Abstract
Rhabdoid tumors (RT) are aggressive tumors characterized by genetic loss of SMARCB1 (SNF5, INI-1), a component of the SWI/SNF chromatin remodeling complex. No effective treatment is currently available. This study seeks to shed light on the SMARCB1-mediated pathogenesis of RT and to discover potential therapeutic targets. Global gene expression of 10 RT was compared with 12 cellular mesoblastic nephromas, 16 clear cell sarcomas of the kidney, and 15 Wilms tumors. 114 top genes were differentially expressed in RT (p<0.001, fold change >2 or <0.5). Among these were down-regulation of SMARCB1 and genes previously associated with SMARCB1 (ATP1B1, PTN, DOCK4, NQO1, PLOD1, PTP4A2, PTPRK). 28/114 top differentially expressed genes were involved with neural or neural crest development and were all sharply down-regulated. This was confirmed by Gene Set Enrichment Analysis (GSEA). Neural and neural crest stem cell marker proteins SOX10, ID3, CD133 and Musashi were negative by immunohistochemistry, whereas Nestin was positive. Decreased expression of CDKN1A, CDKN1B, CDKN1C, CDKN2A, and CCND1 was identified, while MYC-C was upregulated. GSEA of independent gene sets associated with bivalent histone modification and polycomb group targets in embryonic stem cells demonstrated significant negative enrichment in RT. Several differentially expressed genes were associated with tumor suppression, invasion and metastasis, including SPP1 (osteopontin), COL18A1 (endostatin), PTPRK, and DOCK4. We conclude that RTs arise within early progenitor cells during a critical developmental window in which loss of SMARCB1 directly results in repression of neural development, loss of cyclin dependent kinase inhibition, and trithorax/polycomb dysregulation.
Keywords: Gene expression, rhabdoid tumor, pediatric renal tumors, neural crest, INI-1, SMARCB1
Rhabdoid tumors (RT) are highly malignant neoplasms first described in the kidney of young children. RTs also arise in a variety of extra-renal sites including soft tissues and the central nervous system, where they are often called atypical-teratoid/rhabdoid tumors.1,2 3 RTs at all sites have a common genetic abnormality, the mutation or deletion of the SMARCB1/ hSNF5/INI-1 gene located at chromosome 22q11.4 3,5 This gene, which will be referred to as SMARCB1, encodes a component of the SWI/SNF chromatin remodeling complex that plays an important role in transcriptional regulation (reviewed in6). This has resulted in considerable scientific attention to RTs in recent years, as they represent a potent investigational model for the investigation of this important chromatin remodeling complex. Clinically, over 70% of children with RT present with non-localized disease, and chemotherapy alone is rarely curative.1 With an overall survival of 23%, new therapeutic options are needed. The overall goals of this study are to identify genetic pathways that will clarify the nature of RTs and to identify therapeutic targets.
Materials and Methods
Patient samples
Frozen tissue samples were obtained from the Renal Tumor Bank of the Children’s Oncology Group (COG). Specimens showing <80% tumor cellularity on frozen sections were excluded. A total of 53 tumors were analyzed: 10 RT, 12 cellular mesoblastic nephromas (CMN), 16 clear cell sarcomas of the kidney (CCSK), 15 favorable histology blastemal predominant Wilms tumors (WT). These samples were previously examined for the purpose of developing diagnostic signatures.7 Relatively equal numbers of tumors from each category were analyzed so that no single group biased the statistical analysis. All RT demonstrated loss of nuclear staining for BAF47, the protein encoded by SMARCB18, and all cellular CMNs demonstrated the ETV6-NTRK3 fusion transcript characteristic of this entity.9 Two fetal kidneys (16-18 weeks gestation) were analyzed for comparison.
Gene expression analysis
RNA was extracted and hybridized to Affymetrix U133A arrays (Affymetrix, Santa Clara, CA), scanned, and subjected to quality control parameters and normalization as previously described. 7 The gene expression of RT was compared to the other tumor types using two-sample t-tests. Affymetrix NetAffx™ Analysis Center was utilized to categorize genes and to establish the chromosomal location of differentially expressed genes. Two bioinformatics tools were utilized to analyze the data, Protein ANalysis THrough Evolutionary Relationships (PANTHER), and Gene Set Enrichment Analysis (GSEA). PANTHER classifies genes by their functions using published experimental evidence, and these are grouped into families and subfamilies of shared function. The detection of significant over- or under-represented functional pathways in a preselected gene list is determined by the binomial test. The Bonferroni-corrected p values are calculated to adjust for multiple testing.10 GSEA contains gene sets that relate to common biological function, including those from Gene Ontology groups as well curated gene lists from different pathways and publications. GSEA first ranks the expression of each gene based on its correlation with one of two phenotypes being compared (in this case RT vs non-RT). It then identifies this rank position within the independent gene set being queried. From this ranking it calculates an enrichment score that reflects the degree to which genes within the independent gene set are over represented within those genes most highly correlating with one of the two phenotypes. The normalized enrichment score (NES) takes into account the number of genes within the independent gene set. Permutation on the phenotype class labels is used to obtain the null distribution of NES and the nominal p-value. The false discovery rate is computed by comparing the tails of the observed and null distributions for the NES (http://www.broadinstitute.org/gsea).11
Real-time quantitative RT-PCR
TaqMan Gold and the ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA) were utilized. RT-PCR cycle parameters were 48°C for 30 min, 95°C for 15 min, followed by 40 cycles at 95°C for 15 sec and 59°C for 1 min. Sequences of probes and primers are shown in Table 1. Each threshold cycle (CT) was determined and the CT for the housekeeping gene (β –actin or GAPDH) was subtracted from this for normalization (dCt).
Table 1.
Reagents Utilized for Verification of Gene Expression
| A. Primers and Probes for quantitative RT-PCR | |||
|---|---|---|---|
| Gene Name | Taqman Probe | Forward Primer | Reverse Primer |
| SMARCB1 | ACCATGACCCAGCTGTGATCCATGAGA | CCTTCCCCCTTTGCTTTGA | GCACCTCGGGCTGAGATG |
| PTN | CCAAGCCCAAACCTCAAGCAGAATCTAAGA | CAAGCCCTGTGGCAAACTG | TGTTTCTTGCCTTCCTTTTTCTTC |
| FYN | TTCCCAGCAATTATGTGGCTCCAGTTGA | GGAAGCCCGCTCCTTGAC | AAAGTACCACTCTTCTGCCTGGATAG |
| PTPRK | TCAGATCACGACCCTGGAGAAAAAGCC | AGATGCCCCAAGGTTCCTATATG | GTGAGTGTCGTTCTCCTTCATTGTAG |
| NQO1 | TGGTCAGAAGGGAATTGCTCAGAGAAGGTAA | TCCAATGTTCTATTAATCACCTCTCTGTA | CTTGTGCTAGGCATATTAGTAACTGCTT |
| TFRC | ATCCCAGCAGTTTCTTTCTGTTTTTGCGA | GCTTTCCCTTTCCTTGCATATTC | CATGGTGGTACCCAAATAAGGATAA |
| CCND1 | CGGCGCTTCCCAGCACCAAC | TGGGTCTGTGCATTTCTGGTT | CTTTCATGTTTGTCTTTTTGTCTTCTG |
| Beta-actin | ATGCCCTCCCCCATGCCATCCTGCGT | TCACCCACACTGTGCCCATCTACGA | CAGCGGAACCGCTCATTGCCAATGG |
| GAPDH | AACAGCGACACCCACTCCTCCACCTT | AGAAAAACCTGCCAAATATGATGAC | GTGGTCGTTGAGGGCAATG |
| B. Antibodies for Immunohistochemistry and Immunoblotting | |||
| Antibody | Company (city) | Dilution | |
| BAF47 (SMARCB1) | BD Transduction Lab (San Jose, CA) | 1/500 | |
| NQO1 | SigmaAldrich (St Louis, MO) | 1/50 | |
| CTNNB1 | Zymed/Invitrogen | 1/200 | |
| ID3 | Abcam Inc (Cambridge, MA) | 1/2 | |
| SOX10 | SantaCruz (Santa Cruz, CA) | 1/50 | |
| Musashi | R&D Systems (Minneapolis, MN) | 1/200 | |
| CD133 | Miltenyi Biotec (Auburn, CA) | 1/10 | |
| Nestin | SantaCruz (Santa Cruz, CA) | 1/500 | |
| p53 | Dako (Carpenteria, CA) | 1/100 | |
| p21 | Dako (Carpenteria, CA) | 1/50 | |
| p27 | Dako (Carpenteria, CA) | 1/200 | |
| cyclin E | NeoMarkers (Fremont, CA) | 1/25 | |
| cyclin D1 | Dako (Carpenteria, CA) | 1/300 | |
| PEDF | SantaCruz (Santa Cruz, CA) | 1/50 | |
| SPP1 | Novocastra (Newcastle upon Tyne, UK) | 1/100 | |
| CDH2 | Invitrogen (Carlsbad, CA) | 1/100 | |
| NCAM | SantaCruz (Santa Cruz, CA) | 1/200 | |
| LGALS1 | Vector Labs (Burlingame, CA) | 1/200 | |
| CMYC | SantaCruz (Santa Cruz, CA) | 1/100 | |
| TFRC | Invitrogen (Carlsbad, CA) | 1/500 | |
Immunohistochemistry
Formalin-fixed-paraffin-embedded tissue from 6 of each RT, CCSK, CMN, and WT were tested using the monoclonal antibodies provided in Table 1. This was visualized by a streptavidin-biotin system (Vectastain Elite ABC Kit, Vector Laboratories) followed by ImmunoPure Metal Enhanced DAB Substrate (Therm Scientific, Rockford, IL) and counter-stained with hematoxylin.
Immunoblotting
Three frozen CCSKs, CMNs, RTs, and WTs were homogenized in sample buffer [0.125 mol/L Tris-HCl (pH, 6.8), 2% SDS, 10% glycerol, 0.001% bromophenol blue, and 5% beta-mercaptoethanol]. Equal amounts of protein lysate were run on polyacrylamide SDS gels and transferred to a nitrocellulose membrane (NCAM, CDH2, LGALS1) or PVDF (Immobilon-P) membrane (TFRC, CMYC). Membranes were blocked and incubated overnight at 4C in primary antibody (provided in Table 1). The washed membranes were incubated with alkaline phosphatase antimouse IgG (Vector Laboratories), and color was developed with a 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium alkaline phosphatase substrate kit IV detection kit (Vector Laboratories).
Results
Overall Differential Gene Expression in RTs
To establish a broad list of probesets containing the majority of genes involved in the pathogenesis of RT, the expression of RT was compared with all the other tumor types combined (“non-RT”), resulting in 2921 probesets with p-value <0.001 and false discovery rate of ≤1% (Supplemental Table 1, available at the journal’s website). The raw data from this study has been deposited in GEO, accession number GSE11482. To define a more restrictive list of probesets more uniquely expressed in RT, the gene expression of RT was compared with each of the other tumor types separately. Comparing RT to each of these clinically, pathologically, and genetically different tumor categories minimizes erroneous conclusions that may result from a single comparison. Genes present in each one of the resulting four comparisons (RT vs. CCSK, RT vs. CMN, RT vs. WT and RT vs all non-RT) with a p-value of <0.001 in each are provided in Supplemental Table 2 (426 genes). Of these, 114 genes demonstrated fold change >2 or <0.5 in the comparison between RT and all non-RT and are provided in Table 2, arranged in recurring functional groups indicated by GO and PANTHER. The chromosomal location of all probesets in Supplemental Table 1 was analyzed, and no chromosomal arm was over-represented, including probesets on chromosome 22. The 766 genes differentially expressed between RT and non-RT (p<0.0001) were analyzed in PANTHER, and those groups within the biologic process category over-represented with a Bonferoni corrected p value<0.05 are listed in Supplemental Table 3.
Table 2.
Genes with a p-value of <0.001 in each comparison of RT vs non-RT, RT vs CMN, RT vs CCSK, RT vs WT, and with a fold change of >2 or <0.5 in the comparison between RT and non-RT
| Gene Symbol | Description | Chromosome Location | UniGene ID | Fold Change RT vs non-RT | p-value RT vs non-RT | |||
|---|---|---|---|---|---|---|---|---|
| Genes Associated with INI-1 | ||||||||
| SMARCB1* | INI-1 | 22q11 | 534350 | 0.49 | 9.37E-21 | |||
| ATP1B1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | 1q24 | 291196 | 0.13 | 3.87E-19 | |||
| COL5A2 | collagen, type V, alpha 2 | 2q14-q32 | 445827 | 3.11 | 1.64E-10 | |||
| DOCK4 *, # | dedicator of cytokinesis 4 | 7q31.1 | 654652 | 0.40 | 8.11E-20 | |||
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | 16q22.1 | 406515 | 4.58 | 1.33E-06 | |||
| PLOD1 | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 1p36.3-p36.2 | 75093 | 2.17 | 7.27E-11 | |||
| PTN* | pleiotrophin (heparin binding growth factor 8) | 7q33-q34 | 371249 | 0.33 | 6.74E-12 | |||
| PTP4A2 | protein tyrosine phosphatase type IVA, member 2 | 1p35 | 470477 | 2.14 | 7.47E-07 | |||
| PTPRK *,# | protein tyrosine phosphatase, receptor type, K | 6q22.2-q22.3 | 155919 | 0.22 | 3.76E-23 | |||
| RSU1 # | Ras suppressor protein 1 | 10p13 | 524161 | 4.99 | 1.19E-05 | |||
| SPOCK1* | sparc/osteonectin, (testican) 1 | 5q31 | 654695 | 0.25 | 4.85E-14 | |||
| SPP1# | secreted phosphoprotein 1 (osteopontin) | 4q21-q25 | 313 | 5.42 | 3.50E-07 | |||
| SERPINE2* | serpin peptidase inhibitor, clade E,member 2 | 2q33-q35 | 38449 | 0.21 | 2.63E-09 | |||
| Neural Development | ||||||||
| CDH2 | cadherin 2, type 1, N-cadherin (neuronal) | 18q11.2 | 464829 | 0.23 | 3.30E-06 | |||
| CITED2 | Cbp/p300-interacting transactivator, 2 | 6q23.3 | 82071 | 0.47 | 1.93E-10 | |||
| CRIM1 | cysteine rich transmembrane BMP regulator 1 | 2p21 | 332847 | 0.36 | 3.23E-15 | |||
| CXCL12 | chemokine (C-X-C motif) ligand 12 | 10q11.1 | 522891 | 0.27 | 1.14E-15 | |||
| DPYSL2 | dihydropyrimidinase-like 2 | 8p22-p21 | 173381 | 0.41 | 1.10E-04 | |||
| ENC1 | ectodermal-neural cortex | 5q12-q13.3 | 104925 | 0.27 | 1.29E-16 | |||
| FSTL1 | follistatin-like 1 | 3q13.33 | 269512 | 0.29 | 4.35E-07 | |||
| FYN | FYN oncogene related to SRC, FGR, YES | 6q21 | 390567 | 0.34 | 3.69E-15 | |||
| HES1 | hairy and enhancer of split 1, (Drosophila) | 3q28-q29 | 250666 | 0.49 | 2.97E-07 | |||
| IGFBP2 | insulin-like growth factor binding protein 2 | 2q33-q34 | 438102 | 0.35 | 9.35E-08 | |||
| LAMA4 | laminin, alpha 4 | 6q21 | 654572 | 0.33 | 1.12E-05 | |||
| LAMB1 | laminin, beta 1 | 7q22 | 650585 | 0.29 | 4.15E-10 | |||
| LEF1 | lymphoid enhancer-binding factor 1 | 4q23-q25 | 555947 | 0.21 | 8.22E-18 | |||
| LMO4 | LIM domain only 4 | 1p22.3 | 436792 | 0.25 | 2.38E-16 | |||
| NAV2 | neuron navigator 2 | 11p15.1 | 502116 | 0.41 | 1.01E-11 | |||
| NCAM1 | neural cell adhesion molecule 1 | 11q23.1 | 503878 | 0.29 | 1.12E-18 | |||
| NOTCH2 | Notch homolog 2 (Drosophila) | 1p13-p11 | 487360 | 0.40 | 4.05E-11 | |||
| RND3 | Rho family GTPase 3 | 2q23.3 | 6838 | 0.23 | 1.13E-12 | |||
| SOBP | sine oculis binding protein homolog | 6q21 | 445244 | 0.33 | 3.82E-06 | |||
| SOX11 | SRY (sex determining region Y)-box 11 | 2p25 | 432638 | 0.19 | 2.76E-24 | |||
| SPON1 | spondin 1, extracellular matrix protein | 11p15.2 | 643864 | 0.20 | 4.51E-13 | |||
| TSPAN5 | tetraspanin 5 | 4q23 | 591706 | 0.37 | 1.59E-17 | |||
| MYC-C Targets | ||||||||
| ABCE1 | ATP-binding cassette, sub-family E, member 1 | 1q31.2 /// 4q31 | 571791 | 2.10 | 2.08E-08 | |||
| ACAT1 | acetyl-Coenzyme A acetyltransferase 1 | 11q22.3-q23.1 | 232375 | 2.24 | 8.78E-08 | |||
| AHCY | S-adenosylhomocysteine hydrolase | 20cen-q13.1 | --- | 2.30 | 1.71E-06 | |||
| CORO1C | coronin, actin binding protein, 1C | 12q24.1 | 696037 | 2.25 | 3.88E-05 | |||
| CYCS | cytochrome c, somatic | 7p15.2 | 437060 | 2.14 | 7.26E-06 | |||
| DDX21 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | 10q21 | 696064 | 2.66 | 7.73E-10 | |||
| ENO2 | enolase 2 (gamma, neuronal) | 12p13 | 511915 | 2.59 | 1.67E-06 | |||
| MTHFD2 | methylenetetrahydrofolate dehydrogenase 2 | 2p13.1 | 469030 | 2.40 | 1.03E-09 | |||
| MYO1B | myosin IB | 2q12-q34 | 439620 | 0.40 | 1.44E-06 | |||
| NEFH | neurofilament, heavy polypeptide 200kDa | 22q12.2 | 198760 | 2.22 | 3.09E-07 | |||
| PAM | peptidylglycine alpha-amidating monooxygenase | 5q14-q21 | 369430 | 0.41 | 2.29E-08 | |||
| SLC39A14 | solute carrier family 39, member 14 | 8p21.3 | 491232 | 2.39 | 1.58E-06 | |||
| TFRC# | transferrin receptor (p90, CD71) | 3q29 | 529618 | 3.16 | 1.53E-09 | |||
| Tumor Supression, Invasion, or Metastasis | ||||||||
| COL18A1 | collagen, type XVIII, alpha 1 (endostatin) | 21q22.3 | 517356 | 0.41 | 2.48E-13 | |||
| MMP12 | matrix metallopeptidase 12 | 11q22.3 | 1695 | 4.07 | 6.18E-05 | |||
| NCOA3 | nuclear receptor coactivator 3 | 20q12 | 592142 | 2.43 | 5.12E-06 | |||
| SELENBP1 | selenium binding protein 1 | 1q21-q22 | 632460 | 0.44 | 9.15E-20 | |||
| TES | testis derived transcript (3 LIM domains) | 7q31.2 | 592286 | 0.41 | 1.97E-13 | |||
| ZNF217 | zinc finger protein 217 | 20q13.2 | 155040 | 2.65 | 1.51E-05 | |||
| Transcription Regulation | ||||||||
| AFF1 | AF4/FMR2 family, member 1 | 4q21 | 480190 | 0.39 | 7.15E-09 | |||
| ATF5 | activating transcription factor 5 | 19q13.3 | 9754 | 2.05 | 2.17E-04 | |||
| ATXN1 | ataxin 1 | 6p23 | 434961 | 0.44 | 5.25E-10 | |||
| BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | 10q26.3 | 144873 | 2.40 | 1.65E-06 | |||
| CBX6 | chromobox homolog 6 | 22q13.1 | 592201 | 2.36 | 1.65E-10 | |||
| GPR177 | G protein-coupled receptor 177 | 1p31.3 | 647659 | 0.37 | 1.46E-09 | |||
| GTF3A | general transcription factor IIIA | 13q12.3-q13.1 | 445977 | 2.02 | 2.10E-09 | |||
| HIST1H2BK | histone cluster 1, H2bk | 6p21.33 | 437275 | 2.57 | 5.35E-08 | |||
| HOXA11 | homeobox A11 | 7p15-p14 | 249171 | 0.30 | 9.38E-17 | |||
| KHDRBS3 | KH domain, RNA binding, signal transduction associated 3 | 8q24.2 | 444558 | 0.48 | 3.46E-06 | |||
| NRIP1 | nuclear receptor interacting protein 1 | 21q11.2 | 155017 | 0.44 | 2.26E-06 | |||
| SHOX | short stature homeobox | Xp22.3;Yp11.3 | 105932 | 2.26 | 7.35E-05 | |||
| ZMIZ1 | zinc finger, MIZ-type containing 1 | 10q22.3 | 193118 | 0.40 | 3.11E-09 | |||
| Signal Transduction | ||||||||
| PDE10A | phosphodiesterase 10A | 6q27 | 348762 | 0.23 | 5.21E-17 | |||
| DDEF2 | development and differentiation enhancing factor 2 | 2p25|2p24 | 555902 | 0.43 | 1.88E-10 | |||
| DUSP4 | dual specificity phosphatase 4 | 8p12-p11 | 417962 | 0.48 | 6.89E-17 | |||
| FARP1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 | 13q32.2 | 403917 | 0.43 | 6.83E-10 | |||
| GCG | glucagon | 2q36-q37 | 516494 | 24.89 | 2.51E-05 | |||
| GNAI1 | guanine nucleotide alpha inhibiting activity polypeptide 1 | 7q21 | 134587 | 0.33 | 3.40E-09 | |||
| GNG12 | guanine nucleotide binding protein (G protein), gamma 12 | 1p31.3 | 696244 | 0.42 | 3.05E-06 | |||
| IRS2 | insulin receptor substrate 2 | 13q34 | 442344 | 0.24 | 9.21E-20 | |||
| PLCB1 | phospholipase C, beta 1 (phosphoinositide-specific) | 20p12 | 431173 | 0.33 | 4.87E-13 | |||
| PSD3 | pleckstrin and Sec7 domain containing 3 | 8pter-p23.3 | 434255 | 0.45 | 4.76E-13 | |||
| RIN2 | Ras and Rab interactor 2 | 20p11.22 | 472270 | 0.43 | 1.92E-07 | |||
| Miscellaneous | ||||||||
| ALDH7A1 | aldehyde dehydrogenase 7 family, member A1 | 5q31 | 483239 | 3.45 | 2.93E-04 | |||
| APLP1 | amyloid beta (A4) precursor-like protein 1 | 19q13.1 | 74565 | 2.64 | 1.53E-07 | |||
| B4GALT5 | beta 1,4- galactosyltransferase, polypeptide 5 | 20q13.1-q13.2 | 370487 | 2.04 | 3.45E-04 | |||
| BDH2 | 3-hydroxybutyrate dehydrogenase, type 2 | 4q24 | 124696 | 0.49 | 2.12E-07 | |||
| BLVRB | biliverdin reductase B | 19q13.1-q13.2 | 515785 | 2.14 | 1.42E-06 | |||
| BTBD3 | BTB (POZ) domain containing 3 | 20p12.2 | 696121 | 0.44 | 5.95E-12 | |||
| C12orf24 | chromosome 12 open reading frame 24 | 12q24.11 | 436618 | 2.24 | 1.50E-08 | |||
| C18orf1 | chromosome 18 open reading frame 1 | 18p11.2 | 149363 | 0.46 | 6.59E-21 | |||
| C1orf9 | chromosome 1 open reading frame 9 | 1q24 | 204559 | 2.66 | 1.66E-06 | |||
| CHST1 | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 11p11.2-p11.1 | 104576 | 0.44 | 2.26E-12 | |||
| CHST2 | carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 | 3q24 | 8786 | 0.37 | 1.36E-17 | |||
| COL4A2 | collagen, type IV, alpha 2 | 13q34 | 508716 | 0.26 | 5.75E-13 | |||
| DCHS1 | dachsous 1 (Drosophila) | 11p15.4 | 199850 | 0.40 | 6.00E-07 | |||
| EMP3 | epithelial membrane protein 3 | 19q13.3 | 9999 | 2.14 | 7.03E-07 | |||
| FAM59A | family with sequence similarity 59, member A | 18q12.1 | 444314 | 0.45 | 1.03E-07 | |||
| FER1L3 | fer-1-like 3, myoferlin (C. elegans) | 10q24 | 655278 | 4.77 | 5.83E-05 | |||
| FLJ13197 | hypothetical FLJ13197 | 4p14 | 29725 | 0.34 | 6.73E-17 | |||
| PLBD1 | Putative phospholipase B-like 1 Precursor | 12p13.1 | 131933 | 3.30 | 7.11E-10 | |||
| FXYD6 | FXYD domain containing ion transport regulator 6 | 11q23.3 | 635508 | 0.36 | 3.64E-07 | |||
| GRB10 | growth factor receptor-bound protein 10 | 7p12-p11.2 | 164060 | 0.39 | 3.73E-13 | |||
| GLUL | glutamate-ammonia ligase | 1q31 | 518525 | 0.47 | 1.27E-04 | |||
| GYG1 | glycogenin 1 | 3q24-q25.1 | 477892 | 2.73 | 2.15E-05 | |||
| KIAA0644 | Unknown | 7p15.1 | 21572 | 0.19 | 8.59E-28 | |||
| LIMCH1 | LIM and calponin homology domains 1 | 4p13 | 335163 | 0.39 | 1.27E-13 | |||
| NIT2 | nitrilase family, member 2 | 3q12.2 | 439152 | 2.08 | 5.12E-10 | |||
| PFDN4 | prefoldin subunit 4 | 20q13.2 | 91161 | 2.00 | 3.74E-05 | |||
| PGAM1 | phosphoglycerate mutase 1 | 10q25.3 | 592599 | 2.09 | 5.85E-08 | |||
| PLP2 | proteolipid protein 2 | Xp11.23 | 77422 | 3.36 | 8.00E-12 | |||
| PLTP | phospholipid transfer protein | 20q12-q13.1 | 439312 | 2.79 | 9.52E-06 | |||
| PPP3CA | protein phosphatase 3, catalytic subunit, alpha isoform | 4q21-q24 | 435512 | 0.28 | 9.39E-07 | |||
| SEPT11 | septin 11 | 4q21.1 | 128199 | 0.48 | 1.34E-07 | |||
| SH3BP4 | SH3-domain binding protein 4 | 2q37.1-q37.2 | 516777 | 0.37 | 3.70E-10 | |||
| SNX10 | sorting nexin 10 | 7p15.2 | 571296 | 2.16 | 2.72E-05 | |||
| ST6GAL1 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 3q27-q28 | 207459 | 0.43 | 1.24E-12 | |||
| TNNT1 | troponin T type 1 (skeletal, slow) | 19q13.4 | 631558 | 5.14 | 5.14E-07 | |||
| TWF1 | twinfilin, actin-binding protein, homolog 1 | 12q12 | 189075 | 2.03 | 2.46E-04 | |||
Also involved in neural development
Also involved in tumor invasion/metastasis
Decreased Expression of SMARCB1 and Associated Genes in Rhabdoid Tumors
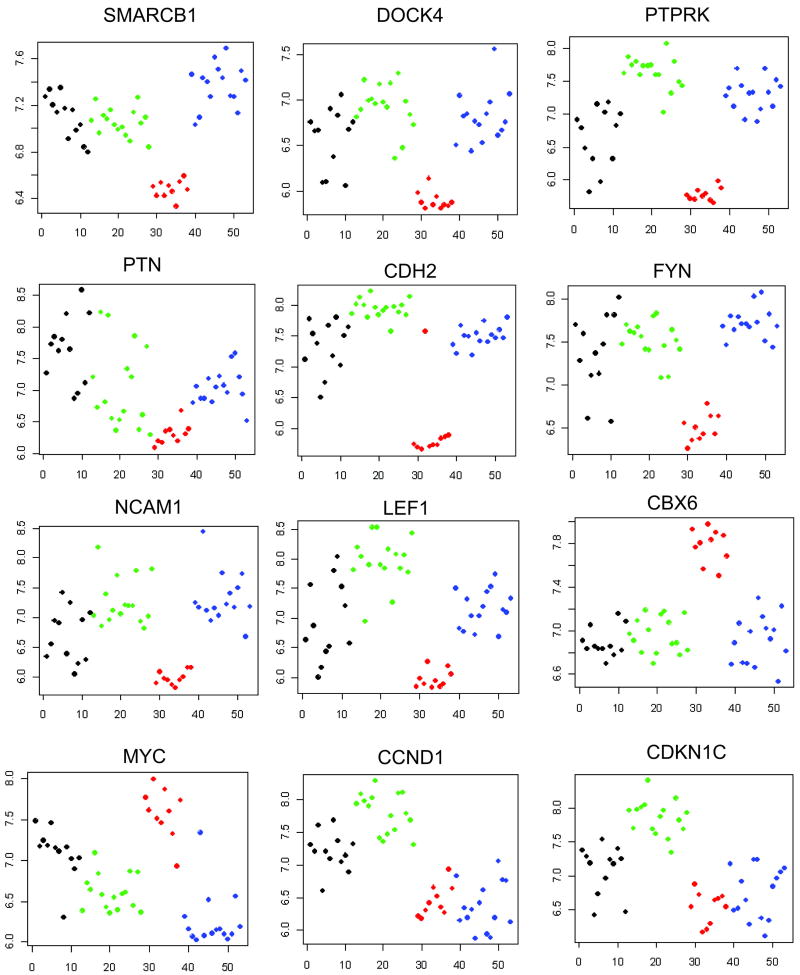
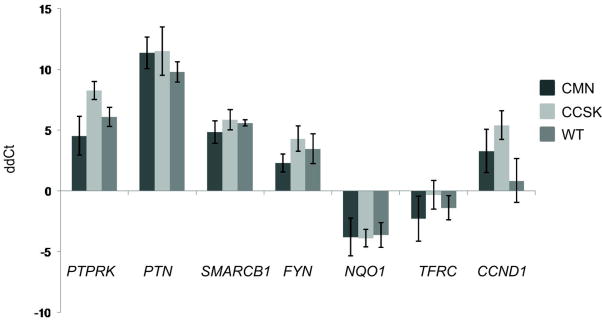
SMARCB1 was within the top five genes most significantly differentially expressed in RT, and 12 of the top 114 genes in Table 2 have been previously shown to be concordantly expressed with SMARCB1 in prior gene expression analyses. These include genes previously demonstrated to be concordantly differentially expressed following induction of SMARCB1 within RT cell lines (ATP1B1, PTN, SPOCK1, DOCK4, SERPINE2), and following inactivation of SMARCB1 within murine embryonic fibroblasts (NQO1, PLOD1, PTP4A2, PTPRK).12 13 14 In addition, Pomeroy et al compared RT with four pediatric central nervous system tumors and provided the top 100 genes characterizing each tumor type. Of the resulting 500 genes, 99 were also found in our Supplemental Table 1 and were concordantly regulated (designated by an asterisk); two genes, COL5A2 and RSUI, were identified in Table 2.15 Lastly, SPP1 was shown to be significantly up-regulated in human RT.16 The gene expression patterns of SMARCB1 and selected SMARCB1 associated genes are illustrated in Figure 1. The down-regulation of SMARCB1, PTN, and PTPRK, and the up-regulation of NQO1 mRNA was confirmed by QRT-PCR (Figure 2). Absence of expression of the SMARCB1 protein was established in all RTs tested by immunohistochemistry, with strong nuclear positivity identified in all CCSK, CMN, and WT tested. In contrast, antibodies against NQO1 demonstrated strong positivity in all RTs and pale to no staining for the remaining tumor types (Supplemental Figure 1).
Figure 1. Patterns of gene expression within the different groups of pediatric renal tumors.
The log expression levels (low to high) are plotted on the Y-axis. The X axis reflects an arbitrary tumor number, grouping the different tumor types starting with the congenital mesoblastic nephromas (infantile fibrosarcomas) in black, followed by clear cell sarcoma of the kidney in green, rhabdoid tumors in red, and Wilms tumors in blue.
Figure 2. Validation of expression of SMARCB1, FYN, PTN, NQO1, PTPRK, TFRC, and CCND1 using quantitative reverse transcriptase polymerase chain reaction.
RNA from 7 examples of each tumor type were analyzed in duplicate using the primers in Table 1. Expression changes are determined by subtracting the Ct value of the housekeeping gene (B-actin or GAPDH) from the Ct of the gene of interest (dCt). The average dCT from each individual tumor was subtracted from the average tumor RT dCt value for each gene (ddCT); these values were averaged for each tumor type and the error bars represent the standard deviation of the averages.
Decreased Expression of Genes Associated with Neural Development
Strikingly, 28 (25%) of the genes in Table 2 are involved with neural development and all were sharply down-regulated. This includes SMARCB1 itself and four genes directly associated with SMARCB1 that have previously been shown to be important in neural development (PTN, DOCK4, SPOCK1, and PTPRK). These findings are supported by analysis in PANTHER, in which the Neurogenesis group was within the top five most enriched groups of biological processes (Supplemental Table 3). To obtain more information regarding specific developmental pathways, GSEA was performed using selected groups in Gene Ontology that pertain to early development pathways.11 Significant enrichment was identified in non-RT compared with RT (consistent with down-regulation in RT) within the following groups: GO:0022008 Neurogenesis (normalized enrichment score (NES) -1.59, nominal p value=0.006, false discovery rate (FDR) 2%, GO:0048666 Neuron Development (NES -1.53, p=0.008, FDR 4%), GO:0030182 Neuron Differentiation (NES -1.48, p=0.008, FDR 5%), GO:0051960 Regulation of Nervous System Development (NES -1.44, p=0.009, FDR 5%). Many genes are redundant within these GO groups, therefore a common gene list containing all genes in all the above groups was developed and is illustrated in order of rank within GSEA in Supplemental Figure 2A.
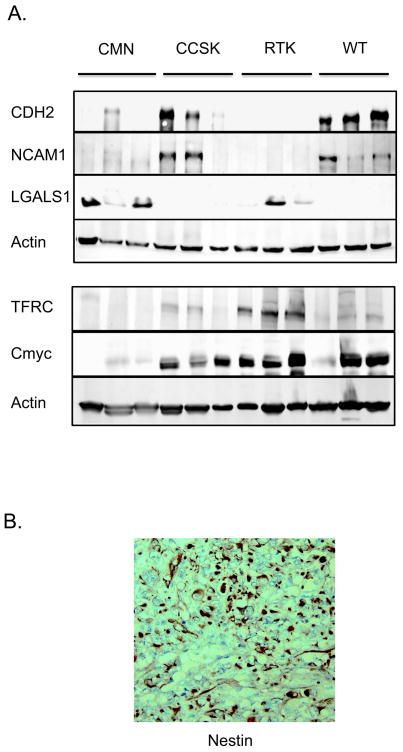
Of particular interest was the highly significant enrichment of the two available GO groups containing genes pertaining to neural crest development in non-RT (down-regulation in RT). This includes GO:0014033 Neural Crest Development (NES -1.65, p<0.001, FDR 2%), and GO:0001755 Neural crest migration (NES -1.60, p<0.001, FDR 2%). A GSEA heatmap of all genes in both lists in rank order is illustrated in Supplemental Figure 3A. There was minimal overlap between the enriched genes of the neural development groups and those of the neural crest group. In contrast, gene sets within other early developmental GO categories (including GO:0045445 Myoblastic Differentiation, GO:0051216 Cartilage Development, GO:0001501 Skeletal development, GO:0048513 Organ development, GO:0042692 Muscle Cell Differentiation) did not show significant enrichment (defined as nominal p value<0.05, FDR<20%) in RT or non-RT. Lastly, a recently published comprehensive list of genes involved in renal development was entered into GSEA and did not show significant enrichment.17 Of note, CCSKs demonstrated increased expression of genes involved with nervous system development (but not neural crest development), as we have previously reported.18 Therefore all the above analyses of RT vs non-RT and fetal kidney were repeated excluding CCSKs, and all groups retained their significance (illustrated in Supplemental Figures 2B, 3B). RNA expression patterns of selected neural developmental genes from Table 2 are shown in Figure 1. Decreased expression of FYN, PTN, and PTPRK was verified by QRT-PCR (Figure 2). Western analysis of key neural regulators CDH2 (N-cadherin) and NCAM (CD56) proteins were entirely negative in all RT tested, with expression in 2/3 CCSKs and in all three WT tested; CMNs showed no to low expression for both markers (Figure 3A).
Figure 3. Protein expression in pediatric renal tumors.
A. Immunoblotting of CDH2, NCAM, LGALS1, MYC-C and TFRC
Protein lysates from 3 examples of each tumor type were analyzed using antibodies provided in Table 1. Decreased expression of neural markers CDH2 and NCAM is identified in RT. Increased expression of oncogene LGALS1 is identified in 2/3 RT tested, as well as in two CMNs. Increased expression of MYC-C and one of its key targets (TFRC) is identified in RT.
B. Nestin expression in RT.
Nestin mRNA was not differentially expressed in the pediatric renal tumors; however Nestin protein cytoplasmic and nuclear expression was identified in all RT tested using immunohistochemistry.
Several genes in Table 2 are involved in Wnt signaling (PTN, DOCK4, CDH2, LEF1, ENC1, FSTL1) or in Notch signaling (HES1, NOTCH2), and all were down-regulated. Both Wnt and Notch pathways are known to be critical to neural development. Significant negative enrichment in RT for the GO:0007219 Notch signaling pathway was identified (NES -1.48, p=0.02, FDR 5%, Supplemental Figure 3C,D). However, independent gene sets available within GSEA that contain targets of Wnt signaling showed no significant enrichment of Wnt targets. When the 84 genes with human homologues on the HU133A array from the Stanford gene set of known Wnt targets was entered into GSEA and analyzed (www.stanford.edu/~musse/Wntwindow.html), there was likewise no significant enrichment in RT. Immunohistochemistry for beta-catenin (CTNNB1) showed no nuclear staining and focal, pale cytoplasmic staining in 2/6 tumors, the remaining RTs were entirely negative (data not shown).
Expression of Neural Stem Cell Markers in Rhabdoid Tumor
The striking down-regulation of neural developmental markers suggests that RTs may arise within neural or neural crest stem cells followed by developmental arrest. We therefore investigated the mRNA expression of markers of known neural and neural crest stem cells, including MSI1 (Musashi 1), CD133 ( Prominen 1), FOXD3, ID3, SOX10, SNAI2, and SNAI1. Of these, only CD133 was found in Supplemental Table 1 and this was down-regulated (fold change 0.3, p=0.0002). Because these transcription factors may be regulated at very low levels, or translationally regulated, those neural crest and neural crest stem cell markers for which robust commercially available antibodies were available were analyzed, including SOX10, ID3, Musashi, and CD133. All were negative in RT with appropriate positive control staining. Of interest, antibodies directed against nestin, a marker of primitive neural as well as mesenchymal stem cells, demonstrated strong cytoplasmic positivity and scattered nuclear positivity in all RTs, with appropriate negative controls (Figure 3B). Embryonic stem cell markers OCT4, NANOG, and SOX2 did not show differential mRNA expression or upregulation in RT.
Expression of MYC-C and its Targets
MYC-C is known to directly interact with SMARCB1,19,20 and was significantly up-regulated in RT (fold change 2.37, p value 1.4 × 10-7, Supplemental Table 1, Figure 1). To validate the potential role for MYC-C upregulation in RT, the independently curated C2 gene sets within GSEA that contain MYC-C targets were analyzed. Significant enrichment was documented using gene sets Schumacher_MYC_UP (NES 1.56, p=0.002, FDR 1%) MYC_TARGETS (NES 1.44, p=0.03, FDR 5%), and MYC_ONCOGENIC SIGNATURES (NES 1.6, p=0.01, FDR 0.1%).21-23 A combined list containing all genes in each list was analyzed and illustrated in Supplemental Figure 4. Within Table 2, 13/114 (11%) genes are known targets of MYC-C and are coordinately expressed in RT (including two genes known to be down-regulated by MYC-C). Lastly, key cell cycle genes regulated by MYC-C show appropriate expression in RT, including CCND1, CCND2, CCNE1, CDKN1A, CDKN1C, and CDKN2B (see below). Confirmation of expression of MYC-C and TFRC (a MYC-C target) proteins was performed and illustrated in Figure 3A; confirmation of mRNA expression of TFRC using QRT-PCR was also performed (Figure 2).
Cell Cycle Gene Expression
Numerous studies have shown that re-expression of SMARCB1 in RT-derived cell lines results in up-regulation of CDKN1A (p21CIP/WAF1) and CDKN2A (p16INK4a) and down-regulation of CCND1 (cyclin D1), CCNE1 (cyclin E), CCNA1 (cyclin A), with secondary phosphorylated RB protein, cellular senescence and apoptosis.24-26 Most of these findings are confirmed in the human RT analyzed in our study, with some exceptions. Of the cyclin dependent kinase inhibitors, both CDKN1A and CDKN2A are expressed at very low levels in all renal tumor types tested, including RT. CDKN1C (p57kip2) mRNA was also significantly down-regulated in RT (fold change 0.4; p=2.15×10-6) (Supplemental Table 1, Figure 1). Immunohistochemistry for CDKN1A and CDKN1B showed staining of fewer than 1% of tumor cells in all renal tumors tested (data not shown). Of the cyclin-dependent kinases, CCND2 and CCNE mRNAs were not differentially expressed but were upregulated in all tumors, and antibodies to both cyclin E and cyclin D2 confirmed strong nuclear staining within the majority of RT cells (data not shown). Immunostaining for p53 was positive in fewer than 1% of RT nuclei. In contrast with prior studies, CCND1 mRNA was down-regulated in RT in our study (FC 0.4, p=1.2×10-5, Figure 1). This was confirmed by quantitative RT-PCR (Figure 2) and immunohistochemistry showed protein expression in fewer than 1% of RT cells in the face of strong nuclear positivity in CCSKs, WT and CMN (Supplemental Figure 1). (Of note, cyclin D2 and cyclin E are activated by CMYC, 27 whereas CMYC has been shown to repress CCND128, CDKN1A, and CDKN2B.21)
Differential Expression of Genes Associated With Tumor Suppression, Invasion, and Metastasis
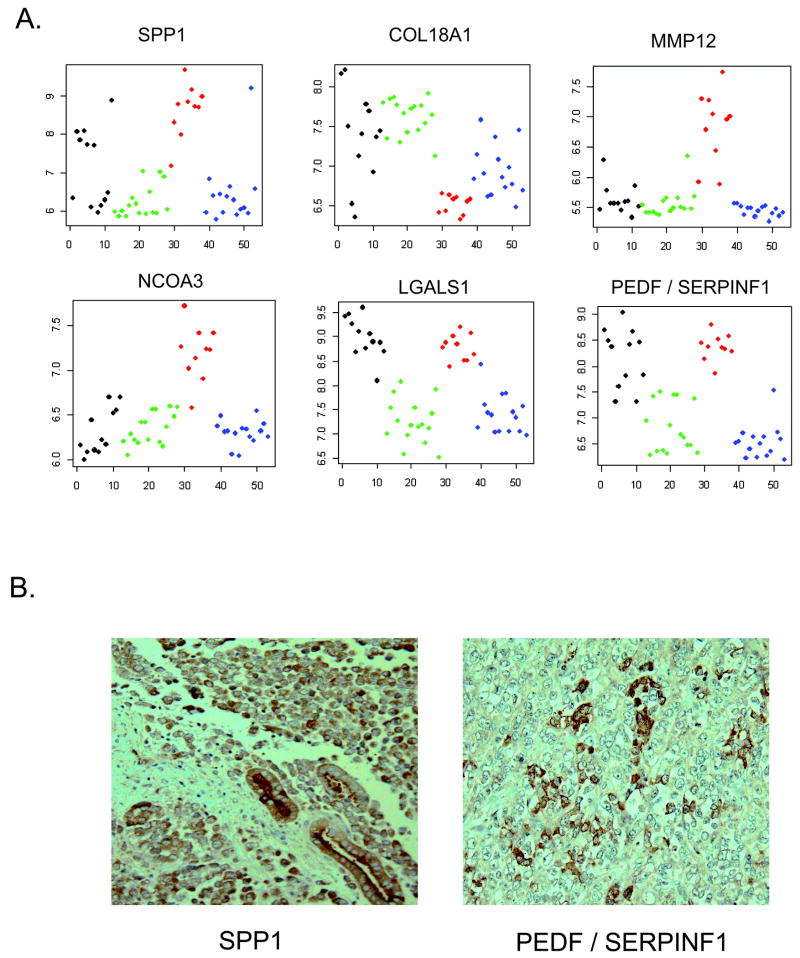
Many of the top 114 genes in Table II are recognized oncogenes yet were down-regulated in RT. Conversely, ten genes associated with tumor suppression, invasion, or metastasis demonstrated regulation in RT concordant with their activity and therefore represent potential therapeutic targets. SPP1 (osteopontin), MMP12, NCOA3, TFRC, RSU1, and ZNF217 are associated with tumor invasion and metastasis and were up-regulated; SELENBP, COL18A1 (endostatin), PTPRK, and DOCK4 have tumor suppression functions and were down-regulated (Table 2, Figures 1 and 4). Of these, PTPRK and DOCK4 have been previously directly associated with SMARCB1 expression.12 13 Additional potential therapeutic targets found in Supplemental Table 1 include LGALS1 (fold change 1.7, p 1.07×10-5) and SERPINF1 (PEDF) (fold change 2.3, p 3.4×10-11). These were not included in the top gene list because of their high expression in CMN as well as RT (Figure 4). Both LGALS1 and SPP1 were significantly up-regulated in previous gene expression analysis studies.15,16 Differential expression of TFRC and PTPRK were validated by QRT-PCR (Figure 2). Expression of SPP1 (osteopontin), PEDF, and TFRC proteins in RT were demonstrated by immunohistochemistry or immunoblotting (Figures 3, 4).
Figure 4. Expression of selected genes involved in tumor invasion and metastasis.
A) Pattern of RNA expression of key genes. The expression levels (low to high) are plotted on the Y-axis. The X axis groups the different tumor types starting with the congenital mesoblastic nephromas (infantile fibrosarcomas) in black, followed by clear cell sarcoma of the kidney in green, rhabdoid tumors in red, and Wilms tumors in blue.
B) Protein expression of SPP1 and PEDF (SERPINF1). The majority of cells within all RT show moderate positivity for SPP1. Note the striking positivity of non-neoplastic renal tubules for SPP1. Clusters of cells within RT show strong positivity for PEDF.
Expression of Genes Associated with Transcription Regulation
In addition to MYC-C, a number of genes involved in transcription regulation and signal transduction were upregulated in RT. Of particular note, CBX6 was highly upregulated (Figure 1, Table 2). CBX6 is a member of the polycomb family which is responsible for transcriptional repression mediated by the specific H3K27 histone methylation. Our data show loss of expression of GRB10 and CDKN1C in RT, both of which are epigenetically regulated by the specific H3K27 trimethylation mediated by the polycomb group.29,30 RTs also showed loss of expression of multiple HOX genes, long known to be repressed by polycomb group proteins.31 (In Supplemental Table 1, HOXA10, HOXA11, HOXC4, HOXD3, HOXD4, HOXD9, HOXD10, HOXD11 are all downregulated). In contrast to the polycomb group, the trithorax family is responsible for transcriptional activation mediated by the specific H3K4 methylation. MLL1, the predominant member of the trithorax family in humans, is known to achieve transcriptional activation by interacting directly with SMARCB1.32 Of further interest in this regard is the newly described role of ZNF217 as an organizer of repressive histones. ZNF217 (an oncogene significantly upregulated in RT, Table 2) has been show to demethylate H3K4me3 and to methylate H3K27 through interaction with EZH2, a member of the polycomb repressive complex 2.33 Therefore, the transcriptional repression seen in RT may be associated with histone 3 modifications.
To investigate this hypothesis, we relied on the availability of whole genomic surveys of methylation markers within embryonic stem cells (ESC). Zhao et al classified the H3K4me3 and H3K27me3 markers associated with over 17,000 genes in ESC and established three groups of genes: Group 1 with neither H3K4 nor H3K27me3 modification, Group 2 with only H3K4me3 modification, and Group 3 with both H3K4 and H3K27me3 (bivalent).34 We placed the gene list from each group in GSEA to determine if their expression was enriched in RT or non-RT. The expression of genes in Groups 1 and 2 did not show significant enrichment in either RT or non-RT. However, the 1141 genes in Group 3 (those bivalently modified in ESC) showed significant enrichment in non-RT, indicating decreased expression in RT (NES -1.5, p=0.01, FDR 5%, Supplemental Figure 5A). When the other pediatric renal tumors were similarly analyzed (leaving RT out), no significant enrichment was identified in either CMN or WT. In contrast, CCSK showed significant positive enrichment for Group 3 genes. Therefore we again analyzed Group 3 genes comparing RT to non-RT leaving out the CCSKs, and significant enrichment non-RT was retained (p<0.001, Supplemental Figure 5B). Of note, 11% of genes in Supplemental Table 1 are Group 3 genes, and 90% of these genes show decreased expression in RT. Zhao et al. also demonstrated the top GO categories differentially represented through PANTHER in Group 3 to be genes important for developmental processes, ectoderm development, neurogenesis, transcription regulation, and signal transduction.34 Within RT, with the exception of transcription regulation, using PANTHER we found the same top categories at the top of our list (Supplemental Table 3). Lastly, we performed a similar in silico analysis of 512 targets of Polycomb Repressive Complex 2 in ESC.35 GSEA analysis again revealed significant enrichment in non-RT (NES -1.4, p=0.05, FDR 9%).
Discussion
Rhabdoid tumors (RT) are rare tumors that are virtually confined to infancy and are characterized by loss of the SMARCB1/hSNF5/INI-1 gene.4,36 SMARCB1 is one of over ten non-catalytic subunits of the highly evolutionarily conserved SWI/SNF ATP-dependent chromatin-remodeling complex expressed in all normal cells at all stages of development.6 SMARCB1 is recognized as a tumor suppressor gene due to its biallelic involvement in the development of RT, reviewed by Biegel et al. 36 The SWI-SNF complex appears to relieve repressive chromatin structures by disrupting the DNA-histone interaction within nucleosomes, allowing the transcriptional machinery to access its targets more effectively, facilitating transcriptional activation and repression (previously reviewed 37,38). The current hypothesis is that SMARCB1, through its ability to bind to several proteins, serves to recruit the SWI/SNF complex to specific target sites thereby mediating transcription regulation.38,39 However, precisely how SMARCB1 accomplishes this is largely unknown. Proteins that have been demonstrated by others to physically interact with SMARCB1 or to be direct targets of SMARCB1 include MLL, MYC-C, PTN, HESR1, ATP1B1, and FZD7.12,20,32 Our gene expression analysis demonstrates key roles for these genes and others in the development of human RT, as discussed below.
SMARCB1 loss results in repression of genes critical to neural and neural crest development
Genes involved with neural and neural crest development were sharply down-regulated in RT. A number of recent studies have shown that SWI/SNF itself is required for normal vertebrate neurogenesis.40,41 Pleiotrophin, PTN has been shown to directly interact with SMARCB112 and to regulate the balance between differentiation and proliferation within neural stem cells by inhibiting proliferation and enhancing differentiation.42 The Notch signaling pathway is also critical for controlling the induction of neural development and for maintaining the neural progenitor population.43 Within the Notch pathway, HESR1 (HEY1) is a known direct target of SMARCB1,12 and recruitment of SMARCB1 to the HES1 and HES5 promoters has been demonstrated. NOTCH2 and HES1 (the ligand of HESR1) were both down-regulated in RT (Table 2) and GSEA analysis confirmed the down-regulation of the Notch signaling pathway in RT. Lastly, a large number of important downstream transcription factors involved with neural and neural crest development were likewise sharply down-regulated, including NCAM (neural cell adhesion molecule, regulated by HES1), CDH2 (neural-cadherin), FYN (a downstream target of PTN), and SOX11, which induces differentiation within neural stem cells and regulates various aspects of neural crest development.44 In summary, RTs show striking repression of neural differentiation, suggesting the possibility that RTs may arise in progenitor cells in which SMARCB1 loss results in prevention of neural development. This is supported by neural differentiation that develops within cell lines derived from both CNS and extra-CNS soft tissue RTs when SMARCB1 is re-introduced.45
Neural development begins within the embryonic ectoderm from which the neural plate differentiates. The neural plate folds to form the neural tube which then differentiates into the structures of the central nervous system. Cells of the neural crest arise at the border between the neural plate and the ectoderm for the entire length of the neuraxis (previously reviewed46). Following induction, the neural crest cells migrate as undifferentiated precursor cells to various parts of the embryo where they differentiate into many cell types including cells of sensory neurons and glia of the peripheral nervous system, bone, cartilage and melanocytes. Therefore, the ability to show divergent differentiation, the ability to migrate over considerable distances, and presence in both CNS and extra-CNS sites are features shared by both neural crest stem cells and RTs. Our study reveals significant down-regulation of both neural and neural crest developmental genes. To address the hypothesis that RT develop within neural or neural crest stem cells with arrested development, we investigated the mRNA and protein expression of key neural and neural crest stem cell markers.46-50 Low to no expression of early neural and neural crest stem cell markers SNAIL1, SLUG, FOXD3, SOX10, ID3, CD133, Musashi was identified. Embryonic stem cell markers OCT4, NANOG, SOX2 were also not upregulated in RT. In contrast, strong expression of nestin was identified in RT. Nestin has been shown to be abundantly expressed in embryonic stem cell-derived progenitor cells that have the potential to develop into neuroectodermal, endodermal and mesodermal lineages.51 Our data therefore suggest that RTs do not arise in neural or neural crest stem cells per se, but instead may arise in early progenitor cells following the differentiation trigger (and following loss of ESC markers) in cells destined to become neural and/or neural crest stem cells. Loss of SMARCB1 at this critical period of time may suppress neural and/or neural crest stem cell development.
Transcriptional repression may play a role in RT development
Clues toward the mechanisms involved in this developmental repression are provided by evidence that SMARCB1 loss may result in alterations in the activity of the trithorax and polycomb families which are responsible for maintaining the reciprocal transcriptional states of key developmental regulators.52 MLL1, the key member of the trithorax group in humans, is a ubiquitously expressed nuclear protein that has been shown to interact directly with SMARCB1 through the MLL SET domain, resulting in remodeling of chromatin and transcriptional activation.32 MLL1 mediates transcriptional activation by catalyzing the methylation of histone H3 lysine 4 (H3K4me).53 MLL1 has also been shown to be required for neurogenesis.54 In contrast, the polycomb group (Pcg) mediates transcriptional repression through catalyzing the methylation of histone H3 lysine 27 (H3K27me).31 These critical histone 3 modifications have been comprehensively mapped across the genome in both human and mouse embryonic stem cells using varied experimental approaches.34,55,56 These studies, which show striking concordance, demonstrate that the majority of genes in ESC are marked with H3K4 alone (50-58%) or in combination with H3K27 (bivalent, 10-17%), whereas 28-33% show neither marking. The majority of genes with H3K4 methylation alone are associated with basic cellular functions such as housekeeping genes, proliferation, protein and DNA metabolism, etc. In contrast, genes bivalently modified were enriched in GO developmental functions, particularly those involving neurogenesis, ectoderm differentiation, transcriptional regulation and signal transduction. These bivalently marked genes overlapped considerably with genes demonstrated to be polycomb group targets 35. Indeed, bivalent genes appear to be dominated by the H3K27 repressive markers and to be down-regulated in ESC. The presence of the H3K4 methylation in these bivalently modified histones may serve to enable a sharp increase in expression of lineage specific transcription markers following the trigger for differentiation.34,35,55,56
Our study demonstrates that the same developmental pathways and genes that are marked by bivalently modified histones and dowregulated within ESC are likewise significantly down-regulated in RT. These include the processes of neurogenesis, signal transduction, and ectoderm differentiation (Supplemental Table 3). Further, genes bivalently modified in ESC were among those that most strongly differentiated RT from non-RT by GSEA (Supplemental Figure 5). Analyses of independent sets of genes known to be polycomb targets35 likewise demonstrated the same findings. These analyses suggest that RT may arise within early stem cells whose bivalently modified histones remain dominated by the polycomb group histone markings. At the time of differentiation, this repression would generally be overcome by SWI/SNF, but in the absence of SMARCB1 this does not occur. Indeed, there is growing evidence that SWI/SNF may play an integral role in the balance between trithorax and polycomb group activity through the direct interaction between SMARCB1 and MLL1. This is best seen in a series of experiments that focused on the colocalized p16/INK4a and p14/ARF tumor suppressor genes. P16 is known to be repressed due to polycomb group-mediated H3K27 methylation, whereas the nearby p14 is not.57 Re-expression of SMARCB1 within RT cell lines has been shown to result in induction of p16 but not p14.24,58 Kia et al. performed chromatin immunoprecipitation demonstrating that SMARCB1 bound strongly to the p16 promoter, but not to the p14 promoter. Further, in the absence of SMARCB1, polycomb group silencers (PRC1, PRC2) were present in large amounts on the p16 promoter, but in low amounts on the p14 promoter. Re-expression of SMARCB1 resulted in the following: 1)reduced binding of PRC1 and PRC2 and increased MLL1 binding at the p16 promoter, 2) reduced H3K27 and increased H3K4 methylation at the p16 promoter, and 3) activated p16.59 Based on these CDKN2A studies, Kia et al. concluded that polycomb group silencers are evicted by SWI/SNF, thereby allowing for chromatin modification. In the absence of SMARCB1, this did not occur. Our data suggest that this mechanism may occur more widely within bivalently modified genes in development. Indeed, a broader role of the SWI/SNF chromatin remodeling complex in development is suggested by studies showing that SMARCB1 is required for hepatocyte and adipocyte development.60,61
It should be noted that there are a number of different histone modifications in addition to H3 methylation that impact on gene expression. Furthermore, many histone modifications interact with one another, creating extreme complexity. To add to this potential complexity withn RT is the presence of MYC-C activation. MYC-C is a transcription factor that has long been known to drive a large number of diverse biological activities during development and oncogenesis. In recent years, it has become apparent that MYC-C may modulate transcription, in part at least, through H3 and H4 acetylation.62. A physical interaction between SMARCB1 and MYC-C proteins has been documented by several laboratories and the SNF complex acts directly to repress MYC-C during differentiation.19-20,27,63 Lastly, CMYC has been shown to be critical regulator of neural crest formation.64 It is clear that further investigation is needed in order to clarify the interface between the two broad processes involved in chromatin remodeling, namely histone modification and the ATP-dependent nucleosomal remodeling mediated by SWI/SNF.
RT show decreased cyclin dependent kinase inhibition
Numerous publications have implicated multiple members of the SWI/SNF complex in cell cycle control. Several laboratories have demonstrated down-regulation of cyclin-dependent kinase inhibitors CDKN1A and/or CDKN2A(p16/INK4a), and their up-regulation upon reintroduction of SMARCB1, accompanied by growth arrest.24-26,59 Mutation analysis of these genes in RT is not available in the literature. These observations would predict up-regulation of MYC, down-regulation of both CDKN1A and CDKN2A, and up-regulation of cyclins A and E within RT, all of which are confirmed in our study. We also demonstrate down-regulation of CDKN1C (p57Kip2), a major regulator of embryonic growth that has recently been shown to be a downstream target of SMARCB1.65 Of interest, both CDKN1C and CDKN2A are regulated in part through the polycomb group by methylation of H3K27.30 Therefore, our data supports that of other studies that indicate a proliferative advantage of RT through loss of cylin-dependent kinase inhibition directly linked to SMARCB1 loss.
Several previous studies based primarily on RT cell lines have suggested that cyclin D1 is a direct target of SMARCB1 and may be a possible therapeutic target.25-66,67 It was therefore surprising that neither our data nor gene expression following SMARCB1 induction in RT derived cell lines12 support these findings. Instead, our study demonstrates decreased expression of CCND1 at both the RNA and protein levels. A likely mechanism for down-regulation of CCND1 in RT is up-regulation of MYC-C, which is known to repress CCND1 expression at its promoter.28
Potential Therapeutic Targets
RTs are highly malignant and lethal tumors, and the ultimate goal of this study is to improve their clinical outcome. Analysis of gene expression reveals differential expression of a number of genes known to cause tumor suppression or tumor progression. In particular, two putative tumor suppressor genes have been previously shown to be directly associated with SMARCB1 and are down-regulated in RTs, PTPRK and DOCK4. PTPRK (protein-tyrosine phosphatase receptor-kappa) dephosphorylates EGFR, the prototypic receptor protein tyrosine kinase, and thereby regulates growth and survival.68 DOCK4 promotes beta-catenin stability and is disrupted during tumorigenesis through mutation and deletion in several human cancers.69,70 In addition, two known therapeutic targets differentially expressed in RT include SPPI and COL18A1. SPP1 (osteopontin) is a secreted phosphoprotein involved in immunity, angiogenesis, cell migration, and cell survival. Overexpression of SPP1 is associated with aggressiveness and metastasis in many different tumor types and is an independent predictor of behavior in melanomas, a tumor derived from the neural crest.71 Inhibition of SPP1 expression can reverse this phenotype.72 Previous reports have demonstrated elevated SPP1 plasma levels and expression levels in RT.73 SPP1 is cleaved into biologically active fragments by MMP12.74 Our study demonstrates both SPP1 and MMP12 to be upregulated in RT. C-terminal cleavage of COL18A1 (down-regulated in RT) results in the protein endostatin, a potent endogenous inhibitor of angiogenesis, migration and invasion (previously reviewed75). In clinical trials, recombinant endostatin inhibited the growth of a variety of tumors while exhibiting no apparent toxic side effects.76
Other genes involved in tumorigenesis or tumor progression were differentially expressed in RT. PEDF (pigmented epithelium-derived factor, or SERPINF1), has neuroprotective and antiangiogenic effects, suppresses cell-cycle progression, and supports neural stem-cell self-renewal.77 Notably, PEDF protein is broadly expressed in the nervous system and circulates in plasma and cerebrospinal fluid and therefore may be useful as a diagnostic marker. LGALS1 (upregulated in RT in both our study as well as that of Pomeroy et al 15) also has a well-documented role in tumorigenesis, metastasis, and invasion.78
Conclusions
The data presented suggest that RT arise within an early progenitor population during a critical developmental window. In these cells, loss of SMARCB1 results in 1) repression of neural development, 2) transcriptional repression that may be mediated by dysregulation of, or loss of interaction with, the trithorax/polycomb groups, 3) silencing of cyclin dependent kinase inhibitors, and 4) differential expression of a number of prominent genes that are known to promote malignant behavior and represent potential therapeutic targets.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the expertise of Donna Kersey, Patricia Beezhold, and Debra Haftl, who contributed greatly to this effort.
Work supported by NIH U01 CA114757 (EJP, CCH), and NIH U10 CA98543 (EJP).
Footnotes
Supplementary information is available at Laboratory Investigation’s website.
Disclosure/Duality of Interest: None
Reference List
- 1.Tomlinson GE, Breslow NE, Dome J, Guthrie KA, Norkool P, Li S, et al. Rhabdoid tumor of the kidney in the National Wilms Tumor Study: age at diagnosis as a prognostic factor. J Clin Oncol. 2005;23(30):7641–7645. doi: 10.1200/JCO.2004.00.8110. [DOI] [PubMed] [Google Scholar]
- 2.Rorke LB, Packer R, Biegel J. Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood. J Neurooncol. 1995;24(1):21–28. doi: 10.1007/BF01052653. [DOI] [PubMed] [Google Scholar]
- 3.Burger PC, Yu IT, Tihan T, Friedman HS, Strother DR, Kepner JL, et al. Atypical teratoid/rhabdoid tumor of the central nervous system: a highly malignant tumor of infancy and childhood frequently mistaken for medulloblastoma: a Pediatric Oncology Group study. Am J Surg Pathol. 1998;22(9):1083–1092. doi: 10.1097/00000478-199809000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive pediatric cancer. Nature. 1998;394(6689):203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 5.Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB. Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res. 2002;8(11):3461–3467. [PubMed] [Google Scholar]
- 6.Imbalzano AN, Jones SN. Snf5 tumor suppressor couples chromatin remodeling, checkpoint control, and chromosomal stability. Cancer Cell. 2005;7(4):294–295. doi: 10.1016/j.ccr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Huang CC, Cutcliffe C, Coffin C, Sorensen PH, Beckwith JB, Perlman EJ. Classification of malignant pediatric renal tumors by gene expression. Pediatr Blood Cancer. 2006;46(7):728–738. doi: 10.1002/pbc.20773. [DOI] [PubMed] [Google Scholar]
- 8.Hoot AC, Russo P, Judkins AR, Perlman EJ, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am J Surg Pathol. 2004;28(11):1485–1491. doi: 10.1097/01.pas.0000141390.14548.34. [DOI] [PubMed] [Google Scholar]
- 9.Argani P, Fritsch M, Kadkol SS, Schuster A, Beckwith JB, Perlman EJ. Detection of the ETV6-NTRK3 chimeric RNA of infantile fibrosarcoma/cellular congenital mesoblastic nephroma in paraffin-embedded tissue: application to challenging pediatric renal stromal tumors. Mod Pathol. 2000;13(1):29–36. doi: 10.1038/modpathol.3880006. [DOI] [PubMed] [Google Scholar]
- 10.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medjkane S, Novikov E, Versteege I, Delattre O. The tumor suppressor hSNF5/INI1 modulates cell growth and actin cytoskeleton organization. Cancer Res. 2004;64(10):3406–3413. doi: 10.1158/0008-5472.CAN-03-3004. [DOI] [PubMed] [Google Scholar]
- 13.Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci U S A. 2005;102(49):17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morozov A, Lee SJ, Zhang ZK, Cimica V, Zagzag D, Kalpana GV. INI1 induces interferon signaling and spindle checkpoint in rhabdoid tumors. Clin Cancer Res. 2007;13(16):4721–4730. doi: 10.1158/1078-0432.CCR-07-0054. [DOI] [PubMed] [Google Scholar]
- 15.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(6870):436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 16.Ma HI, Kao CL, Lee YY, Chiou GY, Tai LK, Lu KH, et al. Differential expression profiling between atypical teratoid/rhabdoid and medulloblastoma tumor in vitro and in vivo using microarray analysis. Childs Nerv Syst. 2009 doi: 10.1007/s00381-009-1016-2. [DOI] [PubMed] [Google Scholar]
- 17.Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, et al. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15(5):781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutcliffe C, Kersey D, Huang CC, Walterhouse D, Perlman EJ. Clear cell sarcoma of the kidney: upregulation of neural markers with activation of the sonic hedgehog and Akt pathways. Clin Cancer Res. 2005;11(22):7986–7994. doi: 10.1158/1078-0432.CCR-05-1354. Ref Type: Journal (Full) [DOI] [PubMed] [Google Scholar]
- 19.Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66(3):1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 20.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22(1):102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 21.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29(2):397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 24.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21(34):5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- 25.Zhang ZK, Davies KP, Allen J, Zhu L, Pestell RG, Zagzag D, et al. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22(16):5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65(22):10192–10198. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- 27.Amati B, Frank SR, Donjerkovic D, Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim Biophys Acta. 2001;14713:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- 28.Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, et al. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14(6):4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamasaki-Ishizaki Y, Kayashima T, Mapendano CK, Soejima H, Ohta T, Masuzaki H, et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol Cell Biol. 2007;27(2):732–742. doi: 10.1128/MCB.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36(12):1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 31.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 32.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, et al. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci U S A. 1998;95(8):4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banck MS, Li S, Nishio H, Wang C, Beutler AS, Walsh MJ. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics. 2009;4(2):100–106. doi: 10.4161/epi.4.2.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1(3):286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 36.Biegel JA, Kalpana G, Knudsen ES, Packer RJ, Roberts CW, Thiele CJ, et al. The role of INI1 and the SWI/SNF complex in the development of rhabdoid tumors: meeting summary from the workshop on childhood atypical teratoid/rhabdoid tumors. Cancer Res. 2002;62(1):323–328. [PubMed] [Google Scholar]
- 37.Stojanova A, Penn LZ. The role of INI1/hSNF5 in gene regulation and cancer. Biochem Cell Biol. 2009;87(1):163–177. doi: 10.1139/O08-113. [DOI] [PubMed] [Google Scholar]
- 38.Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4(2):133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 39.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13(2):136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 40.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132(1):105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 41.Aigner S, Denli AM, Gage FH. A novel model for an older remodeler: the BAF swap in neurogenesis. Neuron. 2007;55(2):171–173. doi: 10.1016/j.neuron.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Hienola A, Pekkanen M, Raulo E, Vanttola P, Rauvala H. HB-GAM inhibits proliferation and enhances differentiation of neural stem cells. Mol Cell Neurosci. 2004;26(1):75–88. doi: 10.1016/j.mcn.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 43.Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem. 2003;278(45):44808–44815. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- 44.Hong CS, Saint-Jeannet JP. Sox proteins and neural crest development. Semin Cell Dev Biol. 2005;16(6):694–703. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Albanese P, Belin MF, Delattre O. The tumour suppressor hSNF5/INI1 controls the differentiation potential of malignant rhabdoid cells. Eur J Cancer. 2006;42(14):2326–2334. doi: 10.1016/j.ejca.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 46.Barembaum M, Bronner-Fraser M. Early steps in neural crest specification. Semin Cell Dev Biol. 2005;16(6):642–646. doi: 10.1016/j.semcdb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38(1):17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 48.Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130(3):483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- 49.Light W, Vernon AE, Lasorella A, Iavarone A, LaBonne C. Xenopus Id3 is required downstream of Myc for the formation of multipotent neural crest progenitor cells. Development. 2005;132(8):1831–1841. doi: 10.1242/dev.01734. [DOI] [PubMed] [Google Scholar]
- 50.Corti S, Nizzardo M, Nardini M, Donadoni C, Locatelli F, Papadimitriou D, et al. Isolation and characterization of murine neural stem/progenitor cells based on Prominin-1 expression. Exp Neurol. 2007;205(2):547–562. doi: 10.1016/j.expneurol.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 51.Wiese C, Rolletschek A, Kania G, Blyszczuk P, Tarasov KV, Tarasova Y, et al. Nestin expression--a property of multi-lineage progenitor cells? Cell Mol Life Sci. 2004;61(19-20):2510–2522. doi: 10.1007/s00018-004-4144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103(17):6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL Targets SET Domain Methyltransferase Activity to Hox Gene Promoters. Molecular Cell 2 A D. 10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 54.Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458(7237):529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6(11):846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 58.Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LM, Mohd-Sarip A, et al. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279(5):3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 59.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28(10):3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24(18):3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caramel J, Medjkane S, Quignon F, Delattre O. The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene. 2008;27(14):2035–2044. doi: 10.1038/sj.onc.1210847. [DOI] [PubMed] [Google Scholar]
- 62.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes and Development. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bagnasco L, Tortolina L, Biasotti B, Castagnino N, Ponassi R, Tomati V, et al. Inhibition of a protein-protein interaction between INI1 and c-Myc by small peptidomimetic molecules inspired by Helix-1 of c-Myc: identification of a new target of potential antineoplastic interest. FASEB J. 2007;21(4):1256–1263. doi: 10.1096/fj.06-7082com. [DOI] [PubMed] [Google Scholar]
- 64.Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev Cell. 2003;4(6):827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 65.Algar EM, Muscat A, Dagar V, Rickert C, Chow CW, Biegel JA, et al. Imprinted CDKN1C Is a tumor suppressor in rhabdoid tumor and activated by restoration of SMARCB1 and histone deacetylase inhibitors. PLoS ONE. 2009;4(2):e4482. doi: 10.1371/journal.pone.0004482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsikitis M, Zhang Z, Edelman W, Zagzag D, Kalpana GV. Genetic ablation of Cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc Natl Acad Sci U S A. 2005;102(34):12129–12134. doi: 10.1073/pnas.0505300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alarcon-Vargas D, Zhang Z, Agarwal B, Challagulla K, Mani S, Kalpana GV. Targeting cyclin D1, a downstream effector of INI1/hSNF5, in rhabdoid tumors. Oncogene. 2006;25(5):722–734. doi: 10.1038/sj.onc.1209112. [DOI] [PubMed] [Google Scholar]
- 68.Xu Y, Tan LJ, Grachtchouk V, Voorhees JJ, Fisher GJ. Receptor-type protein-tyrosine phosphatase-kappa regulates epidermal growth factor receptor function. J Biol Chem. 2005;280(52):42694–42700. doi: 10.1074/jbc.M507722200. [DOI] [PubMed] [Google Scholar]
- 69.Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112(5):673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 70.Upadhyay G, Goessling W, North TE, Xavier R, Zon LI, Yajnik V. Molecular association between beta-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/beta-catenin signaling. Oncogene. 2008;27(44):5845–5855. doi: 10.1038/onc.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rangel J, Nosrati M, Torabian S, Shaikh L, Leong SP, Haqq C, et al. Osteopontin as a molecular prognostic marker for melanoma. Cancer. 2008;112(1):144–150. doi: 10.1002/cncr.23147. [DOI] [PubMed] [Google Scholar]
- 72.Shevde LA, Samant RS, Paik JC, Metge BJ, Chambers AF, Casey G, et al. Osteopontin knockdown suppresses tumorigenicity of human metastatic breast carcinoma, MDA-MB-435. Clin Exp Metastasis. 2006;23(2):123–133. doi: 10.1007/s10585-006-9013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kao CL, Chiou SH, Chen YJ, Singh S, Lin HT, Liu RS, et al. Increased expression of osteopontin gene in atypical teratoid/rhabdoid tumor of the central nervous system. Mod Pathol. 2005;18(6):769–778. doi: 10.1038/modpathol.3800270. [DOI] [PubMed] [Google Scholar]
- 74.Gao YA, Agnihotri R, Vary CP, Liaw L. Expression and characterization of recombinant osteopontin peptides representing matrix metalloproteinase proteolytic fragments. Matrix Biol. 2004;23(7):457–466. doi: 10.1016/j.matbio.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 75.Abdollahi A, Hlatky L, Huber PE. Endostatin: the logic of antiangiogenic therapy. Drug Resist Updat. 2005;8(1-2):59–74. doi: 10.1016/j.drup.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Herbst RS, Hess KR, Tran HT, Tseng JE, Mullani NA, Charnsangavej C, et al. Phase I study of recombinant human endostatin in patients with advanced solid tumors. J Clin Oncol. 2002;20(18):3792–3803. doi: 10.1200/JCO.2002.11.061. [DOI] [PubMed] [Google Scholar]
- 77.Ramirez-Castillejo C, Sanchez-Sanchez F, Andreu-Agullo C, Ferron SR, Aroca-Aguilar JD, Sanchez P, et al. Pigment epithelium-derived factor is a niche signal for neural stem cell renewal. Nat Neurosci. 2006;9(3):331–339. doi: 10.1038/nn1657. [DOI] [PubMed] [Google Scholar]
- 78.Rabinovich GA. Galectin-1 as a potential cancer target. Br J Cancer. 2005;92(7):1188–1192. doi: 10.1038/sj.bjc.6602493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.