Abstract
RNA viruses are the main agents of emerging and re-emerging diseases. It is therefore important to reveal the evolutionary processes that underpin their ability to jump species boundaries and establish themselves in new hosts. Here, I discuss how comparative genomics can contribute to this endeavor. Arguably the most important evolutionary process in RNA virus evolution, abundant mutation, may even open up avenues for their control through “lethal mutagenesis.” Despite this remarkable mutational power, adaptation to diverse host species remains a major adaptive challenge, such that the most common outcome of host jumps are short-term “spillover” infections. A powerful case study of the utility of genomic approaches to studies of viral evolution and emergence is provided by influenza virus and brought into sharp focus by the ongoing epidemic of swine-origin H1N1 influenza A virus (A/H1N1pdm). Research here reveals a marked lack of surveillance of influenza viruses in pigs, coupled with the possibility of cryptic transmission before the first reported human cases, such that the exact genesis of A/H1N1pdm (where, when, how) is uncertain.
Keywords: evolution, influenza, RNA virus, lethal mutagenesis, mutation rate
The recent appearance of swine-origin H1N1 influenza A virus (A/H1N1pdm) in humans serves as a pointed reminder of the global burden of morbidity and mortality caused by influenza viruses. More generally, A/H1N1pdm highlights the ability of RNA viruses to jump species barriers and emerge in new hosts, in this case transferring from pigs to humans. Although the H1N1 subtype of influenza A virus is a familiar one, most famously as the cause of the devastating influenza pandemic of 1918–1919 (1), the current A/H1N1pdm epidemic is notable in that it is caused by a viral lineage that is phylogenetically distinct from the other swine influenza viruses sampled over the last 20 years (2, 3). Hence, despite the considerable effort that has gone into the surveillance and characterization of influenza viruses, and especially in wild birds since the appearance of highly pathogenic avian A/H5N1 viruses (4–6), there has been a marked gap in our surveillance of these viruses in pigs. This represents a serious oversight as swine viruses are already adapted for transmission in mammalian populations and spill-over infections from pigs to humans are relatively commonplace (7).
Another important aspect of the A/H1N1pdm epidemic is how quickly genome sequence data for this virus was generated and placed into the public domain. Indeed, so rapid was the generation of sequence data that analysis was effectively undertaken in real time and laudably often with full public access (see http://tree.bio.ed.ac.uk/groups/influenza for an excellent example). More generally, the rapid generation of genome sequence data represents a very powerful way to determine cause of diseases of unknown etiology. This is illustrated by the case of colony collapse disorder (CCD), in which honey bees leave the hive and seemingly die, such that the hive community eventually collapses. Here, comparative genome sequencing was quickly able to identify the RNA virus Israeli acute paralysis virus (IAPV) as the most likely agent for CCD (8), although this is yet to satisfy Koch’s postulates. As an added bonus, because all of the DNA present in infected hives was sequenced, rather than just that of IAPV, this metagenomics approach to pathogen discovery allowed characterization of much of the microbial flora carried by honey bees, encompassing viruses, bacteria, fungi, and others. In short, the generation and analysis of complete genome sequence data are close to becoming the default way of characterizing new viral pathogens (9).
The aim of this article is to demonstrate how new genomic-scale approaches are able to provide unique insights into the processes that govern the emergence and evolution of RNA viruses. In doing so I make general statements about the nature of RNA virus evolution and highlight some of the key evolutionary lessons learned from the ongoing A/H1N1pdm pandemic in particular. As a sidebar, this work illustrates the increasingly important role played by evolutionary biology in the study of infectious disease.
The Evolutionary Genetics of Viral Emergence
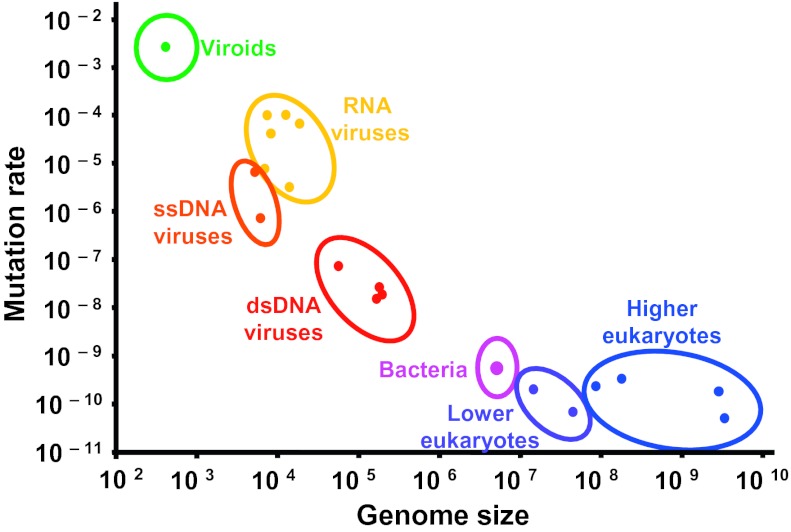
Even allowing for their relative abundance, RNA viruses seem particularly prone to causing emerging diseases in humans and other animals (10). Although these infectious agents have defining characteristics, perhaps the most important from the perspective of their evolution is their capacity for mutation. The vast majority of estimates of mutation rates in RNA viruses are in the range of 0.1 to 1.0 mutations per genome, per replication (11), several orders of magnitude higher than those in most DNA-based organisms (Fig. 1). Such remarkable error rates are evidently a function of replication with a low-fidelity RNA-dependent RNA polymerase, the only protein shared by all RNA viruses and which lacks any of the proof-reading abilities associated with the higher-fidelity DNA polymerases. Although there is an ongoing debate as to what selective forces (if any) are responsible for such high error rates (11, 12), it is likely that RNA viruses survive this enormous mutational burden by an equally remarkable reproductive power, manifest as many progeny in each infected cell, infected cells, and infected hosts (13).
Fig. 1.
Relationship between mutation rate per nucleotide site and genome size for different genomic systems including viruses. [Reproduced with permission from ref. 19 (Copyright 2009, AAAS).]
High mutation rates, coupled with rapid replication, are also the basis for the high rates of nucleotide substitution (fixation) recorded in RNA viruses. Mean substitution rates in this case are usually in the realm of 10−3 to 10−4 nucleotide substitutions per site per year (subs/site/year) and hence some six orders of magnitude higher than those seen in eukaryotes (11, 14). Clearly, evolutionary rates of this magnitude are a major reason clinically important traits, such as drug resistance, escape from vaccine coverage, and host range expansion, appear so readily in some RNA viruses. In the case of A/H1 N1pdm, estimates of genomewide substitution rates, at ≈5 × 10−3 subs/site/year (15), are broadly similar to those seen in other human influenza viruses (16), and hence at the upper end of those seen in RNA viruses generally (14). Whether this means that A/H1N1pdm will generate similar levels of antigenic variation to that seen in other human influenza A viruses will require a longer sampling period.
The high evolutionary rates associated with RNA viruses have several important implications. First, it is likely that high mutation rates limit the genome size of RNA viruses to a median value of ≈10 Kb (and the largest RNA viruses are the coronaviruses at only 29–32 Kb). Specifically, if mutation rates are constant per nucleotide, then the error-prone replication of longer RNA molecules will lead to the accumulation of excessive numbers of deleterious mutations and hence major fitness losses (17). Although a number of theories have been proposed to explain the constrained genome sizes of RNA viruses, such as an inability to package excessively large genomes, the power of the mutation rate hypothesis is that it can be extended to other microbial organisms. In particular, ssDNA viruses, such as the vertebrate parvoviruses, exhibit mutation rates that fall closer to those seen in RNA viruses than dsDNA viruses, and all have genomes that are <12 Kb in length (Fig. 1). Similarly, rates of nucleotide substitution in ssDNA viruses fall within the range seen in RNA viruses (11). Although the reasons that underpin the high evolutionary rates of ssDNA viruses are unclear, one possibility is that mutations in ssDNA viruses are largely the result of postreplicatory processes, such as deamination (18). At the other extreme, recent work has revealed that viroids, small (< 500 nt), highly structured RNA elements that cause a variety of diseases in plants, experience mutation rates that exceed even those seen in RNA viruses (ref. 19 and Fig. 1).
Another important implication of the high mutation rates of RNA viruses, and one that at first glance seems rather paradoxical, is that they may represent an Achilles’ heel for their treatment. The background to this idea is that RNA viruses sit close to what can be notionally thought of as an “error threshold,” beyond which major fitness losses are likely (17). Although the complexity of fitness landscapes means that there is unlikely to be an absolute threshold value per se (20), the general association between viral mutation rates and genome sizes strongly suggests that overly large genomes are subject to severe fitness costs. Given this upper bound on mutation rates (and genome sizes), it then follows that artificially increasing error rates through exposure to mutagens such as ribavirin or 5′-fluorouracil would also lead to major fitness losses (21). Although there is some debate over the precise mechanistic basis to this “lethal mutagenesis” (22), the available experimental data offer strong support to the applicability of this exciting form of antiviral therapy, particularly when the mutagens are combined with more standard replication inhibitors (23). In addition, if there is indeed a fundamental relationship between genome size and error rate then the methods of lethal mutagenesis should equally apply to ssDNA viruses and viroids. The complication, of course, is the evolution of resistance. Although there have been claims that lethal mutagenesis will not be subject to all of the mechanisms of resistance that plague other forms of microbial control (24), in reality there is nothing in the biology of lethal mutagenesis that suggests it is “evolution proof,” particularly as single point mutations can result in improved polymerase fidelity (25).
Although RNA viruses have an enhanced capacity for mutation, which must in part underpin their ability to jump species boundaries and successfully emerge in new hosts, rapid mutation is not the only evolutionary process that needs to be discussed when considering their emergence. Of obvious importance is the fitness distribution of new mutations (26). As with most systems, the majority of mutations that arise in RNA viruses are deleterious and simply act to reduce fitness, particularly in the absence of high rates of recombination and heterozygosity (13). Measures of mutational fitness in single-cell assays suggest that as many as 40% of all mutations in vesicular stomatitis virus may be lethal, with many of the remainder falling into the slightly deleterious class (27). When this effect is magnified across all of the cell types a virus may infect, and considering the complexities of the viral life cycle during which natural selection can act at a variety of times and levels (28), it is evident that the vast majority of mutations will reduce fitness. As a case in point, only ≈1% of poliovirus virions released from a cell are able to complete a full replication cycle (28). Hence, most mutations that will ultimately aid adaptation to a new host species are likely to be strongly deleterious in the donor host (29).
As deleterious mutations are expected to be young (30), the preponderance of deleterious and slightly deleterious mutations is also manifest in phylogenetic analyses as an excess of nonsynonymous mutations on the tips of trees (31). It is this phenomenon that explains the increasingly common observation that rates of nucleotide substitution are higher in the short term, such as among sequences sampled from within single hosts or from individuals directly connected by transmission, than in the long term, such as between epidemics (32, 33). This may also be true of A/H1N1pdm; rate estimates based on the analyses of A/H1N1pdm sequences alone, collected over very short time scales (days to weeks), are greater than those estimated over longer time scales when A/H1N1pdm sequences are combined with other (swine) H1N1 lineages separated by years of evolution (3).
RNA viruses therefore seem to occupy a region of evolutionary parameter space that is acutely different to that where higher eukaryotes reside (13). That genome sizes are constrained by a high mutation rate means that RNA viruses may be less adaptable then is often envisaged, because there will be strong selection against evolutionary processes that act to increase genome size, such as gene duplication and lateral gene transfer (13). Such constraints are also reflected in the fact that the majority of cross-species transmissions of RNA viruses result in transient spillover infections rather than fully endemic pathogens (34). Hence, even though there is frequent exposure, the majority of RNA viruses are unable to fully adapt to new host species (35). A/H1N1pdm is again an important exemplar: between 2005 and 2009 11 human patients in the United States experienced swine influenza infections, yet only A/H1N1pdm has resulted in epidemic spread (7). Understanding the factors that determine whether a new infection will simply spill-over or spread on a larger scale is critical to predicting the future of any new emerging disease.
There are two important generalities about the nature of viral emergence that shed some light on the evolutionary mechanisms at play. First, vector-borne RNA viruses are subject to stronger selective constraints than those viruses transmitted by other routes (36) and correspondingly are less able to establish productive infections in new host species (10). This effect is likely because vector-borne viruses are subject to strong antagonistic pleiotropy, such that mutations favored in one host type are injurious in another, and which greatly limits adaptability after host jumps (37, 38), although different levels of diversity may be generated in the vertebrate and invertebrate components of the transmission cycle (39, 40). Second, a simple (but often broken) rule of thumb is that the more closely donor and recipient host species are related, the easier it will be for any virus (and likely any other pathogen) to jump between them and establish a productive infection (13, 41). Such a tendency arises because the cell types in these host species, their receptors, and likely other key components of the virus–host interaction will diverge along with their hosts to eventually reach a point where they become unrecognizably different for any RNA virus. For example, dengue viruses from nonhuman primates seem to able to replicate in human cells without any additional mutations (42), and only a single mutation appears responsible for the successful transfer of Venezuelan equine encephalitis virus from rodents to horses (43). In contrast, 13 mutations may be required for avian influenza viruses to establish productive infections in humans (44). This point also emphasizes how swine influenza viruses are in some sense “preadapted” to replicate in humans because they already contain the suite of key mutations required for productive replication in mammals. Hence, a simple rule of emergence is that viruses that have achieved this feat once have an inherent capacity to do it again.
The Evolutionary Genomics of Influenza Virus
One virus where genome sequence data has already had a profound impact on evolutionary studies is influenza. The key event in the genomics revolution for influenza A virus was the instigation of the Influenza Genome Sequencing Project (IGSP) in 2005 (ref. 45; see www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). Although influenza viruses have long been the under the gaze of evolutionary biologists, there were surprisingly few complete genome sequences available for analysis before the start of the IGSP. Today, however, >4,000 complete genome sequences of influenza viruses have been generated from a diverse array of avian and mammalian hosts.
One of the most important observations stemming from the data generated under the IGSP is that intrasubtype (i.e., within the A/H3N2 or A/H1N1 human subtypes) reassortment occurs very frequently. As a corollary, this also means that the mixed infection of individual hosts with multiple viral strains is also commonplace, which in turn raises questions about the extent of cross-protective immunity (46). Although the importance of reassortment for the cross-species transmission of influenza virus has a long history (47), complete genome sequence data provide the only clear insight into the frequency and determinants of this process (48). In addition, complete genome sequence analysis reveals how by placing gene segments in new genomic configurations, reassortment can sometimes generate isolates with altered antigenic properties, which in turn may lead to vaccine failure (49, 50). In short, reassortment seems to be a more important process in the day-to-day evolution of influenza A virus than previously realized, and attempts to predict future antigenic evolution without a consideration of reassortment are unlikely to be successful.
A second key insight stemming from the IGSP data is that specific human populations, such as that of New York State, where sampling has been particularly widespread in time and space, are characterized by the circulation of multiple viral lineages during any single season (16, 48–51). Not only does the cocirculation of lineages ensure that there is abundant raw material for reassortment, but it means that viral lineages must be continually imported during the time course of the influenza season (16, 49, 50). It is therefore not the case that a single viral lineage enters a population at the start of the influenza season (winter in the Northern Hemisphere), gradually diffuses through the population over the subsequent 6 months, before dying out the next summer. The same can be expected of A/H1N1pdm in the years that follow. The genetic diversity within a single population is also extremely well mixed spatially. For example, across the United States as a whole, phylogeographic analysis reveals that even relatively geographically isolated communities harbor similar amounts of viral diversity as major cities with more expansive travel networks (51), highlighting how rapidly this virus is able to spread through populations. Influenza virus is clearly readily able to exploit human contact networks, so that the coinfection that fuels reassortment could occur in any number of locations.
The expanse of genomic information on influenza also sheds new light on the genesis of drug-resistant viruses. One of the most important, and unexpected, outcomes of these studies is that direct drug-selection pressure is not always responsible for drug resistance. This is clearly the case with the adamantanes (amantadine and rimantadine), a group of antivirals to which subtype A/H3N2 viruses have shown a global rise in resistance in recent years (52). The most common cause of adamantane resistance is a single amino acid change (Ser31Asn) in the M2 protein. What is most striking in this case is that the Ser31Asn mutation has increased abruptly in frequency in populations where adamantanes are rarely used, such as the United States. Therefore, rather than being caused by direct selection pressure, it is more likely that the Ser31Asn mutation has become fortuitously linked to an antibody escape mutation located on another genome segment (53, 54). That this might be a more general phenomenon is suggested by the fact that the same hitch-hiking process may now be taking place with the neuraminidase inhibitor oseltamivir. In this case, there has been a dramatic rise in oseltamivir resistance in A/H1N1 viruses in many locations, including the United States where nearly all viruses are oseltamivir resistant (55, 56). Because these drugs are not widely used in many populations, linkage to another beneficial mutation again seems the most probable explanation for the rise of oseltamivir resistance, although there is no evidence for reassortment in this case. Although swine-origin A/H1N1 viruses are currently generally sensitive to oseltamivir (but resistant to adamantanes) it is possible that future reassortment among cocirculating human and swine-origin H1N1 viruses will change this picture.
Despite the genomic revolution, aspects of the evolution and epidemiology of influenza A virus remain opaque. In particular, although there is some evolutionary evidence for interaction between the A/H1N1 and A/H3N2 influenza viruses, which experience distinctive out-of-phase dynamics (15), and between influenza viruses A and B (57), the exact cause of these interactions remains elusive. For example, does this competition involve some form of nonspecific cross-immunity or ecological interference? The appearance of A/H1N1pdm adds a new urgency to revealing these interactions: it is obviously important to determine whether this newly emerged virus will outcompete the other influenza viruses circulating in human populations, particularly as estimates of its basic reproductive number (R0) indicate that it has the capacity to spread widely (58). Similarly, it is unclear why the “seasonal” A/H1N1 and A/H3N2 viruses that currently cocirculate in human populations have such different epidemiological dynamics. Seasonal A/H1N1 viruses are characterized by relatively slow antigenic evolution (such that relatively few mutations accumulate at antigenic sites), yet greater circulating genetic diversity, whereas A/H3N2 viruses are characterized by lower levels of circulating genetic diversity, but more rapid antigenic change (50), manifest as the fact that the A/H3N2 vaccine component needs to be updated on a regular basis (59). Finally, what epidemiological and evolutionary processes determine the phylodynamics of the influenza HA protein, manifest as “ladder-like” phylogenies and regular changes in antigenic type, is still the source of considerable debate (60–62).
As noted above, a key goal for the future must be to track the potential reassortment of the cocirculating influenza viruses, including the sporadic cases of highly pathogenic A/H5N1 avian influenza virus that have appeared in humans since 2003. Particular attention should be paid to viral surveillance in East and Southeast Asia, which seems to act as the global source population for influenza viruses (63), and may eventually prove to be the case for A/H1N1pdm. Indeed, the recent emergence of A/H1N1pdm means that three distinct lineages of influenza A virus are currently circulating in human populations, an event that is unprecedented in modern human history. Similarly, it will also be of fundamental importance to determine the spatial dynamics of A/H1N1pdm in human populations, and particularly whether they follow the same general pathways as identified for other influenza A viruses (63).
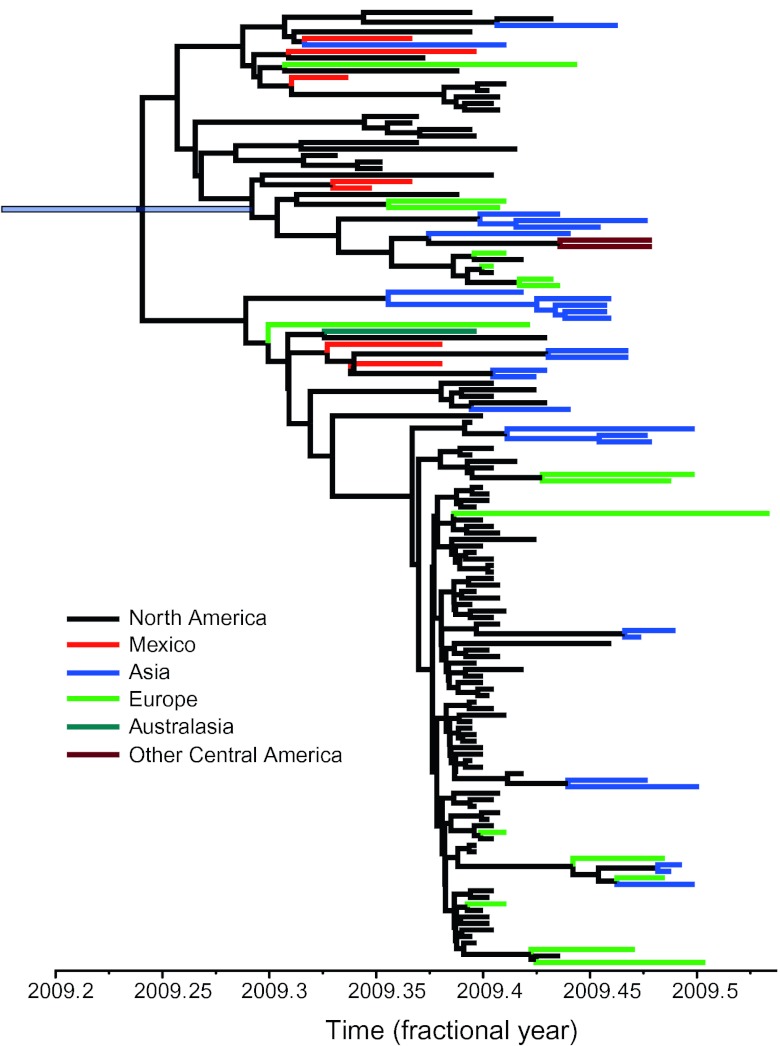
Those evolutionary analyses of A/H1N1pdm undertaken to date have shed light on a number of key issues. Phylogenetic analyses of complete genome sequence data have revealed the series of reassortment events responsible for the origin of A/H1N1pdm and how this virus has spread rapidly in both time and space (3). For example, since its first appearance A/H1N1pdm has spread to >160 countries (ref. 64; www.who.int/csr/don/2009_09_11/en/index.html), including multiple introductions into both Asia and Europe from the Americas (Fig. 2 and ref. 15). As this spatial diffusion continues it will also be essential to track the antigenic evolution of A/H1N1pdm and determine whether it bears more resemblance to the slow antigenic drift of A/H1N1 or the speedier evolution of A/H3N2. More generally, it will be important to understand why the A/H1N1pdm lineage was able to successfully emerge in human populations, and the determinants of this process, when most of the other lineages of swine influenza virus that periodically spill over into human populations fail to become established as endemic pathogens. In addition, although A/H1N1pdm was first reported in Mexico, whether the reassortment events that generated this virus occurred in that country (or continent) is less clear.
Fig. 2.
Maximum clade credibility tree of a sample of 147 complete genomes (13,130 nt) of A/H1N1pdm showing the spatial diffusion of this virus. The tree was estimated by using the Bayesian Markov Chain Monte Carlo method available in the BEAST package (67). The data were analyzed by assuming a relaxed (uncorrelated lognormal) molecular clock under the HKY85 model of nucleotide substitution with a different substitution rate for each codon position and a Bayesian skyline coalescent prior. The chain was run for 200 million generations (with a 10% burn-in). Branches are color-coded by place of origin. The bar at the root node represents the 95% highest probability density for the age of that node (x axis). In all cases tip times reflect the time of sampling. Although there is a heavy sampling bias toward American strains, there have clearly been multiple exportation events to localities such as Asia and Europe. Data kindly provided by Andrew Rambaut (University of Edinburgh, Edinburgh, Scotland, UK).
Finally, molecular clock estimates of the time of origin of H1N1pdm date it to a period spanning the end of 2008 through the first 2 months of 2009 (15), even allowing for a change of rate as the virus spreads in a new host (3). Although these estimates clearly depend on the sample of viruses used in the analysis, such that inclusion of earlier isolates from Mexico (or elsewhere) may push back times of ancestry to some extent, they are compatible with a period of “cryptic” viral transmission during which A/H1N1pdm went unnoticed by health authorities for several months. Interestingly, the identification of periods of cryptic viral transmission appears to be a common observation when using molecular clocks to date the onset of viral epidemics. Noteworthy examples include the genotype C rhinoviruses, first described in 2007 although molecular clock estimates place their ancestry at >250 years ago (65), and HIV in the Americas, where the virus was not identified until the early 1980s even though molecular clocks place its time of emergence in the Americas to the late 1960s or early 1970s, long before the earliest AIDS cases (66). As such, molecular evolutionary analyses offer a way to explore the early stages of emergence characterized by hidden viral transmission.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Evolution in Health and Medicine” held April 2–3, 2009, at the National Academy of Sciences in Washington, DC. The complete program and audio files of most presentations are available on the NAS web site at www.nasonline.org/Sackler_Evolution_Health_Medicine.
The author declares no conflict of interest.
This article is a PNAS Direct Submission. S.C.S. is a guest editor invited by the Editorial Board.
References
- 1.Taubenberger JK, Reid AH, Frafft AE, Bijwaard KE, Fanning TG. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 2.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith GJ, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 4.Dugan VG, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obenauer JC, et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 6.Olsen B, et al. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 7.Shinde V, et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 8.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 9.Lipkin WI. Microbe hunting in the 21st century. Proc Natl Acad Sci USA. 2009;106:6–7. doi: 10.1073/pnas.0811420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woolhouse MEJ. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 2002;10:S3–S7. doi: 10.1016/s0966-842x(02)02428-9. [DOI] [PubMed] [Google Scholar]
- 11.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: Patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 12.Elena SF, Sanjuán R. Adaptive value of high mutation rates of RNA viruses: Separating causes from consequences. J Virol. 2005;79:11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes EC. The Evolution and Emergence of RNA Viruses. In: Harvey PH, May RM, editors. Oxford Series in Ecology and Evolution. Oxford, UK: Oxford Univ Press; 2009. pp. 6–8. [Google Scholar]
- 14.Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J Mol Evol. 2002;54:152–161. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- 15.Rambaut A, Holmes EC. The early molecular epidemiology of the swine-origin A/H1N1 human influenza pandemic. PLoS Currents Influenza Aug. 2009;18:RRN1003. doi: 10.1371/currents.RRN1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rambaut A, et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eigen M. Steps Toward Life. New York: Oxford Univ Press; 1992. [Google Scholar]
- 18.Duffy S, Holmes EC. Validation of high rates of nucleotide substitution in geminiviruses: Phylogenetic evidence from East African cassava mosaic viruses. J Gen Virol. 2009;90:1539–1547. doi: 10.1099/vir.0.009266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gago S, Elena SF, Flores R, Sanjuán R. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323:1308. doi: 10.1126/science.1169202. [DOI] [PubMed] [Google Scholar]
- 20.Wiehe T. Model dependency of error thresholds: The role of fitness functions and contrasts between the finite and infinite sites models. Genet Res. 1997;69:127–136. [Google Scholar]
- 21.Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleo-sides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 22.Bull JJ, Sanjuán R, Wilke CO. Theory of lethal mutagenesis for viruses. J Virol. 2007;81:2930–2939. doi: 10.1128/JVI.01624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pariente N, Sierra S, Lowenstein PR, Domingo E. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J Virol. 2001;75:9723–9730. doi: 10.1128/JVI.75.20.9723-9730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin V, Grande-Perez A, Domingo E. No evidence of selection for mutational robustness during lethal mutagenesis of lymphocytic choriomeningitis virus. Virology. 2008;378:185–192. doi: 10.1016/j.virol.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2005;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new muta-tions. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 27.Sanjuán R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci USA. 2004;101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krakauer DC, Komarova NL. Levels of selection in positive-strand virus dynamics. J Evol Biol. 2003;16:64–73. doi: 10.1046/j.1420-9101.2003.00481.x. [DOI] [PubMed] [Google Scholar]
- 29.Duffy S, Turner PE, Burch CL. Pleiotropic costs of niche expansion in the RNA bacteriophage φ6. Genetics. 2006;172:751–757. doi: 10.1534/genetics.105.051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen R, Weinrich DM. The age of nonsynonymous and synonymous mutations in animal mtDNA and implications for the mildly deleterious theory. Genetics. 1999;153:497–506. doi: 10.1093/genetics/153.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pybus OG, et al. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 32.Ramsden C, et al. High rates of molecular evolution in hantaviruses. Mol Biol Evol. 2008;25:1488–1492. doi: 10.1093/molbev/msn093. [DOI] [PubMed] [Google Scholar]
- 33.Holmes EC. Patterns of intra- and inter-host nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parrish CR, et al. Cross-species viral transmission and the emergence of new epidemic diseases. Micro Mol Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woelk CH, Holmes EC. Reduced positive selection in vector-borne RNA viruses. Mol Biol Evol. 2002;19:2333–2336. doi: 10.1093/oxfordjournals.molbev.a004059. [DOI] [PubMed] [Google Scholar]
- 37.Coffey LL, et al. Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci USA. 2008;105:6970–6975. doi: 10.1073/pnas.0712130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greene IP, et al. Effect of alternating passage on adaptation of sindbis virus to vertebrate and invertebrate cells. J Virol. 2005;79:14253–14260. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jerzak GV, Bernard K, Kramer LD, Shi PY, Ebel GD. The West Nile virus mutant spectrum is host-dependent and a determinant of mortality in mice. Virology. 2007;360:469–476. doi: 10.1016/j.virol.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasilakis N, et al. Mosquitoes put the brake on arbovirus evolution: Experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog. 2009;5:e1000467. doi: 10.1371/journal.ppat.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies TJ, Pedersen AB. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc Biol Sci. 2008;275:1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasilakis N, et al. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology. 2007;358:402–412. doi: 10.1016/j.virol.2006.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anishchenko M, et al. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc Natl Acad Sci USA. 2006;103:4994–4999. doi: 10.1073/pnas.0509961103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finkelstein DB, et al. Persistent host markers in pandemic and H5N1 influenza viruses. J Virol. 2007;81:10292–10299. doi: 10.1128/JVI.00921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghedin E, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 46.Ghedin E, et al. Mixed infection and the genesis of influenza diversity. J Virol. 2009;83:8832–8841. [Google Scholar]
- 47.Bean WJ, Jr, Cox NJ, Kendal AP. Recombination of human influenza A viruses in nature. Nature. 1980;284:638–640. doi: 10.1038/284638a0. [DOI] [PubMed] [Google Scholar]
- 48.Holmes EC, et al. Whole genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson MI, et al. Stochastic processes are key determinants of the short-term evolution of influenza A virus. PLoS Pathog. 2006;2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 2007;3:e131. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson MI, et al. Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 2008;4:e1000133. doi: 10.1371/journal.ppat.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. J Am Med Assoc. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 53.Simonsen L, et al. The rapid global spread of reassortant human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol. 2007;24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 54.Nelson MI, Simonsen L, Miller MA, Viboud C, Holmes EC. The origin and global emergence of adamantane-resistant A/H3N2 influenza viruses. Virology. 2009;388:270–278. doi: 10.1016/j.virol.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rameix-Welti MA, Enouf V, Cuvelier F, Jeannin P, van der Werf S. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 2008;4:e1000103. doi: 10.1371/journal.ppat.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinstock DM, Zuccotti G. The evolution of influenza resistance and treatment. J Am Med Assoc. 2009;301:1066–1069. doi: 10.1001/jama.2009.324. [DOI] [PubMed] [Google Scholar]
- 57.Chen R, Holmes EC. The evolutionary dynamics of human influenza B virus. J Mol Evol. 2008;66:655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraser C, et al. Pandemic potential of a strain of influenza A (H1N1): Early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 60.Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 61.Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylo-dynamics of interpandemic influenza A (H3N2) in humans. Science. 2006;314:1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- 62.Recker M, Pybus OG, Nee S, Gupta S. The generation of influenza outbreaks by a network of host immune responses against a limited set of antigenic types. Proc Natl Acad Sci USA. 2007;104:7711–7716. doi: 10.1073/pnas.0702154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Russell CA, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 64.WHO . Pandemic (H1N1) 2009: Update 65. Geneva: WHO; 2009. [Google Scholar]
- 65.Briese T, et al. Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 2008;14:944–947. doi: 10.3201/eid1406.080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilbert MT, et al. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci USA. 2007;104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drummond AJ, Rambaut A BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]