Summary
2009 marks not only the 200th anniversary of Darwin's birth but also publication of the first scientific evolutionary theory, Lamarck's Philosophie Zoologique. While Lamarck embraced the notion of the inheritance of acquired characters, he did not invent it [1]. New phenomena discovered recently offer molecular pathways for the transmission of several acquired characters. Ciliates have long provided model systems to study phenomena that bypass traditional modes of inheritance. RNA, normally thought of as a conduit in gene expression, displays a novel mode of action in ciliated protozoa. For example, maternal RNA templates provide both an organizing guide for DNA rearrangements in Oxytricha and a template that can transmit spontaneous mutations that may arise during somatic growth to the next generation, providing two such mechanisms of so-called Lamarckian inheritance. This suggests that the somatic ciliate genome is really an "epigenome", formed through templates and signals arising from the previous generation. This review will discuss these new biological roles for RNA, including noncoding "template" RNA molecules. The evolutionary consequences of viable mechanisms in ciliates to transmit acquired characters may create an additional store of heritable variation that contributes to the cosmopolitan success of this diverse lineage of microbial eukaryotes.
Introduction and background
The subject of cytoplasmic inheritance has long been associated with ciliated protozoa because of myriad examples of non-Mendelian inheritance. In 1937 [2], Paramecium became one of the first single-celled eukaryotic model systems, with the observation that mating type determination was non-Mendelian. The same genotypes can produce two opposite mating types, and mating type determinants were cytoplasmically transmitted to sexual progeny. Sonneborn introduced the concept of “plasmagenes” [3], which together with nuclear genes were thought to determine all phenotypic characters.
The exceptional variety of ciliate characters under cytoplasmic inheritance raises the question of why so many examples of unconventional transmission appear in these organisms. Because sex in ciliates (Figure 1) is not reproduction, which occurs asexually, ciliate conjugation imparts to a new zygote not only a diploid genome derived from both parents but also the entire protoplasm of one parent. Though multicellular organisms may also transmit large amounts of protoplasm to their progeny, this usually contributes nutrients, rather than fully differentiated functions, although miRNAs and mRNAs are also present [4–7]. In ciliate exconjugants, a fully functional and integrated organelle system is only gradually taken over by a new nuclear genome. Given this situation in ciliates, the abundance of non-Mendelian characters that persist after conjugation should be less surprising.
Figure 1. Oxytricha’s sexual cycle [25].
All vegetative cells (stages 1 and 10) contain one macronucleus (MAC) and two micronuclei (MIC). The two MIC are genetically identical, but for simplicity we show only one here. (2) When deprived of food, two cells of different mating-types partially fuse, to initiate conjugation. (3) Both vegetative micronuclei in each partner enter meiosis I. (4) One product from each meiosis I is promoted to meiosis II, and one of the four meiosis II products is promoted to a post-meiotic mitosis. (5) The sister products of one mitosis are promoted to develop into gametic nuclei: a posterior stationary, and an anterior migratory nucleus; this happens in both partners, so that both partners emerge (6) with identical zygotic genotypes when the exchanged migratory nucleus fuses with the retained stationary nucleus (7), resulting in genetically-identical exconjugant cells. (8) The newly-formed zygotic nucleus divides twice: one daughter nucleus is destroyed, two become the new MICs, and (9) one differentiates into the new MAC. (Details of Oxytricha’s nuclear events are described in ref. [42]). Inside the circle are representative MIC and MAC versions of the O. trifallax actin I gene (MDSs in purple; IESs and flanking DNA in yellow).
Classic examples include "cortical inheritance" [8]. For instance, after conjugation, Paramecium cells sometimes fail to separate and produce abnormal doublets that only occasionally revert back to “singlets”. Sonneborn found that the genetic determinants of the doublet character were located not in the nucleus or in the cytoplasm, but in the cell cortex itself, or outer layer. Cortical inheritance also exists in Oxytricha [9–11]. Environmentally induced or acquired phenotype changes in organisms can therefore transmit to further generations, even if they are not determined by nucleic acids. This, of course, does not exclude a role in cortical organization of genes, since they encode the proteins that comprise the structures. Genetic mutations do exist that result in altered cortical organization (reviewed in [12]). However, among epigenetic phenomena in ciliates, cortical structural inheritance is the least understood at the molecular level, and begs further investigation.
Homology-dependent epigenetic inheritance in ciliates
Recently there have been expanding cases of non-Mendelian inheritance termed homology-dependent, i.e. sequence-dependent. Like in many other eukaryotes, sequence-dependent silencing of a gene can occur during vegetative growth; in addition, these mechanisms can participate in the programming of genome rearrangements during development. One particular distinguishing feature in ciliates is the decoupling of germline and somatic functions by two distinct nuclei, the diploid micronucleus (MIC) and a DNA-rich macronucleus (MAC). Micronuclear meiosis initiates sexual events and after haploid nuclei exchange, new micro- and macronuclei develop from the zygotic nucleus; the old MAC is eventually lost and replaced by the new one.
Advances in ciliate molecular biology have shown that very similar developmental alternatives can be determined by programmed genome rearrangements. Indeed, the development of a new macronucleus involves extensive rearrangements of the germline genome, including elimination of transposons and other repeated sequences and the precise excision of numerous single-copy Internal Eliminated Sequences (IESs) from coding and non-coding sequences. Genome-wide rearrangements discard nearly all non-genic DNA, resulting in streamlined gene-rich genomes. In Paramecium, for instance, ~40,000 genes occupy only 72 Mb [13]. In some cases, for example in the ciliate Oxytricha, reordering, or "unscrambling", of the remaining DNA segments (Macronucleus Destined Segments, or MDSs) also occurs by translocation or inversion.
The amazingly high degree of specificity and reproducibility suggests a general mechanism for programming rearrangements. The parental ciliate cell must provide sufficient amount of information in order to produce a fully functional new macronucleus. Details of the mechanism that allows the cell to perfectly recognize hundreds of thousands of DNA sequences for elimination or rearrangement remain largely unknown. However, the discovery of homology-dependent maternal effects that can modify rearrangements patterns has shed some light on this process. In Paramecium, both types of developmentally-regulated genome rearrangements—precise excision of IESs and imprecise DNA elimination—can be controlled by homology-dependent maternal effects that score the presence or absence of a gene in the macronucleus [14–18]. Microinjection of a specific DNA sequence into the parental macronucleus can prevent that sequence from being eliminated from the progeny’s somatic genome. This appears to be general for a subset (approximately one third) of Paramecium IESs [19–20] termed maternally controlled IESs, or mcIESs. These observations imply trans-nuclear genome comparison during development.
The earliest model for epigenetic programming of DNA rearrangements, proposed by Paramecium researchers, assumed that the whole genome of the parental MAC is transcribed, and that transcripts are exported to the developing MAC, where they regulate rearrangements in a homology-dependent manner. Two possibilities for regulating IES excision were proposed: either the maternal transcripts simply inhibit excision of homologous sequences or they provide homologous templates for repair after constitutive elimination [19]. The latter hypothesis was rejected after demonstrating that mutations engineered in the sequence injected into the old MAC were not copied in the IES maintained in the new MAC; this was recently confirmed by showing that cleavage does not occur in engineered cell lines containing the IES permanently present in the MAC [21]. On the other hand, there has been recent evidence in support of a protective role of parental MAC transcripts in Paramecium: use of RNAi to degrade specific transcripts results in elimination of homologous DNA sequences in the new MAC for both genes [22] and IESs [23]. However, a template role for maternal transcripts remains an attractive model to explain the specificity of rearrangements in other ciliates, including Oxytricha, which is likely over one billion years distant from Paramecium and requires more complex DNA rearrangements via descrambling, in addition to removal of hundreds of thousands of intervening IESs, to construct functional, transcriptionally-active genes in the macronucleus. In this case, the maternal transcripts would provide templates for cleavage and repair during the DNA descrambling process.
RNA template-guided genome unscrambling
A recent study [24] demonstrates that maternal RNA templates can orchestrate massive genome rearrangements in Oxytricha, supporting an epigenetic model for sequence-dependent comparison between germline and somatic genomes (Figure 2). This work revealed the existence of maternal RNA that have the potential to guide DNA assembly. Furthermore, disruption of specific RNA molecules disables rearrangement of the corresponding gene, while injection of artificial templates reprograms the DNA rearrangement pathway. These observations suggest that RNA molecules guide genome rearrangement. The current data also suggest that the entire somatic genome might be equally transcribed during a narrow window of development, and the resulting RNA copy or cache of the genome would then be transported to the developing somatic nucleus of the sexual progeny to guide rearrangement. This implies the existence of a new system of inheritance, independent of the germline genome, where somatic genetic information is bequeathed through a transient RNA genome. This new system of inheritance in Oxytricha appears to be different from, though related to, the system described in Paramecium and Tetrahymena. These two distant ciliate lineages therefore acquired different variations on an apparently RNAi-like mechanism.
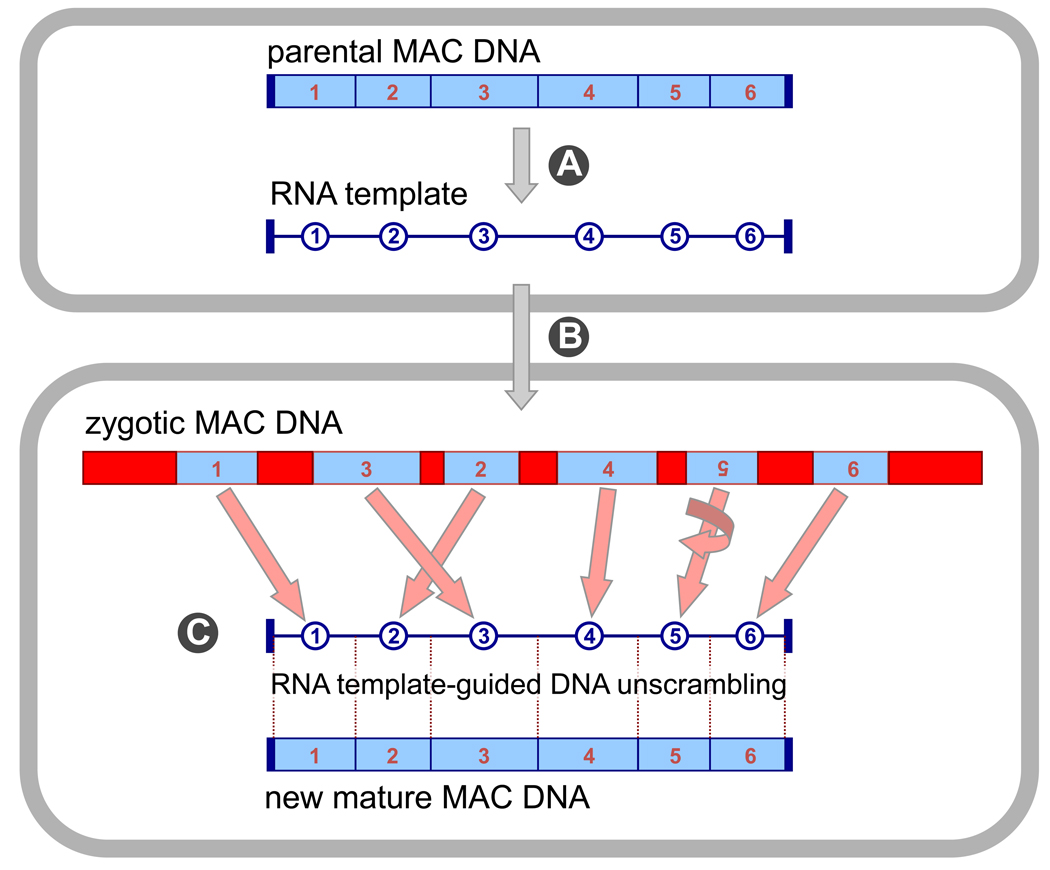
Figure 2. Model for RNA-templated genome rearrangements in Oxytricha [24].
Bidirectional RNA transcription of all DNA nanochromosomes in the old, maternal macronucleus (MAC) produces an RNA genome cache (A). Transport of these RNA transcripts to the newly developing macronucleus (B), where they may act as scaffolds to guide rearrangements (deletion, permutation and inversion) of corresponding micronuclear (MIC) DNA sequences (C). In this illustration, segments 2 and 3 are switched and segment 5 is inverted (number upside down) relative to the final macronuclear order.
The opportunity for RNA-guided DNA repair is most profound in the ciliate Oxytricha, because it deletes 95% of its germline genome through global DNA rearrangements. These severely fragment its germline chromosomes and then sort and reorder the hundreds of thousands of MDS pieces remaining. Because the programmed information for reordering comes from transiently-expressed maternal template RNAs, changes that arise in the maternal somatic genome, whether at the level of point mutations or new DNA rearrangement patterns, have an opportunity to be passed on to the next generation, bypassing the usual mode of inheritance via DNA. Furthermore, the occasional transfer of point mutations in these RNA templates to the F1 generation provides a mechanism for stable inheritance of acquired, spontaneous somatic substitutions, without altering the germline genome. This suggests that the somatic macronuclear genome is really an "epigenome", formed through templates and signals arising from the previous generation. This mechanism of inheritance beyond the conventional DNA genome is capable of epigenetically transferring rearrangement information across multiple generations, at least to the F3, hinting at the power of RNA molecules to reshape genomic information. While details of the mechanism are unknown, a recent study in our lab [25] implicates a role for germline-limited DNA transposons in the DNA rearrangement process, and the epigenetic transfer of specific point substitutions [24] points to an involvement of RNA-directed DNA proofreading.
Small RNA-guided programming of DNA rearrangements
In ciliates, at least two distinct RNAi pathways appear to mediate different types of homology-dependent effects. Post-transcriptional gene silencing (PTGS) was first shown to be induced in Paramecium by untranslatable transgenes [26,27], or by feeding cells with E. coli producing double-stranded RNA [28]. In both cases, silencing of targeted genes correlates with the accumulation of homologous ~23-nt siRNAs [22,29]. The same siRNAs are also involved in vegetative silencing and the deletions induced when Paramecium cells are fed with bacteria producing long dsRNA corresponding to a target gene. In Tetrahymena, a class of ~23–24-nt RNAs is produced endogenously from a limited number of loci [30]. Experimentally induced hairpin transcripts are processed into siRNAs and cause the silencing of homologous genes [31]. In both Tetrahymena and Paramecium, a Dicer-related protein is involved in the biogenesis of siRNAs [32,33]. However, the 23-nt siRNAs cannot explain other epigenetic effects, such as the maternal inheritance of alternative DNA rearrangements that do not depend on experimental silencing.
A second RNAi-like pathway, described in Tetrahymena thermophila, is restricted to conjugation. During developmental DNA rearrangements, elimination of micronuclear-specific DNA in this organism appears targeted by histone H3 K9 methylation [34,35]. Recent advances have shown a requirement for meiosis-specific Dicer- and Piwi-like proteins in the accumulation of scnRNAs (scan RNAs), a complex class of ~26–31-nt RNAs that appear in meiotic micronuclei [36–38]. These Dicer- and Piwi-like proteins are also required later for DNA elimination in the developing macronucleus, suggesting that rearrangement patterns in Tetrahymena are determined by scnRNA-directed methylation of histone H3 on lysine positions 9 (H3K9) and 27 (H3K27) marking germline-specific sequences in the developing MAC [35,37,39]. Histone methylation-dependent DNA elimination in ciliates suggests analogy to heterochromatin-related transcriptional gene silencing in other eukaryotes. This analogy is important because similar proteins might be involved in both phenomena. The observation that Tetrahymena scnRNAs appear to migrate from parental to zygotic macronucleus and become progressively enriched in micronucleus-specific sequences during conjugation [39] suggests that they may mediate the trans-nuclear comparison that is required to mimic the parental genome organization in the zygotic macronucleus. ScnRNAs that are able to find and possibly pair with homologous sequences in the parental macronucleus would be degraded along with destruction of the old macronucleus, whereas germline-specific RNAs would be exported to the newly developing macronucleus to target DNA elimination [40].
Current experimental data point to a combination of the scan RNA and the maternal transcript models: scnRNAs may instead be compared to an RNA copy of the maternal MAC genome, rather than to the MAC DNA itself (Figure 3). The elusive mechanism by which ciliates recognize germline sequences for elimination may thus involve a natural genomic subtraction, computed in vivo by RNA-RNA pairing interactions. Both Tetrahymena and Paramecium researchers have turned to a model proposing scnRNA selection through their interaction with long maternal RNAs [23,41]. The interaction between these two novel types of noncoding RNAs during development is the key to our understanding the large-scale genome reconstruction that is glorified in ciliates.
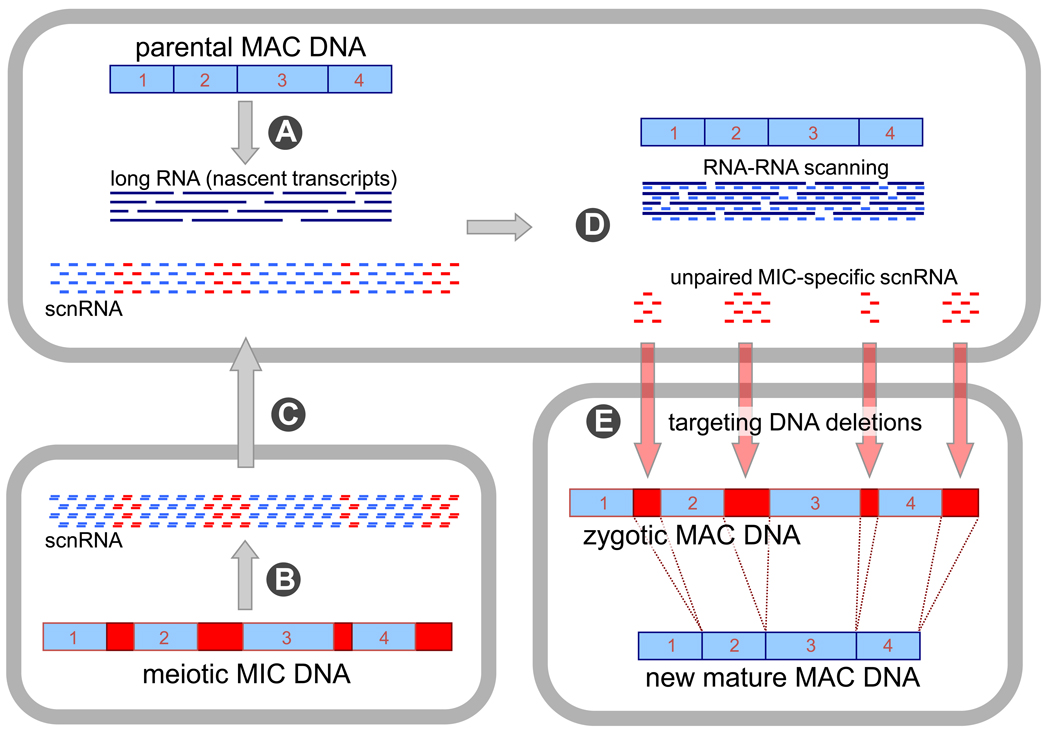
Figure 3. Scan RNA model for programmed DNA deletions in Paramecium and Tetrahymena [23,41].
Like in Oxytricha, the parental somatic genome (MAC DNA) is entirely transcribed during conjugation (A). The meiotic micronucleus produces short, 25–30-nt dsRNA, so-called scnRNA that derive from the entire germline genome (B), and are subsequently transported to the parental nucleus (C). Pairing between an RNA copy of the parental genome and scnRNA takes place prior to new MAC development, allowing selection of unpaired MIC-specific scnRNA (D). MIC-specific scnRNAs target deletions of homologous DNA sequences in the developing MAC (E), leading to a mature new MAC. Blue boxes represent MAC-destined DNA, which in the MIC are separated from each other by germline-specific sequences (red boxes) like IESs or transposons.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Burkhardt RW. The Zoological Philosophy of J. B. Lamarck. University of Chicago Press; 1984. Introduction. [Google Scholar]
- 2.Sonneborn TM. Sex, Sex Inheritance and Sex Determination in P. aurelia. Proc Nat Acad Sci USA. 1937;23:378–395. doi: 10.1073/pnas.23.7.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonneborn TM. Beyond the gene. American Scientist. 1949;37:33–59. [PubMed] [Google Scholar]
- 4.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 5.Tadros W, Lipshitz HD. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 2005;232:593–608. doi: 10.1002/dvdy.20297. [DOI] [PubMed] [Google Scholar]
- 6.Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 8.Sonneborn TM. Does preformed cell structure play an essential role in cell heredity? In: Allen JM, editor. The nature of biological diversity. New York: McGraw- Hill; 1963. [Google Scholar]
- 9.Aufderheide KJ, Frankel J, Williams NE. Formation and positioning of surface-related structures in protozoa. Microbiol Rev. 1980;44:252–302. doi: 10.1128/mr.44.2.252-302.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimes GW. An analysis of the determinative difference between singlets and doublets of Oxytricha fallax. Genet Res. 1973;21:57–66. [Google Scholar]
- 11.Grimes GW, McKenna ME, Goldsmith-Spoegler CM, Knaupp EA. Patterning and Assembly of Ciliature are Independent Processes in Hypotrich Ciliates. Science. 1980;209:281–283. doi: 10.1126/science.209.4453.281. [DOI] [PubMed] [Google Scholar]
- 12.Beisson J. Preformed cell structure and cell heredity. Prion. 2008;2:1–8. doi: 10.4161/pri.2.1.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 14.Epstein LM, Forney JD. Mendelian and non-mendelian mutations affecting surface antigen expression in Paramecium tetraurelia. Mol Cell Biol. 1984;4:1583–1590. doi: 10.1128/mcb.4.8.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jessop-Murray H, Martin LD, Gilley D, Preer JR, Jr, Polisky B. Permanent rescue of a non-Mendelian mutation of Paramecium by microinjection of specific DNA sequences. Genetics. 1991;129:727–734. doi: 10.1093/genetics/129.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You Y, Aufderheide K, Morand J, Rodkey K, Forney J. Macronuclear transformation with specific DNA fragments controls the content of the new macronuclear genome in Paramecium tetraurelia. Mol Cell Biol. 1991;11:1133–1137. doi: 10.1128/mcb.11.2.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim CS, Preer JR, Jr, Polisky B. Identification of DNA segments capable of rescuing a non-mendelian mutant in paramecium. Genetics. 1994;136:1325–1328. doi: 10.1093/genetics/136.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You Y, Scott J, Forney J. The role of macronuclear DNA sequences in the permanent rescue of a non-mendelian mutation in Paramecium tetraurelia. Genetics. 1994;136:1319–1324. doi: 10.1093/genetics/136.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- 20.Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21. Gratias A, Lepère G, Garnier O, Rosa S, Duharcourt S, Malinsky S, Meyer E, Bétermier M. Developmentally programmed DNA splicing in Paramecium reveals short-distance crosstalk between DNA cleavage sites. Nucleic Acids Res. 2008;36:3244–3251. doi: 10.1093/nar/gkn154. This work demonstrates that the maternal epigenetic signal for DNA elimination directly targets the ends of the excised DNA (IES) by inducing dsDNA breaks. The authors show that a mutation in one IES end blocks DNA cleavage of not only the mutated but also the wild-type end. This suggests an interaction between both IES ends during their cleavage and helps us to understand the mechanism DNA processing during IES excision in Paramecium.
- 22.Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23. Lepere G, Bétermier M, Meyer E, Duharcourt S. Maternal non-coding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. This study demonstrates direct evidence for scnRNA-mediated DNA elimination in Paramecium, as well as the presence of maternal transcripts that antagonize the action of the DNA deletion-inducing scnRNA. The results support an RNA/RNA scanning process that leads to elimination of the germline sequences that lack the protection offered by the maternal transcripts.
- **24. Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. This work suggests that a complete RNA cache of the maternal somatic genome provides a template for correct and precise DNA rearrangement in Oxytricha, including DNA deletion and unscrambling. Injection of artificial templates, containing segment translocations, reprograms the DNA rearrangement pathway, suggesting that RNA molecules directly guide genome rearrangements. Furthermore, the occasional transfer of point mutations from RNA templates to the rearranged DNA molecules provides evidence for RNA-directed DNA proofreading. These phenomena offer possible mechanisms for the stable inheritance of acquired, spontaneous somatic mutations (whether in DNA sequence or alternative splicing pattern) without altering the germline genome.
- **25. Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. In Oxytricha, germline-limited transposase genes appear to mediate genome-wide DNA rearrangements, providing a tantalizing role for thousands of functional transposase proteins encoded in the germline-limited fraction of a large eukaryotic genome. This study shows that transposase gene expression occurs during germline-soma differentiation and its silencing leads to abnormal DNA rearrangement in the offspring.
- 26.Ruiz F, Vayssié L, Klotz C, Sperling L, Madeddu L. Homology-dependent gene silencing in Paramecium. Mol Biol Cell. 1998;9:931–943. doi: 10.1091/mbc.9.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galvani A, Sperling L. Transgene-mediated post-transcriptional gene silencing is inhibited by 3' non-coding sequences in Paramecium. Nucleic Acids Res. 2001;29:4387–4394. doi: 10.1093/nar/29.21.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galvani A, Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- 29.Nowacki M, Zagorski-Ostoja W, Meyer E. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr Biol. 2005;15:1616–1628. doi: 10.1016/j.cub.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 30.Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howard-Till RA, Yao MC. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol Cell Biol. 2006;26:8731–8742. doi: 10.1128/MCB.01430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32. Lepere G, Nowacki M, Serrano V, Gout JF, Duharcourt S, Duret L, Meyer E. Silencing-associated and meiosis-specific small RNAs in the ciliate Paramecium tetraurelia. Nucleic Acids Res. 2009;37:903–915. doi: 10.1093/nar/gkn1018. A functional analysis of Dicer and Dicer-like genes and the sequencing of small RNAs provides evidence for the existence of two distinct small RNA pathways in Paramecium. Both are associated with homology-dependent effects, the siRNA pathway involved in post-transcriptional gene silencing, and the scan RNA pathway responsible for homology-dependent regulation of genome rearrangements after sexual reproduction.
- 33.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 34.Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci USA. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 37.Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- **41. Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. This study focuses on a Piwi-interacting RNA helicase involved in the interaction between scnRNA and chromatin. The authors suggest that the Piwi-associated scnRNAs scan the somatic genome via pairing with nascent transcripts in the maternal nucleus. Similarly, RNA-RNA pairing is believed to occur in the developing nucleus, where the selected scnRNAs presumably pair with nascent transcripts. These events are thought to target DNA elimination by triggering histone modification on the corresponding chromatin.
- 42.Adl SM, Berger JD. Timing of life cycle morphogenesis in synchronous samples of Sterkiella histriomuscorum. II. The sexual pathway. J Eukaryot Microbiol. 2000;47:443–449. doi: 10.1111/j.1550-7408.2000.tb00073.x. [DOI] [PubMed] [Google Scholar]