Abstract
Neo-Darwinian evolutionary theory is based on exquisite selection of phenotypes caused by small genetic variations, which is the basis of quantitative trait contribution to phenotype and disease. Epigenetics is the study of nonsequence-based changes, such as DNA methylation, heritable during cell division. Previous attempts to incorporate epigenetics into evolutionary thinking have focused on Lamarckian inheritance, that is, environmentally directed epigenetic changes. Here, we propose a new non-Lamarckian theory for a role of epigenetics in evolution. We suggest that genetic variants that do not change the mean phenotype could change the variability of phenotype; and this could be mediated epigenetically. This inherited stochastic variation model would provide a mechanism to explain an epigenetic role of developmental biology in selectable phenotypic variation, as well as the largely unexplained heritable genetic variation underlying common complex disease. We provide two experimental results as proof of principle. The first result is direct evidence for stochastic epigenetic variation, identifying highly variably DNA-methylated regions in mouse and human liver and mouse brain, associated with development and morphogenesis. The second is a heritable genetic mechanism for variable methylation, namely the loss or gain of CpG dinucleotides over evolutionary time. Finally, we model genetically inherited stochastic variation in evolution, showing that it provides a powerful mechanism for evolutionary adaptation in changing environments that can be mediated epigenetically. These data suggest that genetically inherited propensity to phenotypic variability, even with no change in the mean phenotype, substantially increases fitness while increasing the disease susceptibility of a population with a changing environment.
Keywords: DNA methylation, epigenetics, evolution, stochastic variation
A key tenet of Origin of Species argues that phenotype is the result of many discrete traits that are individually and exquisitely selected, to quote Darwin, “detecting the smallest grain in the balance of fitness,” which has been described as Newtonian in its dependence on static forces acting in consistent ways (1). This concept is the basis for quantitative trait loci proposed by R. A. Fisher (2). This concept has led to the modern basis of population genetics that continuous variation exists within a population, yet selection is on individuals, which has led to models of balancing or purifying selection at the extremes of phenotype (1). The classic model also has significant limitations in explaining common human disease; common variants can explain only a small fraction of a given disease phenotype, even the most well understood, such as adult-onset diabetes and height (3).
Epigenetics, the study of nonsequence-based changes in DNA and associated proteins, was first suggested by Jablonka to play a role in evolution through Lamarckian inheritance, that is, direct modification of the genome by the environment, which is then transmitted transgenerationally (4). Two examples are commonly cited: changes in coat color caused by dietary modifications of DNA methylation of the agouti gene in mice (5, 6) and methylation of the axin-fused allele in kinked tail mice (6, 7). Both of these examples involve methylation of a retrotransposon LTR sequence, and thus fit into various genetic exceptions to classical Darwinian thinking, including anticipation due to trinucleotide repeat expansion and lateral gene transfer in the evolution of influenza strains (8). But they have not been shown to be general mechanisms for either speciation or developmental differences across species, so-called “evo-devo,” or for canalization, a term coined by Waddington to refer to a mechanism by which environmental perturbations during development are corrected by the genetic program, leading to a consistent developmental plan (9). Indeed, canalization remains a “black box,” as noted by West-Eberhard (8). Others have discussed the potential role for Lamarckian inheritance in disease; for example, Slatkin proposed a model of transgenerational epigenetic Lamarckian inheritance and noted that such modifications must persist for many generations to contribute substantially to average risk (10), which has implications for public health management (11). Although not disputing an important contribution of Lamarkcian inheritance, here we propose an alternative view in which genetic modification could provide stochastic phenotypic variation favored by selection in changing environments, and also provide an alternative non-Lamarckian role for epigenetics in evolution.
A New Advance Over Darwinism: Stochastic Variation, Not Lamarckian Inheritance
It has occurred to us that increased variability with a given genotype might itself increase fitness. This could arise by genetic variants that do not change the mean phenotype but do change the variability of phenotype. A natural mechanism to use to consider such a model is epigenetic plasticity during development, for example, varying DNA methylation patterns. This idea differs from Lamarckian inheritance, in that in our model the genetic change is inherited, and this change leads to increased epigenetic variation. It also differs from the likely role of epigenetics in modifying mutation rate, both through C to T transition due to deamination of methylcytosine and through modified rates of chromosomal rearrangement (12, 13). As a proof of principle, we revisited previously generated data sets (14) of genome-scale analysis of DNA methylation in human and mouse tissues and explored them in two new ways. First, we investigated whether there were regions of variable methylation across individuals for a given tissue type. Then we explored whether tissue-specific differentially methylated regions (T-DMRs) differed across species and whether the underlying DNA sequence could account for these differences.
Variably Methylated Regions Across Individuals
To assess the degree of intrinsic variability in DNA methylation of a given tissue, we set out to identify the location of the most highly variable regions of DNA methylation in mouse liver from four individuals. We chose this specific tissue because it is relatively homogeneous. We examined newborns in whom polyploidy is minimal, although copy number would not be expected to affect DNA methylation, because our method controls for copy number (15). Environmental effects were minimized by examining inbred mice (indeed, littermates from the same cage). Surprisingly, many loci throughout the genome showed striking variations in DNA methylation, which we term variably methylated regions (VMRs). Surprisingly, these VMRs were significantly enriched in the vicinity of genes with Gene Ontogeny (GO) functional categories for development and morphogenesis (Table 1) when using either all genes for comparison or all regions present on the CHARM array, indicating that enrichment is not explained solely by high CpG content, because the array itself is designed to assay high-CpG regions. Examples of developmental genes with VMRs—Bmp7, involved in early embryogenic programming and bone induction, Pou3f2, involved in neurogenesis and stem cell reprogramming, and Ntrk3, involved in body position sensing—are shown in Fig. 1.
Table 1.
Enrichment scores of GO categories of genes in the vicinity of VMRs in mouse liver
| GOBPID | P value | Odds ratio | Expected count | Count | Size | Term |
| GO:0048699 | 2.8E-05 | 2.0 | 26.9 | 49 | 384 | Generation of neurons |
| GO:0009880 | 8.5E-05 | 4.9 | 2.8 | 11 | 41 | Embryonic pattern specification |
| GO:0030030 | 0.00033 | 2.0 | 19.1 | 35 | 272 | Cell projection organization |
| GO:0021517 | 0.00034 | 8.8 | 1.0 | 6 | 15 | Ventral spinal cord development |
| GO:0035107 | 0.00041 | 2.9 | 6.2 | 16 | 89 | Appendage morphogenesis |
| GO:0048666 | 0.00046 | 2.0 | 17.2 | 32 | 245 | Neuron development |
| GO:0032990 | 0.00050 | 2.2 | 12.3 | 25 | 175 | Cell part morphogenesis |
| GO:0009887 | 0.00052 | 1.6 | 35.9 | 56 | 512 | Organ morphogenesis |
| GO:0021515 | 0.00055 | 6.2 | 1.5 | 7 | 22 | Cell differentiation in spinal cord |
| GO:0048812 | 0.00065 | 2.2 | 11.8 | 24 | 168 | Neurite morphogenesis |
| GO:0060173 | 0.00068 | 2.7 | 6.5 | 16 | 93 | Limb development |
| GO:0007411 | 0.00075 | 2.8 | 5.9 | 15 | 85 | Axon guidance |
| GO:0006270 | 0.00088 | 9.5 | 0.8 | 5 | 12 | DNA replication initiation |
| GO:0001708 | 0.0010 | 4.6 | 2.1 | 8 | 31 | Cell fate specification |
| GO:0000904 | 0.0014 | 2.0 | 13.2 | 25 | 188 | Cell morphogenesis involved in differentiation |
| GO:0048869 | 0.0017 | 1.3 | 86.5 | 112 | 1,231 | Cellular developmental process |
| GO:0007420 | 0.0020 | 1.9 | 15.0 | 27 | 214 | Brain development |
| GO:0048663 | 0.0021 | 3.6 | 2.9 | 9 | 42 | Neuron fate commitment |
| GO:0042415 | 0.0031 | 19.9 | 0.3 | 3 | 5 | Norepinephrine metabolic process |
| GO:0009954 | 0.0033 | 4.9 | 1.5 | 6 | 22 | Proximal/distal pattern formation |
| GO:0042472 | 0.0033 | 3.1 | 3.7 | 10 | 53 | Inner ear morphogenesis |
| GO:0048598 | 0.0035 | 1.7 | 19.4 | 32 | 277 | Embryonic morphogenesis |
| GO:0007417 | 0.0050 | 2.9 | 3.9 | 10 | 57 | Central nervous system development |
| GO:0021846 | 0.0053 | 7.6 | 0.7 | 4 | 11 | Cell proliferation in forebrain |
| GO:0021520 | 0.0058 | 13.2 | 0.4 | 3 | 6 | Spinal cord motor neuron cell fate specification |
| GO:0021521 | 0.0058 | 13.2 | 0.4 | 3 | 6 | Ventral spinal cord interneuron specification |
| GO:0045773 | 0.0058 | 13.2 | 0.4 | 3 | 6 | Positive regulation of axon extension |
| GO:0021536 | 0.0065 | 4.2 | 1.7 | 6 | 25 | Diencephalon development |
| GO:0035116 | 0.0067 | 5.1 | 1.2 | 5 | 18 | Embryonic hindlimb morphogenesis |
| GO:0007275 | 0.0076 | 1.2 | 124.8 | 149 | 1,776 | Multicellular organismal development |
| GO:0007423 | 0.0076 | 1.8 | 13.4 | 23 | 191 | Sensory organ development |
| GO:0030326 | 0.0090 | 2.6 | 4.2 | 10 | 61 | Embryonic limb morphogenesis |
| GO:0035270 | 0.0095 | 2.7 | 3.6 | 9 | 52 | Endocrine system development |
| GO:0006268 | 0.0097 | 9.9 | 0.49 | 3 | 7 | DNA unwinding during replication |
| GO:0021546 | 0.0097 | 9.9 | 0.49 | 3 | 7 | Rhombomere development |
| GO:0048856 | 0.0099 | 1.2 | 106.1 | 128 | 1,538 | Anatomical structure development |
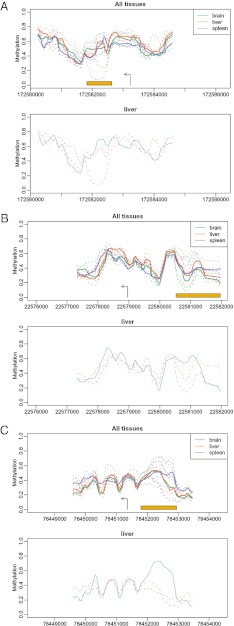
Fig. 1.
Examples of developmental genes with VMRs in livers from isogenic mice raised in the same environment. Shown are Bmp7 (A), Pou3f2 (B), and Ntrk3 (C), involved in early embryogenic programming and bone induction, neurogenesis and stem cell reprogramming, and body position sensing, respectively. In each paired plot, the top panel shows estimated methylation levels from various biological replicates from three different tissues: brain, liver, and spleen (dashed lines). The thicker solid lines represent the average curves for each tissue. The orange bar denotes the region in which our statistical method detected a VMR. The bottom panel highlights the liver. Only the four liver curves are shown. The different line types and colors represent the four individual mice.
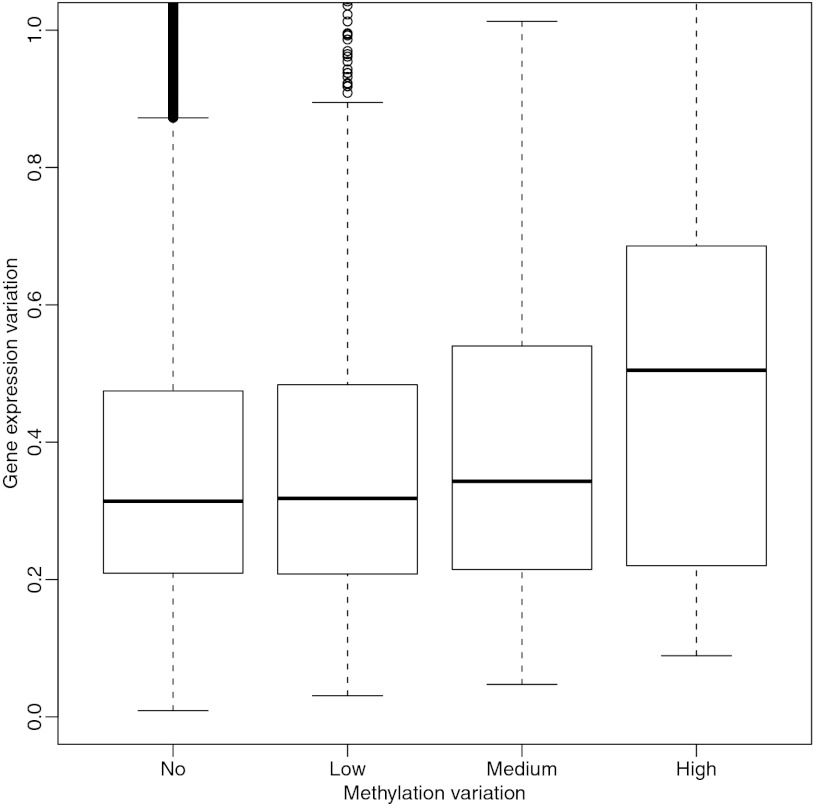
Furthermore, the VMRs were associated with a functional property: expression. As shown in Fig. 2, VMRs within 500 bp of a transcriptional start site (TSS) exhibited a stronger association between gene expression variability and methylation variability.
Fig. 2.
VMRs are associated with variability in gene expression of nearby genes. The human liver VMRs detected with our statistical algorithm were divided into three types: low variation (lowest 70%), high variation (highest 5%), and medium variation (the remainder). The VMRs within 500 bases from a gene’s transcription start site were associated with that gene. The expression measurements were obtained for the same human livers, and the SD across subjects was used to quantify variability. These boxplots show the distribution of this variability stratified by VMR variability. The first boxplot represents genes not associated with a VMR.
We then examined human liver for the presence of VMRs. Similar to our mouse results, we found significant variability. Where the VMRs were near genes, as in the mouse, there was a strong enrichment in the vicinity of genes with GO functional categories for development and morphogenesis when controlled for the mouse CHARM array (Table 2).
Table 2.
Enrichment scores of GO categories of genes in the vicinity of VMRs in human liver
| GOBPID | P value | Odds ratio | ExpCount | Count | Size | Term |
| GO:0009790 | 1.8E-05 | 1.8 | 43.1 | 70 | 320 | Embryonic development |
| GO:0019222 | 2.3E-05 | 1.3 | 319.5 | 379 | 2,372 | Regulation of metabolic process |
| GO:0006355 | 4.0E-05 | 1.3 | 239.6 | 292 | 1,779 | Regulation of transcription, DNA-dependent |
| GO:0032774 | 5.0E-05 | 1.3 | 246.8 | 299 | 1,832 | RNA biosynthetic process |
| GO:0009887 | 5.3E-05 | 1.6 | 54.1 | 82 | 402 | Organ morphogenesis |
| GO:0048704 | 8.4E-05 | 4.0 | 5.2 | 15 | 39 | Embryonic skeletal system morphogenesis |
| GO:0001501 | 8.5E-05 | 1.9 | 27.8 | 48 | 207 | Skeletal system development |
| GO:0051093 | 8.5E-05 | 1.7 | 43.5 | 68 | 323 | Negative regulation of developmental process |
| GO:0016339 | 0.00012 | 7.2 | 2.2 | 9 | 17 | Calcium-dependent cell-cell adhesion |
| GO:0009952 | 0.00013 | 2.5 | 12.3 | 26 | 92 | Anterior/posterior pattern formation |
| GO:0048518 | 0.00017 | 1.3 | 133.2 | 171 | 989 | Positive regulation of biological process |
| GO:0019219 | 0.00025 | 1.2 | 269.0 | 317 | 1,997 | Regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| GO:0007389 | 0.00028 | 2.0 | 22.3 | 39 | 166 | Pattern specification process |
| GO:0010468 | 0.00029 | 1.2 | 272.3 | 320 | 2,029 | Regulation of gene expression |
| GO:0043009 | 0.00032 | 2.1 | 18.7 | 34 | 140 | Chordate embryonic development |
| GO:0031326 | 0.00037 | 1.2 | 279.8 | 327 | 2,077 | Regulation of cellular biosynthetic process |
| GO:0006350 | 0.00038 | 1.2 | 267.6 | 314 | 1,987 | Transcription |
| GO:0001824 | 0.00040 | 4.9 | 3.0 | 10 | 23 | Blastocyst development |
| GO:0010556 | 0.00048 | 1.2 | 271.3 | 317 | 2,014 | Regulation of macromolecule biosynthetic process |
| GO:0050678 | 0.00051 | 3.6 | 4.8 | 13 | 36 | Regulation of epithelial cell proliferation |
| GO:0048863 | 0.00064 | 7.5 | 1.7 | 7 | 13 | Stem cell differentiation |
| GO:0019827 | 0.00076 | 9.6 | 1.3 | 6 | 10 | Stem cell maintenance |
| GO:0007399 | 0.00080 | 1.4 | 84.5 | 112 | 631 | Nervous system development |
| GO:0000165 | 0.00089 | 2.0 | 16.0 | 29 | 119 | MAPKKK cascade |
| GO:0043284 | 0.0011 | 1.2 | 327.0 | 372 | 2,428 | Biopolymer biosynthetic process |
| GO:0043583 | 0.0014 | 2.7 | 7.2 | 16 | 54 | Ear development |
| GO:0042472 | 0.0016 | 3.5 | 4.1 | 11 | 31 | Inner ear morphogenesis |
| GO:0048468 | 0.0016 | 1.4 | 62.6 | 85 | 465 | Cell development |
| GO:0007420 | 0.0017 | 1.8 | 21.2 | 35 | 158 | Brain development |
| GO:0034645 | 0.0017 | 1.2 | 346.4 | 390 | 2,572 | Cellular macromolecule biosynthetic process |
| GO:0001656 | 0.0018 | 3.8 | 3.6 | 10 | 27 | Metanephros development |
| GO:0035239 | 0.0018 | 2.6 | 7.4 | 16 | 55 | Tube morphogenesis |
| GO:0043066 | 0.0019 | 1.7 | 26.9 | 42 | 200 | Negative regulation of apoptosis |
| GO:0045747 | 0.002 | Inf | 0.4 | 3 | 3 | Positive regulation of Notch signaling pathway |
| GO:0045597 | 0.0027 | 1.9 | 15.6 | 27 | 116 | Positive regulation of cell differentiation |
| GO:0043067 | 0.0030 | 1.4 | 58.7 | 79 | 436 | Regulation of programmed cell death |
| GO:0032501 | 0.0037 | 1.2 | 297.8 | 336 | 2,211 | Multicellular organismal process |
| GO:0007156 | 0.0039 | 1.9 | 13.7 | 24 | 102 | Homophilic cell adhesion |
| GO:0021546 | 0.0039 | 12.8 | 0.8 | 4 | 6 | Rhombomere development |
| GO:0065007 | 0.0040 | 1.1 | 633.7 | 677 | 4,704 | Biological regulation |
| GO:0045884 | 0.0043 | 5.5 | 1.7 | 6 | 13 | Regulation of survival gene product expression |
| GO:0048523 | 0.0043 | 1.2 | 129.7 | 157 | 963 | Negative regulation of cellular process |
| GO:0021915 | 0.0044 | 3.2 | 4.0 | 10 | 30 | Neural tube development |
| GO:0001525 | 0.0046 | 1.9 | 14.6 | 25 | 109 | Angiogenesis |
| GO:0048856 | 0.0048 | 1.2 | 202.8 | 235 | 1,525 | Anatomical structure development |
| GO:0048646 | 0.0049 | 2.2 | 8.8 | 17 | 66 | Anatomical structure formation |
| GO:0000122 | 0.0055 | 1.7 | 21.1 | 33 | 157 | Negative regulation of transcription from RNA polymerase II promoter |
| GO:0045595 | 0.0055 | 1.8 | 16.4 | 27 | 123 | Regulation of cell differentiation |
| GO:0007507 | 0.0063 | 1.8 | 16.5 | 27 | 123 | Heart development |
| GO:0000070 | 0.0065 | 4.1 | 2.4 | 7 | 18 | Mitotic sister chromatid segregation |
| GO:0021545 | 0.0067 | 4.8 | 1.8 | 6 | 14 | Cranial nerve development |
| GO:0006366 | 0.0070 | 1.3 | 59.7 | 78 | 448 | Transcription from RNA polymerase II promoter |
| GO:0048869 | 0.0073 | 1.2 | 149.1 | 176 | 1,107 | Cellular developmental process |
| GO:0008284 | 0.0076 | 1.5 | 28.1 | 41 | 209 | Positive regulation of cell proliferation |
| GO:0001708 | 0.0079 | 3.4 | 3.0 | 8 | 23 | Cell fate specification |
| GO:0007020 | 0.0081 | 8.5 | 0.9 | 4 | 7 | Microtubule nucleation |
| GO:0001655 | 0.0083 | 2.2 | 7.8 | 15 | 58 | Urogenital system development |
| GO:0001666 | 0.0083 | 2.2 | 7.8 | 15 | 58 | Response to hypoxia |
| GO:0000281 | 0.0087 | 19.3 | 0.5 | 3 | 4 | Cytokinesis after mitosis |
| GO:0009058 | 0.0088 | 1.1 | 405.0 | 442 | 3,007 | Biosynthetic process |
| GO:0035270 | 0.0093 | 2.5 | 5.7 | 12 | 43 | Endocrine system development |
| GO:0001649 | 0.0094 | 2.6 | 5.1 | 11 | 38 | Osteoblast differentiation |
| GO:0048699 | 0.0096 | 1.4 | 40.4 | 55 | 300 | Generation of neurons |
| GO:0007215 | 0.0099 | 4.2 | 2.0 | 6 | 15 | Glutamate signaling pathway |
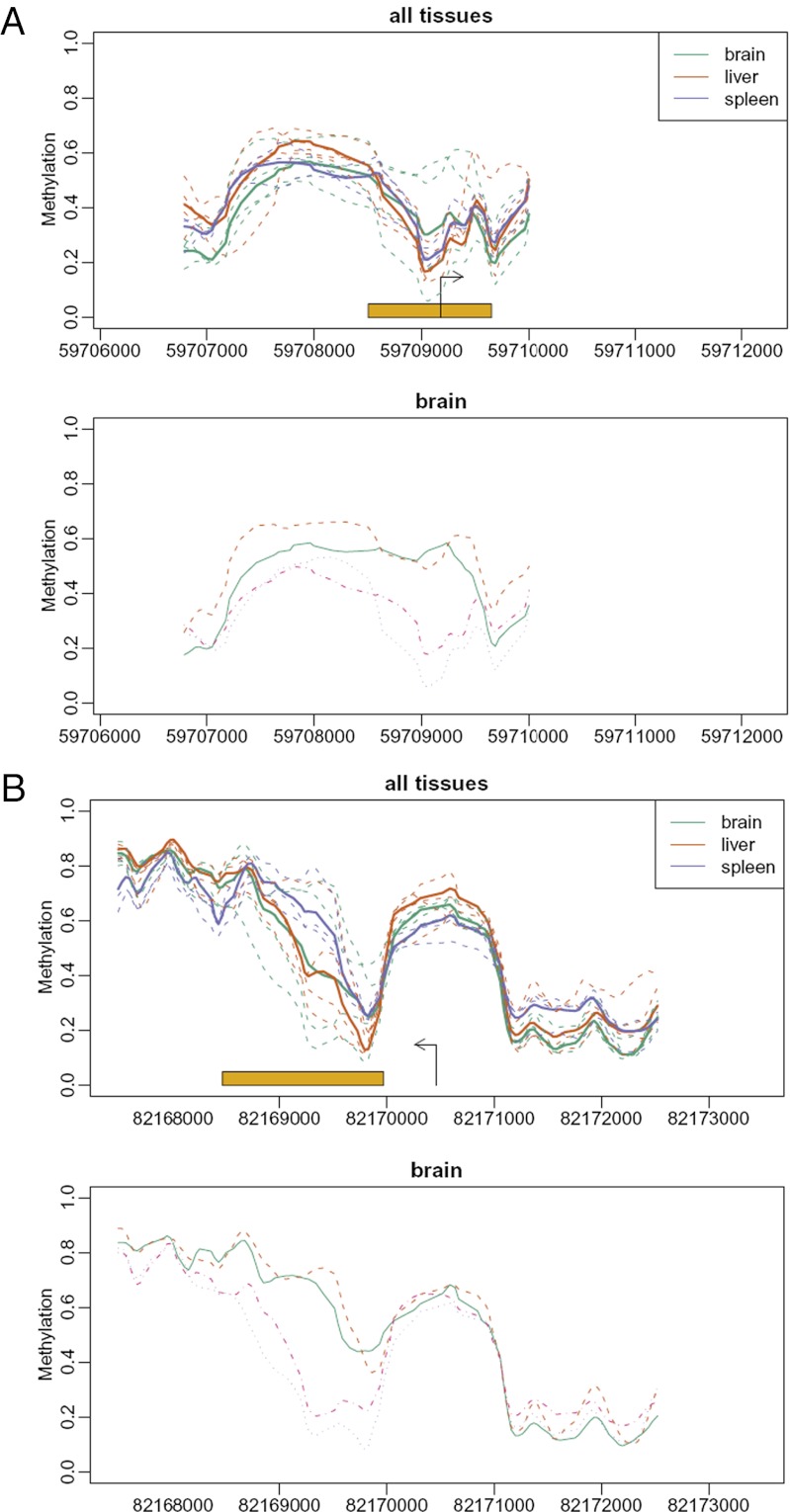
We then performed a similar analysis on mouse brain. The results were even more striking. For example, Fig. 3 shows two examples of VMRs: Bmpr2, the receptor for the morphogenetic BMP protein, and Irs1, a key mediator of insulin-driven differentiation. Our findings indicate that VMRs are present across tissues and species, are enriched in development-related genes, and are related to phenotype, at least at the level of expression of the proximate gene.
Fig. 3.
Examples of developmental genes with VMRs in brains from isogenic mice raised in the same environment. Shown are Bmpr2, the receptor for the morphogenetic BMP protein (A), and Irs1, a key mediator of insulin-driven differentiation (B). Labeling is as in Fig. 1.
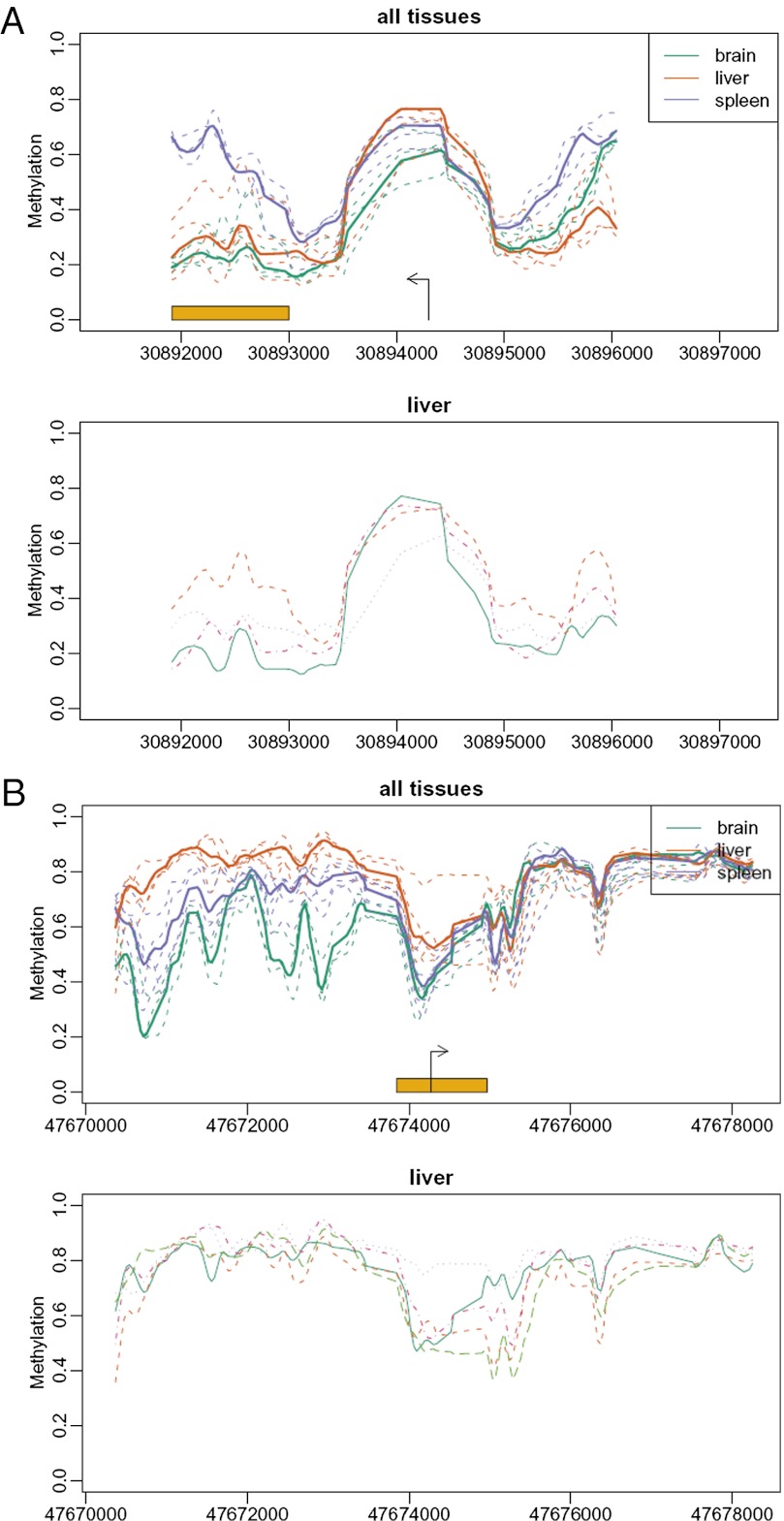
Also note that VMRs often are located near tissue-varying DMRs (T-DMRs), suggesting a mechanism by which they might evolve into each other over time. This is illustrated in Fig. 4 for mouse Ptp4a1, a protein tyrosine phosphatase involved in maintaining differentiated epithelial tissues, and for human FOXD2, a forkhead transcription factor involved in embryogenesis.
Fig. 4.
VMRs are often located near T-DMRs. Shown are mouse Ptp4a1, a protein tyrosine phosphatase involved in maintaining differentiated epithelial tissues (A), and human FOXD2, a forkhead transcription factor involved in embryogenesis (B). Labeling is as in Fig. 1. In (A), the VMR and T-DMR coincide, whereas in (B), they are adjacent.
Tissue-Specific Differentially Methylated Regions Across Species
Next, we were interested in whether changes in differential methylation across species (mouse and human) could be traced back to an underlying genetic basis. To address this question, we focused on T-DMRs, given the wealth of data gathered in previous studies and their relevance to human diseases, such as cancer. Previously we reported that DMRs that distinguish colorectal cancer from normal colonic mucosa (C-DMRs) are enriched for T-DMRs, and this finding was validated in a large independent set of samples. In many cases, the loss of differential methylation in one species was related to an underlying loss of CpGs at the corresponding CpG island or nearby CpG island shore (14). A typical example of an evolutionary change in differential methylation involved LHX1, a transcriptional regulator essential for vertebrate head organization and mesoderm organization, (shown in Fig. 5). Note the T-DMR in human that is not in mouse on the left of the TSS. The human has gained CpGs at a CpG island shore (with the island shown in orange tick marks in the bottom panel). In contrast, both species have a moderate CpG count to the right of the TSS, and both have DMRs in this region. This is an example of how a genetic variation (i.e., gain of CpGs) allows for development-relevant tissue-specific differences in a highly conserved gene. Thus, differential methylation that itself differs across species may be due to underlying sequence variation at the site of these DMRs. Additional examples of this are available at rafalab.jhsph.edu/evometh.pdf.
Fig. 5.
An underlying genetic basis for species differences in DMRs. A 7,500-bp human region was mapped to the mouse genome. The x-axis shows an index so that mapped bases are on top of one another. (Top) Methylation profiles for each human sample. As in Fig. 1, the dashed lines represent the individuals, and the solid lines represent the tissue averages. (Middle) The same plot for mouse. (Bottom) Ticks representing CpG locations for human and mouse. The orange ticks represent CpGs that were conserved. The curves represent CpG counts in a moving window of size 200 bases. Note that the lack of CpGs in the mouse at the beginning of the regions is associated with a difference in methylation patterns between species. Shown is LHX1, a transcriptional regulator essential for vertebrate head organization and mesoderm organization. Note the DMR in human that is not in mouse on the left of the TSS. The human has gained CpGs at a CpG island shore (orange tick marks). In contrast, both species have a moderate CpG count to the right of the TSS, and both have DMRs in this region.
Increased Stochastic Variation Would Increase Fitness in a Varying Environment
To model the role of epigenetic variation in natural selection, we performed three simulations based on a single quantitative phenotype that contributes to fitness, arbitrarily called Y. We assumed that mutations of eight genomic locations affected the expected value of Y, with four mutations increasing Y and four decreasing Y. For two of the simulations (simulations 1 and 2), we included a novel stochastic element controlled by eight mutations, four of which increased the variance of Y across the population given an identical genotype and four of which decreased this variance. Mathematical details are given in Materials and Methods.
In simulation 1, we emulated natural selection in a fixed environment favoring positive Y but including a novel stochastic epigenetic element, such that eight mutations affect the average of Y and eight mutations affect the variance of Y. As expected, this simulation favored the genotype with the largest expected value and the smallest variance (Fig. 6A). Simulation 2 was the same as simulation 1, but in this case we allowed a changing environment across generations that favor at times large Y and at times small Y. In this simulation, the most highly variable genotype was selected for and dominated by the 1,000th generation (Fig. 6A). In simulation 3, we did not permit the variance to change. In this case, 72% of the iterations resulted in extinction before the 1,000th generation. This occurred because the genotype selected in one environment was not fit for the environment change after a dramatic environmental change. In contrast, when variance was allowed to change (simulation 2), extinction never occurred.
Fig. 6.
Results of simulations demonstrating that increased stochastic variation in the epigenome would increase fitness in a varying environment. (A) Simulations of natural selection. For each simulation, we computed the population average and SD of the phenotype as a function of generation. Two simulations are shown: simulation 1, natural selection in a fixed environment favoring positive Y but including a novel stochastic epigenetic element, such that eight mutations affect average Y and eight mutations affect variance of Y, and simulation 2, similar to simulation 1 but in this case allowing a changing environment across generations that favor at times positive Y and at times negative Y. The top panel shows the average (across all iterations) population average of Y as a function of generation for simulation 1 (green) and simulation 2 (orange). The dashed vertical lines indicate the generations at which the environment was changed in simulation 2. The bottom panel shows the average (across all iterations) population standard deviation of Y. Note that with a changing environment, the average Y fluctuates around a common point, but the SD of Y increases consistently. (B) Emulation of GWAS analysis based on simulation 2 (varying variance of Y). Observed odds ratios are for SNPs that change the mean phenotype.
In addition, we also emulated genome-wide association studies (GWAS) for Y. The individuals that did not survive were considered diseased, and the survivors were considered controls. An interesting finding was that the odds ratios for association between the genes known to affect fitness with disease hovered around 1.10 (Fig. 6B). The reason for this is because many of the diseased individuals were unfit only because of the affect of SNPs on variation, not because of the usual SNP-defined genetic change that directly affects function. This is simply a result of the low heritability that results from a large variance. Thus, the results of the epigenetic variation model are in agreement with results from current GWAS studies that explain very little attributable risk of disease.
Discussion
Here we have proposed a model in which increased variability with a given genotype might increase fitness not by changing mean phenotype, but rather by changing the variability of phenotype with a given genotype. We also have provided a possible mechanism by which such enhanced variability could be genetically inherited and lead to increased stochastic epigenetic variation during development. Note that the genomic loci for such variation would be well defined in our model; we have provided examples of these loci. Although these loci do not represent the primary engine of development, they do provide plasticity in the developmental program by virtue of the stochastic variation that they impart through the genes in their proximity.
Our model differs from that of a transgenerational epigenetic effect on phenotypic variation and disease risk (16), in that in our model, the genetic variant is inherited and contributes to enhanced phenotypic variation, which can be mediated epigenetically in each generation. It also differs from a hypermutable genetic-switching model, in which the genotype itself changes from generation to generation, increasing phenotypic plasticity (17).
Our model provides a mechanism for developmental plasticity and evolutionary adaptation to a fluctuating environment. Although the model is general and does not necessitate epigenetic variation, we have demonstrated the existence of VMRs that affect phenotype (i.e., gene expression) in isogenic mice raised in an identical environment, and have shown that similar VMRs exist in humans as well. We also have reported a potential genetic mechanism for differences in tissue-specific methylation across species—namely, the gain or loss of a CpG island or the associated shore. The localization near a specific gene would provide specificity of the effect of variation, but the mechanism for variation could entail the relationship to tissue-specific promoters, transcription factor binding sites, population variation in CpG density in these regions, or a combination of such factors. Distinguishing among these possibilities will require further experimentation.
Nonetheless, our model makes a specific prediction: that heritable genetic variation affects stochastic phenotypic variation. Thus, one should be able to identify SNPs that contribute to variance but not mean phenotype. Such SNPs do not necessitate an epigenetic mechanism for their influence, but at least some of them would be predicted to be in linkage disequilibrium to VMRs, such as those described above. The VMRs provide a possible mechanism for phenotypic variation in a given genetic background, and we have direct evidence for this at least at the level of expression of the proximate gene. Waddington (9) also proposed that in a given environment, phenotypes eventually become genetically assimilated, and that the sequence differences in CpG islands and shores could provide a mechanism for both gain and loss in evolution of developmental variation mediated by DNA methylation.
Our model and our data differ from Lamarckianism, which argues that the environment modifies the genome. While not disputing the existence of such inheritance, here we propose a genetic mechanism that may underlie this ability to vary epigenetically. We also depart from the neo-Darwinian and classical population genetics principle that heritable quantitative phenotypic variation is due entirely to the additive effect of individual trait loci. Here the heritable component is in part be a propensity to variation itself, adding an element of randomness to the phenotypic outcome. Thus, selection would be determined in part by the ability to vary around a setpoint, rather than by the setpoint itself. This notion is consistent with the idea of “order for free” of Stuart Kaufmann (18). Although Kaufman did not anticipate a role for epigenetics in evolution, inherent epigenetic variation itself will create new possibilities for ordered function—a question that now might be addressable mathematically, given our identification of a possible measurable substrate for this variation, namely DNA methylation. Of course, we do not know how much variation can be tolerated; at some point of increased variation, the individual species “identity” might deteriorate.
Our model also may help explain observations in the evolutionary and epigenetic literature that have seemed paradoxical. In epigenetics, the apparent high degree of instability in the fidelity of epigenetic marks is puzzling. For example, cell lines propagated clonally show a high frequency of random monoallelic expression (19). This epigenetic instability may have been first described while observing individual cancer cells (20), and data show clear epigenetic differences between identical twins (21). In evolutionary biology, social insects show environment-mediated phenotypic differences in social castes, and the distribution of those differences can be selected for (22), leading those authors to speculate that an epigenetic mechanism might be involved (23); the bee would be an outstanding model for testing these ideas. Finally, substantial variations in phenotype of crayfish from an identical genotype have been reported (24). The authors also observed variable global DNA methylation, but as a phenotype, not a mechanism, and found no relationship between methylation and phenotype; they did not examine individual genes (24). We suggest that the mechanism for phenotypic variation is epigenetic, and that increased variation would promote fitness.
Finally, not only variable phenotypes in normal tissue, but also variable disease phenotypes, might be obtained through inherent epigenetic variation. This is because a genetic variant providing a higher variance in phenotype also will increase the tails at both ends of the phenotype; that is, the same variant increasing fitness in one environment will increase the risk of decreasing fitness in a different environment. In support of this idea, we analyzed DMRs that are present in human but not in mouse, and found that many of these genes are associated with human disorders of development as well as common complex diseases, including TAL1 (leukemia), FOXD3 (several disorders), HHEX (diabetes), PLCE1 (nephrotic syndrome), NKX2 (heart trunk malformation), TLX1 (leukemia), FEZ1 (esophageal cancer), ALX4 (forebrain absence), SHANK3 (brain/immune defect), NKX2 (heart malformations), and IGF2 (colorectal and other cancers). We also note that in cancer the high degree of epigenetic variation (the mechanism of which has proved elusive) would follow directly from our evolutionary model. Thus, rather than arising from a varying environment acting across generations, cancer may arise in part from a repeatedly changing microenvironment due to, for example, repeated exposures to carcinogens, which would select for epigenetic heterogeneity, and thus the ability of cells to grow outside of their normal milieu.
Materials and Methods
Tissue Samples and CHARM.
Human tissues were obtained from the Stanley Foundation, and mouse tissues from C57BL/6 wild-type mice were obtained from Jackson Laboratory. Sample preparation and the CHARM DNA methylation analysis from which the data sets were derived are described in more detail elsewhere (14, 15).
VMRs.
First, the microarray raw data from CHARM arrays (14) were transformed into estimated methylation percentages for each genomic location represented by a probe. These values were then smoothed (14) to obtain estimated methylation profiles for each sample. Then for each tissue, the SD for each location was computed. A region of locations surpassing a 99.95% percentile of all of the variances was designated a VMR.
Simulations.
To create the simulation, we expanded the Fisher-Wright neutral selection model. In the neutral model, we started with N individuals and to create the next generation, we selected N individuals at random with replacement. This implies that the number of children for each individual follows a multinomial distribution, with population size remaining fixed at N. To introduce selection, we permitted each individual to die with probability 1-pn, with the survival probability pn depending on a phenotype, Yn. For the next generation, we selected N individuals, with replacement, from those that survived. For the simulation shown here, we quantified this relationship with a simple logistic function, log{ pn /(1-pn) }= a + bYn. Note that if b is positive, then positive Y individuals are more fit, and if b is negative, then negative Y individual are more fit. We then assumed the existence of M SNPs, Xm, m = 1,…,M, that affect the phenotype. We assumed two possible polymorphisms, designated 0 and 1, and denoted the expected change on the phenotype by βj, j = 1,…,M. We refer to (X1,…,XM) as the genotype. Note that there are 2M different genotypes.
We followed Fisher’s additive model for complex traits and assumed that the phenotype was a random variable with
Here e represents variation not explained by the standard genetic model and assumed to be a Gaussian random quantity with mean 0 and standard deviation s. Note that each genotype will have a different average Y value, determined by the effects β. We then added an epigenetic variation term caused by sequence changes (e.g., the addition of a CpG island that allows the presence of a VMR or T-DMR). We modeled this by incorporating another feature; we assumed the existence of M SNPs that altered the individual’s variability (i.e., changed s). This is the epigenetic scenario, in which we are incorporating sequence variation that affects the variability of the phenotype, without altering the mean of the phenotype. This would be analogous to the earlier examples of loss or gain of CpGs that lead to the loss or gain of differentially methylated regions. We denote this epigenetic variation-inducing sequence change by Z and the effects by γ, and assume that
Simulation 1.
We started this simulation with an isogenic population and permit mutations to occur independently and at random at rate r. We ran this simulation with n = 10,000, a = -4, b = 4, M = 8 with (β1,…, β8) = (-1,-1,-1,-1, 1, 1, 1), s = 1, and r = 10−4. Note that these values of a and b imply that a average individual (Y = 0) has about a 1% chance of surviving. In contrast, an individual with the (0,0,0,0,1,1,1,1) genotype has about a 99% chance of surviving. For the epigenetic part of our model, we used (γ1,…, γ8)=(-1,-1,-1,-1,1,1,1,1)/2. This implies that some mutations increase phenotype variance by 50% and others decrease it by 50%. We ran 1,000 generations 250 times.
Simulation 2, environment changing.
We repeated simulation 1 except that we imitated dramatic environmental changes that changed the environment and its relationship with phenotype and fitness. The occurrence of these events was assumed to be random at a rate of 1 per 25 generations. Such a change resulted in b changing from 4 to -4. This implies that after the first event, smaller-than-average individuals were more fit than taller-than-average individuals. To check whether the outcome was stable, we considered a more skewed initial condition. Specifically, we reran the original simulation using 12 different sets of initial parameters. We first increased the number of iterations to 5,000. We then varied the environment changing rate to be 1 per 5, 1 per 10, 1 per 25, or 1 per 50 generations. Finally, we varied the number of mutating SNPs to be 2, 8, or 16. The conclusions from these simulations were as expected: Variability increased fitness, particularly in a changing environment (see Fig. S1).
Simulation 3.
Simulation 3 was the same as simulation 1, except we did not permit mutations to affect the variance of Y.
Acknowledgments
We thank Elisabet Pujadas for providing helpful discussions and comments on the manuscript, Simon Tavaré for pointing out evolution papers containing simulations, and Sarah Wheelan for help with BLAST. This work was supported by National Institutes of Health Grants P50 HG003233 and R01 GM083084.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Evolution in Health and Medicine,” held April 2–3, 2009, at the National Academy of Sciences in Washington, DC. The complete program and audio files of most presentations are available on the NAS web site at www.nasonline.org/Sackler_Evolution_Health_Medicine.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906183107/DCSupplemental.
References
- 1.Weiss KM. The smallest grain in the balance. Evol Anthropol. 2004;13:122–126. [Google Scholar]
- 2.Barton NH, Briggs DEG, Eisen JA, Goldstein DB, Patel NH. Evolution. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. [Google Scholar]
- 3.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 4.Jablonka E, Lamb MJ. Epigenetic Inheritance and Evolution: The Lamarckian Dimension. New York: Oxford University Press; 1995. [Google Scholar]
- 5.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132(Suppl 8):2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 6.Waterland RA, Jirtle RL. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakyan VK, et al. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West-Eberhard MJ. Developmental Plasticity and Evolution. New York: Oxford Univ Press; 2003. [Google Scholar]
- 9.Waddington CH. How Animals Develop. London: Allen & Unwin; 1935. [Google Scholar]
- 10.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handel AE, Ramagopalan SV. Public health implications of epigenetics. Genetics. 2009;182:1397–1398. doi: 10.1534/genetics.109.106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone L, et al. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 2009;5:e1000538. doi: 10.1371/journal.pgen.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janion C. Influence of methionine on the mutation frequency in Salmonella typhimurium. Mutat Res. 1982;94:331–338. doi: 10.1016/0027-5107(82)90295-0. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadeau JH. Transgenerational genetic effects on phenotypic variation and disease risk. Hum Mol Genet. 2009;18(R2):R202–R210. doi: 10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salathé M, Van Cleve J, Feldman MW. Evolution of stochastic switching rates in asymmetric fitness landscapes. Genetics. 2009;182:1159–1164. doi: 10.1534/genetics.109.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kauffman SA. The Origins of Order: Self-Organization and Selection in Evolution. New York: Oxford Univ Press; 1994. [Google Scholar]
- 19.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 20.He L, et al. Hypervariable allelic expression patterns of the imprinted IGF2 gene in tumor cells. Oncogene. 1998;16:113–119. doi: 10.1038/sj.onc.1201501. [DOI] [PubMed] [Google Scholar]
- 21.Kaminsky ZA, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 22.Page RE, Jr., Scheiner R, Erber J, Amdam GV. 8. The development and evolution of division of labor and foraging specialization in a social insect (Apis mellifera L.) Curr Top Dev Biol. 2006;74:253–286. doi: 10.1016/S0070-2153(06)74008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omholt SW, Amdam GV. Epigenetic regulation of aging in honeybee workers. Sci Aging Knowl Environ. 2004;26:pe28. doi: 10.1126/sageke.2004.26.pe28. [DOI] [PubMed] [Google Scholar]
- 24.Vogt G, et al. Production of different phenotypes from the same genotype in the same environment by developmental variation. J Exp Biol. 2008;211:510–523. doi: 10.1242/jeb.008755. [DOI] [PubMed] [Google Scholar]