Abstract
Our aims were to demonstrate that natural selection is operating on contemporary humans, predict future evolutionary change for specific traits with medical significance, and show that for some traits we can make short-term predictions about our future evolution. To do so, we measured the strength of selection, estimated genetic variation and covariation, and predicted the response to selection for women in the Framingham Heart Study, a project of the National Heart, Lung, and Blood Institute and Boston University that began in 1948. We found that natural selection is acting to cause slow, gradual evolutionary change. The descendants of these women are predicted to be on average slightly shorter and stouter, to have lower total cholesterol levels and systolic blood pressure, to have their first child earlier, and to reach menopause later than they would in the absence of evolution. Selection is tending to lengthen the reproductive period at both ends. To better understand and predict such changes, the design of planned large, long-term, multicohort studies should include input from evolutionary biologists.
Keywords: evolutionary rates, heritability, Homo sapiens, medical traits
Are contemporary humans experiencing natural selection and evolving in response to it? The answer to that question depends on whom one asks. A long tradition in the medical community (1) holds that natural selection does not operate on contemporary human populations because medicine keeps “alive many who otherwise would have perished” (2). No evolutionary biologist would now agree with that claim, for natural selection works through differential reproductive success rather than simple differential survival, and individuals in contemporary human populations vary in lifetime reproductive success (LRS). Selection operates on any trait that varies and is correlated with LRS, and traits respond to selection with change across generations if they vary genetically. But what traits is selection operating on? Do they include the traits treated by physicians? Previous work (e.g., ref. 3) has shown that human life history traits, most significantly age at first reproduction, are currently under selection, but evidence for selection operating on traits of medical importance is scarce. Here, we report estimates of natural selection, and the potential genetic response to selection, in the women of the first two generations of the Framingham Heart Study (FHS) population. The traits we analyzed include traits of medical significance: total cholesterol (TC), systolic blood pressure (SBP), diastolic blood pressure (DBP), and blood glucose (GLU). We had three general aims: first, to correct the still widespread misconception that natural selection is not operating on contemporary humans; second, to make quantitative predictions about future evolutionary change for specific traits with medical significance; and third, to register firmly a point of general cultural interest that follows directly from our first two aims: We are still evolving, and for some traits we can make short-term predictions about our future evolution.
The Framingham Heart Study
The FHS was established in 1948 in Framingham, MA, by the National Heart, Lung, and Blood Institute and Boston University to identify factors that contribute to cardiovascular disease. It is the longest running multigenerational study in medical history. The people originally enrolled in the study were of predominantly European ancestry (20% United Kingdom, 40% Ireland, 10% Italy, 10% Quebec). The original cohort (n = 5,209) has been examined every 2 years, a total of 29 times between 1948 and 2008. The offspring cohort (n = 5,124) has been examined approximately every 4 years, a total of eight times between 1971 and 2008 (4). There is also a third generation cohort (n = 4,095) that is not included in this study because many in it have not yet completed reproduction. At each examination many physical and blood chemistry traits are measured and a questionnaire is administered, yielding data on >70 traits. Data are deidentified by the FHS and delivered to the National Institutes of Health dbGaP database, from which we downloaded them for analysis. In this study, we use only the data on individuals who were measured three or more times.
Measuring Selection in a Multicohort Medical Study
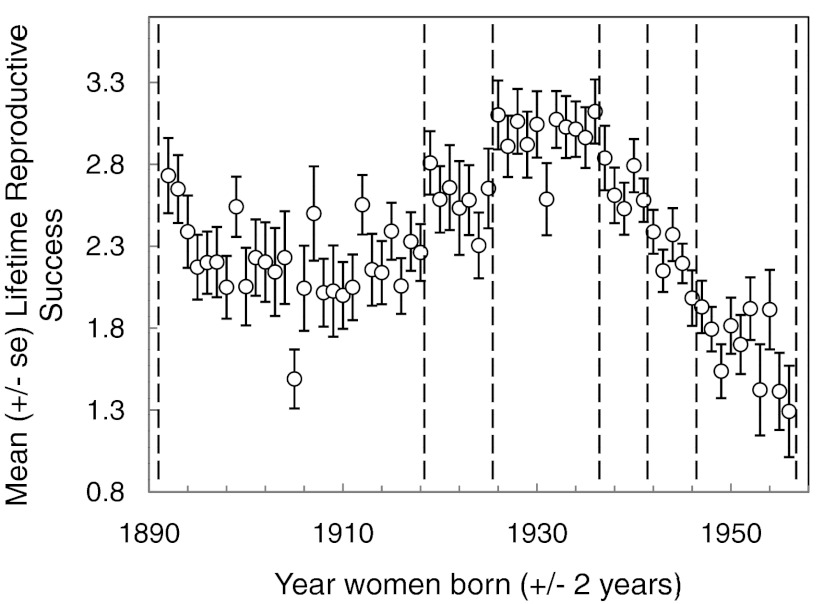
Natural selection has been measured many times in natural populations of animals and plants (5) using methods inspired by Robertson (6), developed by Lande and Arnold (7), and refined by Janzen and Stern (8), Hereford et al. (9), and others. To apply those methods to contemporary human populations requires consideration of several special features of data on humans. Some, such as cultural variation related to education, smoking, and medication, we dealt with as covariates. Others could in principle be measured on natural populations of animals and plants but in practice often are not; these include repeated measures on individuals that establish the developmental trajectories of multiple traits with age and long-term observations of populations across several generations that reflect secular demographic trends. Both make the measurement of traits more complex: at what age and in what portion of a secular trend—a change in conditions across time rather than age—should the expression of the trait be measured? The solution we chose was to calculate the response surface of each trait for age and time and to express the measurement of that trait for each individual as an average deviation from that surface (e.g., Fig. 1). Thus, for several traits we asked whether through their adult years individuals tended to have higher or lower values than other individuals of the same age measured in the same year. Because many individuals have been measured repeatedly in the FHS, the response surface can be estimated accurately. And how should one express relative LRS in a population that goes through a baby boom and a baby bust? We asked whether an individual had relatively high LRS compared with others bearing children during the same time period. To do so we divided the population into six periods of differing average LRS by year of birth and expressed the fitness of women relative to the average for their year of birth (Fig. 2).
Fig. 1.
TC between the ages of 20 and 60, 1955–2003. Because the measurement of TC changes with the age of the individual and the year of measurement, we fitted a surface to all measurements, calculated the residuals of all of the measurements for each individual from this surface, and expressed the trait measurement as a single value: the average of the residuals. We only used individuals who had been measured at least three times (n = 2,227 women measured 16,516 times for TC, with one individual’s measures from each cohort added for reference).
Fig. 2.
LRS, by year of birth, for women in the FHS. To deal with secular demographic change, we divided the data into six periods and divided the relative reproductive success of each woman by the mean reproductive success of the women in her group.
Measuring Evolutionary Change
To study evolutionary change, one conceives of the process as occurring in two steps. First, selection acts through differences in LRS. With multitrait phenotypes there are both direct effects on the individual traits and indirect effects mediated by the phenotypic correlations among the traits. Thus, a trait may be favored by selection only because it is correlated with other traits that are directly associated with greater LRS. The direct effects of selection are captured by the linear selection gradients (β) estimated as partial regression coefficients (Table 1); the indirect effects are mediated by the phenotypic variance/covariance matrix P (Table 2).
Table 1.
Mean values and direct and total selection gradients
| TC, mg/100 mL | WT, kg | HT, cm | SBP, mmHg | DBP, mmHg | GLU, mg/100 mL | Age at menopause | Age at first birth | |
| Means | 2.350 | 1.811 | 2.205 | 2.106 | 1.895 | 1.950 | 1.689 | 1.418 |
| β | −0.743 | 0.861 | −3.999 | −0.963 | 0.982 | −0.848 | 1.280 | −1.267 |
| P × β | −2.841 | 2.581 | −0.533 | −0.323 | 0.526 | −2.554 | 1.981 | −6.940 |
Means, − log10. β, linear selection gradient from partial regressions. P × β, direct and indirect selection from P matrix, × 1,000.
Table 2.
Phenotypic variance/covariance matrix
| TC, mg/100 mL | WT, kg | HT, cm | SBP, mmHg | DBP, mmHg | GLU, mg/100 mL | MEN | BIR | |
| TC | 3.860 | 0.022 | −0.089 | 0.336 | 0.278 | 0.300 | −0.180 | 0.112 |
| WT | 0.022 | 5.710 | 0.426 | 1.040 | 1.070 | 1.030 | 0.045 | 0.040 |
| HT | −0.089 | 0.426 | 0.266 | −0.039 | 0.009 | −0.023 | 0.022 | 0.109 |
| SBP | 0.336 | 1.040 | −0.039 | 2.590 | 1.800 | 0.688 | −0.005 | −0.222 |
| DBP | 0.278 | 1.070 | 0.009 | 1.800 | 1.920 | 0.399 | −0.020 | −0.183 |
| GLU | 0.300 | 1.030 | −0.023 | 0.688 | 0.399 | 3.570 | −0.032 | 0.021 |
| MEN | −0.180 | 0.045 | 0.022 | −0.005 | −0.020 | −0.032 | 1.460 | 0.094 |
| BIR | 0.112 | 0.040 | 0.109 | −0.222 | −0.183 | 0.021 | 0.094 | 5.378 |
P matrix, log10, all values × 1,000. MEN, age at menopause; BIR, age at first birth.
Second, the multitrait response to selection also depends on direct and indirect effects of inheritance. Selection differentials will have no long-term consequences if the traits are not passed to the next generation. The direct effects on the response are determined by the additive genetic variances of the individual traits; these are the diagonal elements in the genetic variance–covariance matrix G (Table 3). The indirect effects are mediated by the off-diagonal elements of that matrix, the genetic covariances. Inheritance can either be genetic or cultural. We are not able to fully differentiate the effects of genes and culture with these data.
Table 3.
Additive genetic variance/covariance matrix
| TC, mg/100 mL | WT, kg | HT, cm | SBP, mmHg | DBP, mmHg | GLU, mg/100 mL | MEN | BIR | |
| TC | 2.371 ± 0.097 | −0.126 ± 0.101 | −0.070 ± 0.022 | 0.180 ± 0.068 | 0.108 ± 0.059 | 0.071 ± 0.080 | 0.040 ± 0.081 | −0.095 ± 0.089 |
| WT | −0.126 ± 0.101 | 3.014 ± 0.143 | 0.376 ± 0.021 | 0.250 ± 0.079 | 0.408 ± 0.065 | 0.329 ± 0.090 | 0.030 ± 0.100 | 0.002 ± 0.103 |
| HT | −0.070 ± 0.022 | 0.376 ± 0.021 | 0.225 ± 0.005 | −0.020 ± 0.017 | 0.022 ± 0.015 | −0.035 ± 0.020 | 0.027 ± 0.029 | 0.074 ± 0.024 |
| SBP | 0.180 ± 0.068 | 0.250 ± 0.079 | −0.020 ± 0.017 | 1.386 ± 0.066 | 0.723 ± 0.018 | 0.454 ± 0.060 | −0.090 ± 0.064 | −0.127 ± 0.072 |
| DBP | 0.108 ± 0.059 | 0.408 ± 0.065 | 0.022 ± 0.015 | 0.723 ± 0.018 | 0.950 ± 0.050 | 0.226 ± 0.055 | −0.088 ± 0.056 | −0.173 ± 0.065 |
| GLU | 0.071 ± 0.080 | 0.329 ± 0.090 | −0.035 ± 0.020 | 0.454 ± 0.060 | 0.226 ± 0.055 | 1.235 ± 0.098 | 0.040 ± 0.053 | −0.121 ± 0.080 |
| MEN | 0.040 ± 0.081 | 0.030 ± 0.100 | 0.027 ± 0.029 | −0.090 ± 0.064 | −0.088 ± 0.056 | −0.052 ± 0.053 | 0.695 ± 0.084 | −0.054 ± 0.043 |
| BIR | −0.095 ± 0.089 | 0.002 ± 0.103 | 0.074 ± 0.024 | −0.127 ± 0.072 | −0.173 ± 0.065 | −0.121 ± 0.080 | −0.054 ± 0.043 | 0.537 ± 0.137 |
G matrix, log10, values × 1,000, ± 1. MEN, age at menopause. BIR, age at first birth.
Selection Gradients
Selection gradients measure the extent to which individuals with a given value of a quantitative trait tend to have higher or lower fitness (LRS). If individuals with low levels tend to have higher LRS, then the next generation is apt to have a lower average level of that trait (Fig. 3). We measured significant linear selection gradients acting to reduce total cholesterol (TC), height (HT), SBP, and age at first birth and to increase weight (WT) and age at menopause. These results strongly suggest that natural selection is acting on the women of the FHS through differences in the number of children they had during their reproductive ages (Table 4).
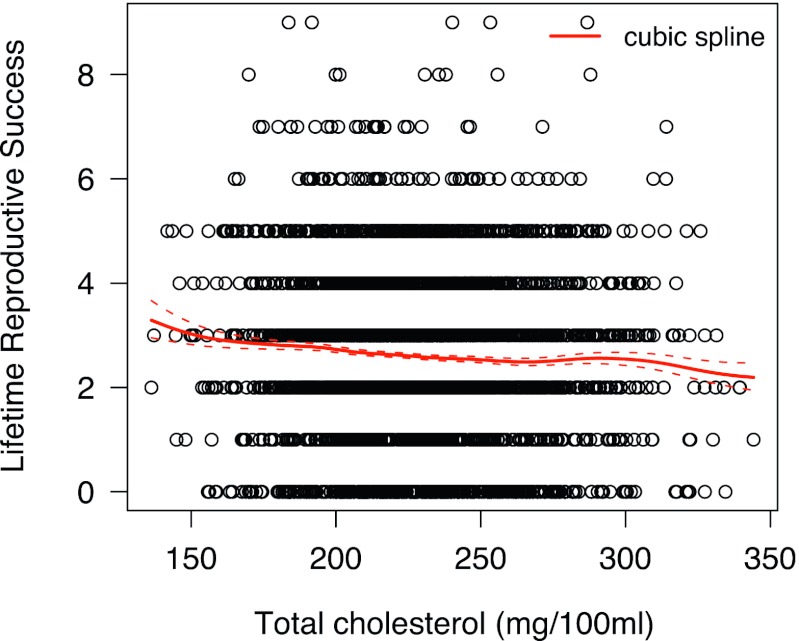
Fig. 3.
Plot of average cholesterol values against LRS. Residuals from the surface in Fig. 1 were converted back to the original metric by adding the global mean to each value and then averaged to yield a single average residual estimate for each woman. A cubic spline [using glms40 (25)] was then fitted to these residuals, which have the effects of age and year of measure removed, to give a visual impression of the variation in the data and the strength of selection. The cubic spline is not the linear selection gradient. Dashed lines are ± 1 SE of fitness associations (solid line).
Table 4.
Selection gradients acting on women in the Framingham population (combined dataset, women born between 1892 and 1956)
| Trait | N | μ | SD | β | β μ | P | % linear | % Quad | |
| TC, mg/100 mL | 2,227 | 223.2 | 48.2 | −0.743 | −1.447 | 0.0011 | 0.295 | ||
| HT, cm | 2,227 | 160.7 | 6.4 | −3.999 | −6.672 | 0.0002 | 0.689 | ||
| WT, kg | 2,227 | 65.7 | 13.3 | 0.861 | 0.985 | <0.0001 | 0.197 | 0.394 | |
| SBP, mmHg | 2,227 | 127.2 | 22.1 | −0.963 | −1.545 | 0.0236 | 0.019 | ||
| DBP, mmHg | 2,227 | 79.1 | 11.9 | 0.982 | 1.317 | 0.0724 | - | ||
| GLU, mg/100 mL | 2,227 | 89.6 | 22.8 | −0.848 | −0.548 | 0.0596 | - | 0.492 | |
| Diabetes | 2,227 | - | - | 0.076 | 0.008 | 0.3473 | - | ||
| Age at first birth | 1,448 | 26.5 | 4.6 | −1.267 | −1.758 | <0.0001 | * | ||
| Age at menopause | 2227 | 49.2 | 4.1 | 1.280 | 2.839 | 0.0035 | 0.689 | 0.098 |
Associations with age at first birth were estimated only for women who reported at least one live birth. N, sample size of individual women; μ, mean; β, unstandardized directional selection gradient from a multiple linear regression model that included interaction and quadratic terms; β μ, mean standardized selection gradient; P, significance estimates from the Poisson model; %, percentage of variation in lifetime reproductive success explained. Presenting the mean and SD for diabetes would be inappropriate because it was recorded as presence/absence.*, The percentage of variation in lifetime reproductive success explained by age at first birth is not strictly comparable to that explained by the other traits because the sample was smaller; in that separate analysis it explained ≈5%. One covariate, level of education, was significant; it explained 0.295% of the variation in lifetime reproductive success. Only one of the 45 two-way interaction terms was significant, estrogen therapy × diastolic blood pressure; it explained 0.197% of the variation in lifetime reproductive success.
In estimating the selection gradients for most of the traits, we controlled for level of education, foreign or native birth, and whether the individual smoked. We also estimated quadratic terms to detect stabilizing selection (Table S1). These terms indicate whether a continuation of past trends will lead a trait to change at a steady or increasing rate or whether it will tend to level off. A total of 5.74% of the variation in LRS was explained by all factors combined (linear, quadratic, and interaction terms). The impact of stabilizing selection is reflected in our projections of evolutionary change (Table 5).
Table 5.
Projected evolutionary change, untransformed values
| TC, mg/100 mL | WT, kg | HT, cm | SBP,mmHg | DBP, mmHg | GLU, mg/100 mL | Age at menopause | Age at first birth | |
| Generation | ||||||||
| 0 | 223.9 | 64.7 | 160.2 | 127.6 | 78.5 | 89.1 | 48.9 | 26.18 |
| 5 | 219.8 ± 1.58 | 65.4 ± 0.58 | 159.3 ± 0.41 | 126.3 ± 0.71 | 78.6 ± 0.34 | 88.4 ± 0.52 | 49.3 ± 0.23 | 25.96 ± 0.23 |
| 10 | 215.9 ± 3.35 | 65.6 ± 1.33 | 158.1 ± 0.90 | 125.2 ± 1.47 | 78.7 ± 0.73 | 88.1 ± 1.03 | 49.7 ± 0.51 | 25.74 ± 0.48 |
| % | 3.6 | 1.4 | 1.3 | 1.9 | 0.3 | 1.1 | 1.6 | 1.7 |
| Haldanes | 0.016 | 0.007 | 0.032 | 0.010 | 0.002 | 0.004 | 0.020 | 0.010 |
The averages for generation 0 are those observed for the Framingham original and offspring cohorts between the ages of 20 and 60 over the years 1955–2003. Values for later cohorts are the means that would be expected given the average conditions over the period of observation and evolutionary change. %, percent change in traits from generation 0 to 10. Haldanes: rate of evolution in SD per generation.
Inheritance of Traits
We estimated the inheritance of traits by taking advantage of the longitudinal, multigeneration, family design for the FHS. It yielded pedigrees, some quite complex, for hundreds of families. We applied a maximum-likelihood method implemented in the software package SOLAR (24) to extract from these pedigrees estimates of additive genetic variance and covariance. The heritability of a trait, perhaps a more familiar term, is its additive genetic variance divided by its total phenotypic variance, and the genetic correlation between two traits is their additive covariance divided by the square root of the product of their additive variances. Thus, in essence we were measuring the heritabilities of and genetic correlations between all of the traits and expressing them in a form suitable for evolutionary projections. This method cannot completely discriminate between vertical cultural transmission and genetic transmission, but the use of all degrees of relationship, not just those between parents and offspring, does to some degree get around this problem. Our estimates of heritabilities were: HT, 0.84 ± 0.01 (SE); TC, 0.61 ± 0.02; SBP, 0.53 ± 0.02; WT, 0.52 ± 0.02; DBP, 0.49 ± 0.02; age at menopause, 0.47 ± 0.05; GLU, 0.34 ± 0.02; and age at first birth, 0.09 ± 0.02. These are similar to heritabilities found in other studies examining phenotypic (10, 11) and life history traits (12) in humans.
Projecting Evolutionary Change
Table 5 presents the projected changes on the assumption that conditions will continue to mirror the averages encountered by this population over the past 60 years. In the next 10 generations mean TC among women is projected to decline from the average of 224 over the past 60 years to 216 (209.3–222.5) mg/100 mL (95% C.I.; see Methods). Because the environment has changed over the past 60 years (Fig. 2) and will continue to change, these results suggest that whatever changes in environment occur evolutionary changes will lead to mean cholesterol levels among women that are ≈0.8 (0.14–1.46) mg/100 mL lower in the next generation than they would be in the absence of evolution.
Similarly, we expect that as a result of evolution, in the next generation mean body WT among women will increase by 0.2 (−0.20 to 0.62) kg then stabilize; HT will decrease a bit, ≈0.2 (0.03–0.39) cm; SBP will decrease by ≈0.25 (−0.05 to 0.53) mmHg; DBP will remain essentially unchanged; blood GLU will decrease slightly by 0.8 (−0.09 to 0.29) mg/100 mL; age at menopause will increase by ≈1.0 (−0.23 to 2.15) months; and age at first birth will decrease by ≈0.5 (−0.6 to 1.7) months. The rates of projected evolution in haldane units (SD per generation) range from 0.032 (HT) to 0.002 (DBP), slower than those estimated for Galapagos finches and Trinidadian guppies but comparable to those estimated for New Zealand chinook salmon and Hawaiian mosquitofish (13). In sum, as a result of evolution future generations of women in this population are predicted to be slightly shorter and stouter, to have lower values for TC and SBP, to have their first child slightly earlier, and to reach menopause slightly later than they would have otherwise. These are small, gradual evolutionary changes in the middle to lower range of those observed in contemporary populations of nonhuman species.
Secular Changes
To see whether selection intensities were changing over the period of the study, we broke the dataset down into three periods defined by year of birth: 1892–1913 (n = 716), 1914–1935 (n = 842), and 1936–1956 (n = 669). Most of the significance in the selection gradients we estimated in Tables 1–3 was contributed by variation among women in the first period. The only trait consistently under significant selection in all three periods was age at first birth (period 1: β = −1.120, P = 0.0037; period 2: β = −1.107, P = 0.0018; period 3: β = −1.488, P = 0.018).
Would Unrecorded Early Mortality Have Changed Any of Our Conclusions?
LRS should be measured from birth to the end of reproduction. However, we could only study individuals who survived to adulthood and had measurements on adult traits. During most of human history, high mortality at early ages was a major factor driving evolution, but now most children survive well into the reproductive ages.
To determine whether excluding early deaths from LRS could have biased our results, we simulated what would happen if a trait was associated with early death and, therefore, no reproduction. For example, we asked what would happen if higher cholesterol was associated with an increased risk of not surviving to age 20 and, therefore, remaining childless? What would the sample look like if we were to include those people who died? We might see a group of childless women whose adult (never actually measured) cholesterol levels were on average higher than other women. We simulated such effects by adding to the dataset a group of phantom women for two levels of preadult mortality, 0.011 (survival to age 20, p20 = 0.989, the average for women in the United States in 2002) and 0.06 (p20 = 0.94, the average for women in the United States in 1939–1940) and for each of 10 traits, by giving each phantom individual a value for the trait equal to the population mean plus 2 SD. For 90% of the 20 cases, the unstandardized β values remained in the same direction, positive or negative, and for 86% of the traits P values did not change significance. For example, selection on cholesterol was always negative and always significant (20 of 20 cases); selection on HT was always negative (20 of 20) and almost always significant (19 of 20); selection on WT was almost always positive (18 of 20) and usually significant (16 of 20); selection on age at menopause was almost always positive (19 of 20) and almost always significant (19 of 20); selection on SBP was usually positive (17 of 20) but only significant approximately half the time (11 of 20); selection on GLU was almost always negative (19 of 20) but rarely significant (2 of 20); patterns in the other traits were mixed. Apparently the proportion not surviving to age 20 is so low that if they were in the dataset our major conclusions would not be likely to change.
Conclusions
Natural selection is acting slowly and gradually on traits of medical importance and on life history traits in the FHS population. Selection varied in intensity, becoming generally less intense over time, but not in direction, and it has only operated consistently over the entire period to reduce age at first birth. Predictions for one generation are fairly reliable, but whether selection will be consistent and sustained enough to bring about significant genetic change can only be answered with longer periods of observation of more traits relevant to human health.
These results suggest slow evolutionary change. It is noteworthy, although not surprising, that both age at first birth and age at menopause appear to be changing so as to lengthen the reproductive period, which is consistent with previous findings (3). Because fertility is the driving force behind evolution in modern populations, we might have found larger effects of evolution on the levels of sex hormones and related traits had they been measured. The impact of fertility on selection could prove especially important now that many couples that would otherwise remain childless can produce offspring with medical assistance.
The traits we studied were those available from the FHS, which was focused on heart disease, not reproduction. To better understand and predict evolutionary changes, the design of planned large, long-term, multicohort studies should include input from evolutionary biologists and, in particular, should consider measuring traits that might be closely associated with reproductive success.
Methods
The FHS continues today. Study design and entry criteria for the FHS are detailed in Dawber et al. (14) and Kannel et al. (15). We included subjects from the original and offspring cohorts; they received physical examinations and questionnaires administered by trained interviewers every 2 and 3–4 years, respectively. The Institutional Review Board requirements have been adequately addressed for all of the participants in the FHS, and formal approval for the data used was obtained from the dbGaP (www.ncbi.nlm.nih.gov/sites/entrez?Db = gap).
Relative Fitness.
We calculated LRS for women who reported at a postmenopausal age how many live births they had had. Of the 5,372 women in the original and offspring cohorts, 739 were removed because of missing values for menopause (or menopause had not been reached) and 852 were removed because the number of live births was not recorded after a postmenopausal age. Sample size was further reduced to 3,224 after excluding an additional 557 women who reached menopause unnaturally (e.g., ovaries removed, hysterectomy, radiation, chemotherapy) before the age of 45 and to 2,238 after excluding those who had been measured <3 times for any of the continuous traits. Accuracy of LRS estimates was improved by using the FHS pedigree file (and other data) to correct for number of live births >5. Twenty-four women (1.0%) who had five or more live births were recorded as having had five as part of the deidentification of the data. We estimate that ≈7 of those 24 women actually had more than the five births (generally only one more) indicated by the dataset; we recorded all 24 as having had five births.
Main Phenotypes.
We used some traits known to be associated with cardiovascular disease. They included TC, WT, HT, SBP, DBP, and serum GLU. TC included both serum (earlier survey rounds) and plasma (later) measures; both were treated as one here. Blood pressure was measured twice in each examination; only the first physician’s measure was used here. Methods for the various cholesterol measures throughout the study have been described (16, 17).
For both cohorts, WT, HT, SBP, DBP, and GLU were measured from examination 1 (1948 and 1971 onward for original and offspring cohorts, respectively). Number of measures per trait varied depending on how many exams a woman attended throughout the study.
We found complex nonlinear changes in traits with age and year of measure that may reflect demographic and developmental changes throughout the study period and a woman’s life. We removed these effects by taking residuals from a 3D surface of a generalized additive model of each trait by age and year of measure, then converted these back into the original metric by adding the overall trait mean (Fig. 2). We used measures across the ages of 20–60, which span the reproductive years of modern human women, and only included women with three or more measures for each trait, because there is considerable intraindividual trait variation over time (18, 19). Residuals were then averaged to obtain one point per trait per woman and log-transformed to correct for deviations from normality.
Because traits may be affected by number of offspring (e.g., women might gain WT with number of births), thereby potentially confounding the covariation between traits and fitness, we compared traits measured at an age when women had not yet reproduced (according to age at first birth estimates from the FHS pedigree) or at the earliest age a trait was measured (up to the age of 30), to the same traits measured at an age when women in the sample had completed fertility (between the ages of 45 and 55). We found no significant difference in an ANOVA for any of the traits examined, suggesting few, if any, cumulative effects of number of offspring on the traits examined.
Covariates.
Demographic covariates included level of education and whether foreign or native born. We assumed that women from the offspring cohort were all born in the United States. Education was coded as number of years of education completed; the few women with missing values here were assumed to have a minimum of 8 years of education.
Other covariates included self-reported use of cholesterol-lowering medication (e.g., statins, fibrates, resins), self-reported use of medication for hypertension or high blood pressure, presence or absence of diabetes mellitus, self-reported smoking status, and estrogen use (hormone therapy or contraceptive use), which can affect cholesterol levels (20), other traits (21, 22), and women’s LRS. We only used information on medication use across the ages of 20–60. Smoking status was recorded as whether a woman was or had been a smoker. Of the remaining 2,238 women, only 11 (or 0.49%) were recorded as having taken cholesterol-lowering medication between the ages of 20 to 60; they were removed from the dataset, yielding a final sample size of 2,227. All covariates (except smoking intensity and education level) were coded as binary (present/absent) in the multiple regression analyses.
Projections.
We estimated the effect of directional selection on the mean value of each trait in the population in the next generation (the response to selection) with this equation:
Δ z is a vector of predicted changes in the population means of characters between the observed and next generation, G is the additive genetic variance- covariance matrix, and β is a vector of the selection gradients. Predicted changes in trait means should be interpreted cautiously for the direction and intensity of selection and the additive genetic variances and covariances all change over time and across generations.
To estimate the cumulative effect over several generations, we projected the mean change between generations using the equation Δ z = Gβ = GP−1 s, where P is the phenotypic variance-covariance matrix and s is the vector of changes in the traits that would occur in one generation as a result of selection alone. The change between generations in the variance–covariance matrix for traits was then estimated as P* − P = PγP − ssT, where γ is the matrix of interaction terms among the traits calculated from the coefficient estimates from a linear regression (7). We assumed that G, s, and γ remain fixed across generations.
Confidence Intervals for Projections.
The confidence intervals for the projection reflect the uncertainty in the β, γ, and G matrixes by using Monte Carlo simulations. Random number generators were used to run 5,000 projections with different estimates for each parameter. The β and γ were handled simultaneously by using a multinomial random number generator and the matrix of variances and covariances among the regression coefficients (including the intercept). The coefficients on the linear and quadratic terms were then used to set up β and γ. Each element of the G matrix was handled separately by using a random number generator and the standard errors produced by SOLAR (24). The projections were all done on the log of traits.
Statistical Analysis.
LRS from women measured for TC, WT, HT, SBP, DBP, GLU, age at menopause, age at first birth, age at death, and level of education were used to assess whether these traits influenced women’s fitness. Multiple linear regression was used to infer the strength and direction of selection, and multiple Poisson regression was used to test statistical significance.
We adjusted LRS to account for fluctuations in fertility over time (Fig. 2) relative to women in the same birth cohort (birth years 1892–1918, 1919–1925, 1926–1936, 1937–1941, 1942–1946, 1947–1956). Relative fitness was calculated by dividing each woman's number of births by the mean for her birth cohort. The Poisson models included binary markers for each cohort. We first used education level, smoking, and country of birth as controls to remove their potentially confounding effects on LRS. When we then assessed selection on cultural traits, we included education level and smoking as covariates. In all analyses we included quadratic terms for the main traits (TC, WT, HT, SBP, DBP, GLU, age at menopause, age at first birth) and two-way interactions between traits and covariates. Quadratic coefficients were doubled to estimate stabilizing/disruptive selection gradients (23). Coefficients from the multiple linear regressions were standardized by using trait means [see Hereford et al. (9)]. Partial regression coefficients, termed directional selection gradients (β), estimate the magnitude of directional selection on a trait i, with the effects of selection on all of the other measured traits removed.
G Matrix Estimates.
We used average residuals (described above) to control for changes in heritability over time and age. Additive genetic variance and covariance estimates were made with a pedigree-based maximum-likelihood method implemented in SOLAR (24). Significance was determined by likelihood ratio tests.
Acknowledgments
We thank the National Evolutionary Synthesis Center for support and the other members of our National Evolutionary Synthesis Center working group (Charles Goodnight, David Houle, Martin Larson, Trudy Mackay, Shamil Sunyaev, Anatoli Yashin, and Sebastian Zoellner) for helpful comments. D.E. was supported by National Institutes of Health (National Institute on Aging) Grant P30 AG12836, the Boettner Center for Pensions and Retirement Security at the University of Pennsylvania, and National Institutes of Health (National Institute of Child Health and Development Population Research Infrastructure Program) Grant R24 HD-044964.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Evolution in Health and Medicine” held April 2–3, 2009, at the National Academy of Sciences in Washington, DC. The complete program and audio files of most presentations are available on the NAS web site at www.nasonline.org/Sackler_Evolution_Health_Medicine.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906199106/DCSupplemental.
References
- 1.Tait L. Has the law of natural selection by survival of the fittest failed in the case of man? Dublin Q J Med Sci. 1869;47:102–113. [Google Scholar]
- 2.Bynum WF. Darwin and the doctors: Evolution, diathesis, and germs in 19th century. Gesnerus. 1983;40:43–53. [PubMed] [Google Scholar]
- 3.Kirk KM, et al. Natural selection and quantitative genetics of life-history traits in Western women: A twin study. Evolution (Lawrence, Kans) 2001;55:423–435. doi: 10.1111/j.0014-3820.2001.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 4.Govindaraju DR, et al. Genetics of the Framingham Heart Study population. Adv Genet. 2008;62:33–65. doi: 10.1016/S0065-2660(08)00602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kingsolver JG, et al. The strength of phenotypic selection in natural populations. Am Nat. 2001;157:245–261. doi: 10.1086/319193. [DOI] [PubMed] [Google Scholar]
- 6.Robertson A. A mathematical model of culling process in dairy cattle. Anim Prod. 1966;8:95–108. [Google Scholar]
- 7.Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution (Lawrence, Kans) 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 8.Janzen FJ, Stern HS. Logistic regression for empirical studies of multivariate selection. Evolution (Lawrence, Kans) 1998;52:1564–1571. doi: 10.1111/j.1558-5646.1998.tb02237.x. [DOI] [PubMed] [Google Scholar]
- 9.Hereford J, Hansen TF, Houle D. Comparing strengths of directional selection: How strong is strong? Evolution (Lawrence, Kans) 2004;58:2133–2143. doi: 10.1111/j.0014-3820.2004.tb01592.x. [DOI] [PubMed] [Google Scholar]
- 10.Brown WM, et al. Age-stratified heritability estimation in the Framingham Heart Study families. BMC Genet. 2002;4(Suppl 1):S32. doi: 10.1186/1471-2156-4-S1-S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathias RA, et al. Comparison of year-of-exam and age-matched estimates of heritability in the Framingham Heart Study data. BMC Genet. 2002;4(Suppl 1):S36. doi: 10.1186/1471-2156-4-S1-S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler HP, Rodgers JL, Christensen K. Is fertility behavior in our genes? Findings from a Danish twin study. Popul Dev Rev. 1999;25:253–288. [Google Scholar]
- 13.Hendry AP, Kinnison MT. Perspective–The pace of modern life: Measuring rates of contemporary microevolution. Evolution (Lawrence, Kans) 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. [DOI] [PubMed] [Google Scholar]
- 14.Dawber TR, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: Framingham Study. Ann NY Acad Sci. 1963;107:539–556. doi: 10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 15.Kannel WB, et al. Investigation of coronary heart disease in families: Framing-ham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 16.Abell LL, Levy BB, Brodie BB, Kendall FE. A simplified method for the estimation of total cholesterol in serum and demonstration of its specificity. J Biol Chem. 1952;195:357–366. [PubMed] [Google Scholar]
- 17.Warnick GR, Benderson J, Albers JJ. Dextran sulfate Mg2+ precipitation procedure for quantitation of high-density-lipoprotein cholesterol. Clin Chem. 1982;28:1379–1388. [PubMed] [Google Scholar]
- 18.Frishman WH, et al. Serum lipids and lipoproteins in advanced age: Intraindividual changes. Ann Epidemiol. 1992;2:43–50. doi: 10.1016/1047-2797(92)90036-p. [DOI] [PubMed] [Google Scholar]
- 19.Bookstein L, Gidding SS, Donovan M, Smith FA. Day-to-day variability of serum cholesterol, triglyceride, and high-density-lipoprotein cholesterol levels: Impact in the assessment of risk according to the National Cholesterol Education Program Guidelines. Arch Intern Med. 1990;150:1653–1657. [PubMed] [Google Scholar]
- 20.Demissie S, et al. Estrogen receptor-α variants are associated with lipoprotein size distribution and particle levels in women: The Framingham Heart Study. Atherosclerosis. 2006;185:210–218. doi: 10.1016/j.atherosclerosis.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Fox CS, et al. Sex-specific association between estrogen receptor-α gene variation and measures of adiposity: The Framingham Heart Study. J Clin Endocrinol Metab. 2005;90:6257–6262. doi: 10.1210/jc.2005-0670. [DOI] [PubMed] [Google Scholar]
- 22.Peter I, et al. Variation in estrogen-related genes and cross-sectional and longitudinal blood pressure in the Framingham Heart Study. J Hypertens. 2005;23:2193–2200. doi: 10.1097/01.hjh.0000188728.66183.92. [DOI] [PubMed] [Google Scholar]
- 23.Stinchcombe JR, et al. Estimating nonlinear selection gradients using quadratic regression coefficients: Double or nothing? Evolution (Lawrence, Kans) 2008;62:2435–2440. doi: 10.1111/j.1558-5646.2008.00449.x. [DOI] [PubMed] [Google Scholar]
- 24.Blangero J, Almasy L, Dyer T, Peterson C. Sequential oligogenic linkage analysis routines: SOLAR. Westerville, OH: Solar Software; 1999. Version 4.2.0. Available at http://solar.sfbrgenetics.org/ [Google Scholar]
- 25.Schluter D, Nychka D. Exploring fitness surfaces. Am Nat. 1994;143:597–616. [Google Scholar]