flanking CTCF gene fragments were included by incorporation in PCR primers. An NdeI site, the sequence encoding amino acids 2–265, a TAA stop codon, and a BamHI site was amplified and inserted between the NdeI and BamHI sites of pET-15b (EMD Biosciences) to generate pSRM10. An NdeI site, the sequence encoding amino acids 580–727, the sequence TGGAGCCACCCGCAGTTCGAAAAA that encodes the Strep-tag WSHPQFEK, a TAA stop codon, and a BamHI site was amplified and inserted between the NdeI and BamHI sites of pET-15b (EMD Biosciences) to generate pSRM8. The construct pSRM10 fuses an N-terminal His-tag to residues 1–265; pSRM8 fuses both an N-terminal His-tag and a C-terminal Strep-tag to residues 580–727. Sequencing of the open reading frames (UCSF Genomics Core Facility) confirmed proper cloning.
Protein expression and purification
The N-terminal CTCF fragment was purified from Escherichia coli BL21(DE3) (EMD Biosciences) transformed with pSRM10 and grown in LB media. Expression was induced in log phase with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37°C for 4 h. Cells were lysed by sonication on ice in 50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% isopropanol, pH 7.5. All subsequent purification steps were performed at 4°C. The lysate was centrifuged and the supernatant incubated in batch with Ni-NTA agarose (Qiagen). The resin was washed with 50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol, pH 7.5, and the bound protein eluted with the same buffer containing 200 mM imidazole. The eluant was concentrated and resolved on a Superdex 200 10/30 column (GE Healthcare) equilibrated in 25 mM HEPES, 250 mM NaCl, 1 mM β-mercaptoethanol, pH 7.4.
The C-terminal CTCF fragment was purified from E. coli BL21(DE3) (EMD Biosciences) transformed with pSRM8 and grown in Terrific Broth (TB) media. Expression was induced in log phase with 1 mM IPTG at 17°C overnight. Cells were lysed by sonication on ice in 50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, 1% isopropanol, pH 7.5. All subsequent purification steps were performed at 4°C. The lysate was centrifuged and the supernatant incubated in batch with Ni-NTA agarose (Qiagen). The resin was washed with 50 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol, pH 7.5, and the bound protein eluted with the same buffer containing 200 mM imidazole. The eluant was incubated in batch with Strep-Tactin Sepharose (IBA). The resin was washed with 50 mM sodium phosphate, 500 mM NaCl, 5 mM β-mercaptoethanol, pH 7.5, and the bound protein eluted with the same buffer containing 250 μM desthiobiotin. The eluant was concentrated and resolved on a Superdex 200 10/30 column (GE Healthcare) equilibrated in 25 mM HEPES, 250 mM NaCl, 1 mM β-mercaptoethanol, pH 7.4.
Protein concentration was determined by measuring UV absorption.42 Protein identity was confirmed with Edman sequencing (Tufts University Core Facility).
Biochemistry
Hydrodynamic radii were measured using size exclusion chromatography with an ÄKTApurifier fast protein liquid chromatography system (GE Healthcare). 500 μL of 200 μg/mL purified protein was resolved on a Superdex 200 10/30 column (GE Healthcare) equilibrated in 25 mM HEPES, 250 mM NaCl, 1 mM β-mercaptoethanol, pH 7.4 at 4°C. The void volume was estimated with blue dextran (GE Healthcare). The column was calibrated with standard proteins (GE Healthcare) of known Stokes radii calculated from intrinsic viscosity measurements and molecular weight:22 19.7 Å RNAse, 28.7 Å ovalbumin, 46.4 Å aldolase, and 67.9 Å ferritin. The reciprocal of elution volume was plotted versus the Stokes radius to obtain a linear regression curve used to calculate the radii of experimental samples.43
Molar masses were determined by multi-angle light scattering coupled with size exclusion chromatography using an Ettan LC fast protein liquid chromatography system (GE Healthcare) coupled to a DAWN HELEOS multi-angle light scattering instrument (Wyatt) and an Optilab rEX refractive index detector (Wyatt). 100 μL of 200 μg/mL purified protein was resolved on a PROTEIN KW-803 column (Shodex) equilibrated in 25 mM HEPES, 250 mM NaCl, 1 mM β-mercaptoethanol, pH 7.4 at 4°C. The weight average molar mass was determined with ASTRA V (Wyatt).
Secondary structure content was calculated by measuring circular dichroism with a J-715 spectropolarimeter (Jasco) equipped with a PTC-348WI Peltier temperature control system (Jasco). Purified protein was exchanged into 25 mM sodium phosphate, 250 mM NaF, 1 mM β-mercaptoethanol, pH 7.5 by repeated concentration and dilution in a centrifugal filter unit. Spectra were acquired with 200 μg/mL protein in a 1 mm cuvette (Hellma) at 20°C and 80°C. For each measurement, the spectrum of the buffer alone was subtracted from the spectrum of the protein solution. Fractional secondary structure content was estimated with the CONTIN/LL algorithm of CDPro.24 Data from 200 to 240 nm was compared to reference set SDP48.
Acknowledgments
We thank Brian K. Shoichet, Veena L. Thomas, and Matthew Merski for use of the circular dichroism spectropolarimeter as well as David A. Agard and Dan R. Southworth for the use of the light scattering system.
References
- 1.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vostrov AA, Quitschke WW. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- 3.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol Cell Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 9.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 10.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 11.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke LJ, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachidanandam R, Sun H, Lobanenkov V, Zhou J. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol. 2009;328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 15.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu TH, Hoffman AR. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 21.Klenova EM, Nicolas RH, U S, Carne AF, Lee RE, Lobanenkov VV, Goodwin GH. Molecular weight abnormalities of the CTCF transcription factor: CTCF migrates aberrantly in SDS–PAGE and the size of the expressed protein is affected by the UTRs and sequences within the coding region of the CTCF gene. Nucleic Acids Res. 1997;25:466–474. doi: 10.1093/nar/25.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uversky VN. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 1993;32:13288–13298. doi: 10.1021/bi00211a042. [DOI] [PubMed] [Google Scholar]
- 23.Greenfield N, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 24.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 25.Venyaminov S, Baikalov IA, Shen ZM, Wu CS, Yang JT. Circular dichroic analysis of denatured proteins: inclusion of denatured proteins in the reference set. Anal Biochem. 1993;214:17–24. doi: 10.1006/abio.1993.1450. [DOI] [PubMed] [Google Scholar]
- 26.Sreerama N, Venyaminov SY, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal Biochem. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- 27.Uversky VN. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 28.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 31.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 32.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 34.Klenova EM, Chernukhin IV, El-Kady A, Lee RE, Pugacheva EM, Loukinov DI, Goodwin GH, Delgado D, Filippova GN, Leon J, Morse HC, 3rd, Neiman PE, Lobanenkov VV. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol Cell Biol. 2001;21:2221–2234. doi: 10.1128/MCB.21.6.2221-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Kady A, Klenova E. Regulation of the transcription factor, CTCF, by phosphorylation with protein kinase CK2. FEBS Lett. 2005;579:1424–1434. doi: 10.1016/j.febslet.2005.01.044. [DOI] [PubMed] [Google Scholar]
- 36.MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, Oshimura M, Feinberg AP, Lobanenkov V, Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 38.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 39.McCarty AS, Kleiger G, Eisenberg D, Smale ST. Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell. 2003;11:459–470. doi: 10.1016/s1097-2765(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 40.Liew CK, Simpson RJ, Kwan AH, Crofts LA, Loughlin FE, Matthews JM, Crossley M, Mackay JP. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc Natl Acad Sci USA. 2005;102:583–588. doi: 10.1073/pnas.0407511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison SC. A structural taxonomy of DNA-binding domains. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 42.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 43.Davis LC. A simplified calibration procedure for elution chromatography on gel-filtration columns. J Chromatogr Sci. 1983;21:214–217. [Google Scholar]


