Abstract
The drive to characterize functions of human genes on a global scale has stimulated interest in large-scale generation of mouse mutants. Conventional germ-cell mutagenesis with N-ethyl-N-nitrosourea (ENU) is compromised by an inability to monitor mutation efficiency, strain1 and interlocus2 variation in mutation induction, and extensive husbandry requirements. To overcome these obstacles and develop new methods for generating mouse mutants, we devised protocols to generate germline chi-maeric mice from embryonic stem (ES) cells heavily mutagenized with ethylmethanesulphonate (EMS). Germline chimaeras were derived from cultures that underwent a mutation rate of up to 1 in 1,200 at the Hprt locus (encoding hypoxanthine guanine phosphoribosyl transferase). The spectrum of mutations induced by EMS and the frameshift mutagen ICR191 was consistent with that observed in other mammalian cells. Chimaeras derived from ES cells treated with EMS transmitted mutations affecting several processes, including limb development, hair growth, hearing and gametogenesis. This technology affords several advantages over traditional mutagenesis, including the ability to conduct shortened breeding schemes and to screen for mutant phenotypes directly in ES cells or their differentiated derivatives.
EMS induces predominantly point mutations in mammalian cells, causing null mutations at frequencies exceeding 1 in 1,000 cells3–5. We treated ES-cell cultures of genotypes 129/Sv and (129×C57BL/6J)F1 with increasing concentrations of EMS, then selected surviving cells in 6-thioguanine (6TG) to measure the mutation rate at the X-linked Hprt locus (Table 1; all three ES lines are XY). We observed mutation frequencies of up to 1 in 1,200, similar to those in Chinese hamster fibroblasts4.
Table 1.
Chimaeras from EMS-treated cells and Hprt mutation rate
| Cells | Genotype | [EMS] | % dead | Hprt– | Chimaeras | Germ line |
|---|---|---|---|---|---|---|
| v6.4* | 129/SvJae×C57BL/6J | 600 | 95% | 1/2,000 | 6 | 4 |
| CT129 c* | 129/Sv +c, +p | 237.5 | 38% | 1/14,600 | 9 | 1 |
| CT129 d* | 129/Sv +c, +p | 475 | 92% | ND | 6 | 1 |
| CJ7 | 129/SvEv | 200 | 41% | ND | 4 | 2 |
| CJ7 | 129/SvEv | 300 | 44% | 1/7,222 | 8 | 7 |
| CJ7 | 129/SvEv | 400 | 64% | 1/3,277 | 14 | 6 |
| CJ7* | 129/SvEv | 400 | 92% | 1/4,524 | 2 | 2 |
| CJ7 | 129/SvEv | 500 | 94% | 1/1,200 | 4 | 1 |
The EMS concentration is in μg/ml. The two classes of CT129 cells (‘c’ and ‘d’) represent different EMS mutagenesis experiments on the same line of cells. ND, not done. Mutation rate at Hprt is expressed as a fraction derived from screens of >5×106 cells, representing >2×104 founder cells.
Cell population used in the phenotype screen. The mutation rate is adjusted for the background of spontaneous 6-TG–resistant colonies. These were 1 in 377,777 for CT129, 1 in 187,500 for CJ7 and <1 in 1.1×106 for v6.4.
To investigate the mutational spectrum, we sequenced Hprt coding regions (amplified by RT–PCR) from clones resistant to 6TG. This revealed 17 classes of mutations (Table 2): 11 G→A transitions, 2 C→T transitions and 4 putative splicing mutations. This spectrum is typical of mammalian cells4. We also induced mutations with ICR191, which induces primarily +1 frameshifts in stretches of guanines6. Indeed, all 6TG-resistant clones we sequenced contained an additional guanine in a stretch of five or six guanines. Together with the characterization of ENU-induced mutations in ES cells in the accompanying paper11, we conclude that most mutagens will induce lesions in ES cells, consistent with other mammalian cell types.
Table 2.
Hprt mutations in EMS-and ICR191-treated ES cells
| Chemical | Cells | Change | Position | Mutation |
|---|---|---|---|---|
| EMS | v6.4 | splicing | – | exon 8 skipped |
| EMS | CJ7 | ? | ? | no PCR product |
| EMS | CJ7 | splicing | – | extra exon betw. exons 5,6 |
| EMS | v6.4 | splicing | – | 66 bp exon betw. exons 5,6 |
| EMS | CJ7 | G→A | 47 | Gly→Asp |
| EMS | CJ7 | G→A | 118 | Gly→Arg |
| EMS | v6.4 | G→A | 119 | Gly→Glu |
| EMS | v6.4 | C→T | 202 | Leu→Phe |
| EMS | v6.4 | G→A | 208 | Gly→Arg |
| EMS | CJ7 | G→A | 209 | Gly→Glu |
| EMS | v6.4 | G→A | 209 | Gly→Glu |
| EMS | CJ7 | G→A | 355 | Gly→Arg |
| EMS | v6.4 | G→A | 418 | Gly→Asn |
| EMS | CJ7 | G→A | 419 | Gly→Asp |
| EMS | CJ7 | G→A | 539 | Gly→Glu |
| EMS | v6.4 | C→T | 551 | Pro→Leu |
| EMS | CJ7 | G→A | 601 | Asp→Asn |
| EMS | v6.4 | G→A | 601 | Asp→Asn |
| EMS | v6.4 | G→A | 610 | His→Tyr |
| ICR191 | v6.4 | +1 G | 207–212 | inc. G run from 6 to 7 |
| ICR191 | v6.4 | +1 G | 333–337 | inc. G run from 5 to 6 |
‘Position’ refers to the coding region nucleotide number; nt 1 is the ‘A’ in the initiation codon. The 10 different mutations in EMS-treated v6.4 cells were distributed among 15 sequenced, 6TG-resistant clones, and the 9 different mutations in CJ7 were distributed among 17 clones. We analysed eight 6TG-resistant, ICR191 clones, revealing two classes of mutations. The CJ7 line that failed to yield a PCR product was analysed by Southern blot, revealing no deletions or rearrangements; it is possible that this is a regulatory mutation. The positions of mutations causing splicing defects were not determined.
To determine if ES cells treated with EMS retain the ability to colonize the germ line, we injected surviving cells into blastocysts to create chimaeras. Germline chimaeras (24) were derived from both 129 and F1 hybrid ES cells exposed to a range of EMS treatments (Table 1).
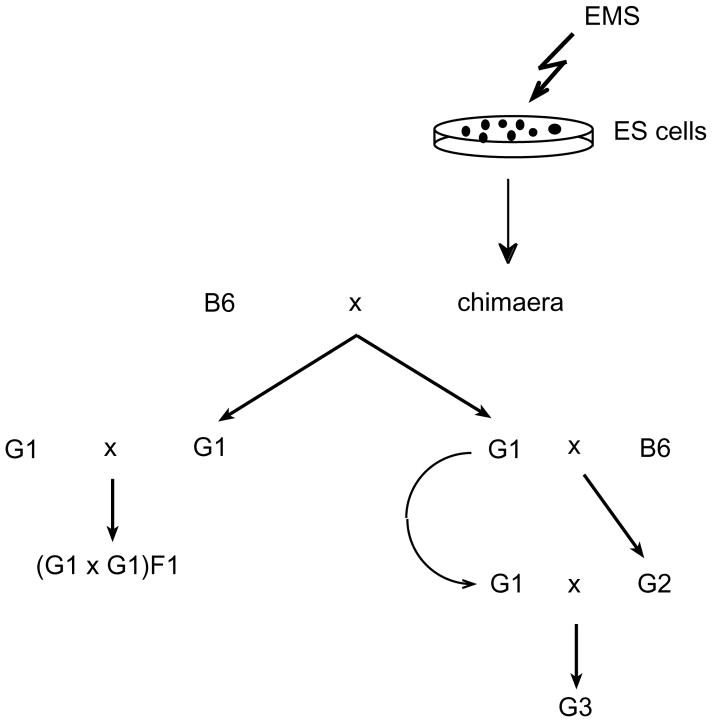
We intercrossed G1 siblings from each of the four germline chimaeras derived from v6.4 cells (Hprt mutation rate=1/2,000) to generate offspring with recessive mutations (Fig. 1, left). Such matings yielded few or no progeny (maximum of 3; Table 3). This low fecundity was not attributable to infertility of either parent, because the average size of their backcross litters was 5.3 (Table 3). This suggests that G1 mice bore a high load of recessive lethal mutations, and that most or all of the G1 mice from each chimaera were derived from a single, mutagenized ES-cell lineage. There was no evidence of a high load of recessive lethal mutations in G1 siblings sired by chimaera CT129-C, which was derived from a culture bearing more than 30% fewer (1/14,600) Hprt mutations (Tables 1 and 3). It is possible, however, that the germ line of this chimaera was derived from multiple independent ES cells. These data suggest that ES cells are tolerant to higher levels of mutagenesis than are practically required, and that two-generation recessive screens should ideally use ES-cell clones mutagenized at an intermediate level.
Fig. 1.
Breeding strategy for recovery of recessive mutations. Chimaeras generated from EMS-treated ES cells were mated with C57BL/6J (B6) partners. Agouti G1 progeny derived from gametes of ES cell origin were mated with either opposite sex G1 siblings (left) or B6 (right). We backcrossed G2 progeny with the opposite sex G1 parent to yield the G3 generation. Because each of the founding chimaeras could transmit two different alleles at each locus (assuming a germ line derived from a single founder ES cell), several G1 progeny from each chimaera were used to initiate these screens. Furthermore, to optimize the likelihood that most mutagenized alleles would be captured in this screen, we sought to generate 6–8 G2 progeny from each G1 male (up to four G2s from a female G1) for backcrossing.
Table 3.
Intercrosses of G1 progeny
| Chimaera | Progeny crosses | Days mated | G1×G1 pups | G1×B6 pups/litter |
|---|---|---|---|---|
| v6.4-C1 | C1B×C1A | 50 | 0 | 6; 7 |
| C1D×C1C | 50 | 0 | 0; 5.7 | |
| C1F×C1E | 11 | 0 | 10; 8.3 | |
| 10 | 2* | |||
| v6.4-C2 | C2A×C2B | 11 | 0 | 7.3; 6.5 |
| 14 | 0 | |||
| C2C×C2B | 20 | 3 | 8; 6.5 | |
| v6.4-C3 | C3A×C3B | 48 | 0 | 6.5; 3 |
| v6.4-C7 | C7B×C7A | 27 | 2* | 6; 6 |
| 2 | ||||
| C7D×C7C | 84 | 2 | 3; − | |
| C7F×C7E | 84 | 3 | 8.5; 2.5 | |
| C7I×C7H | 57 | 0 | 0; 7 | |
| C7J ×C7K | 57 | 3* | 0; 4.2 | |
| CT129-C | TC2I×TC2F | 68 | 6 | 7; 8 |
| TC2J×TC2G | 68 | 5 | −; 7 | |
| 8 | ||||
| TC2K×TC2H | 68 | 1 | −; 7 | |
| 11 |
The animals listed in ‘Progeny crosses’ are G1 siblings of the corresponding chimaera, indicated as dam×sire. The length of time these crosses were performed is listed under ‘Days mated’. In some cases, the animals were mated twice, as indicated by separate line entries. The number of pups from these intercrosses is listed under ‘G1×G1 pups’. The values in ‘G1×B6 pups/litter’ are the average litter sizes of each G1 parent (dam; sire) when mated with C57BL/6J (a dash indicates that the animal was never outcrossed). The average of these averages is 5.3.
Pups died before weaning.
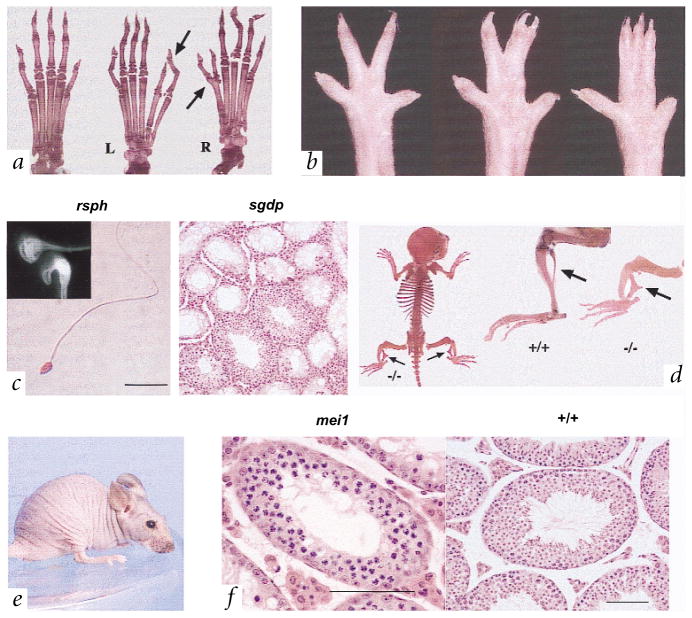
To ‘dilute’ the high load of lethal mutations in the v6.4 cells, we conducted a three-generation screen for recessive mutations (Fig. 1, right, and Table 4). We assessed G3 animals for visible dysmorphology, ocular lesions, hearing and male fertility, and recovered the following mutants: Polydactyly ems (Pde), an autosomal dominant mutation that arose in CJ7 cells (Fig. 2a); syndactyly ems (sne), an autosomal recessive mutation affecting hindpaw digits (Fig. 2b); twisted legs (twl), an apparently recessive mutation causing a bowed tibia and a malformed, triangular shaped fibula (Fig. 2d); and peachfuzz (pfz), in which homozygotes lose nearly all hair by the time of weaning (Fig. 2e).
Table 4.
Mutagenesis screens
| Expansion of families |
Phenotypic screens |
|||||||
|---|---|---|---|---|---|---|---|---|
| chimaera | no. G1 | no. G2 | pups/mating | no. G3 | vis. | fert. | eye | hear. |
| v6.4-C1 | 6 | 38 | 1.4 | 93 | 1 + ser (3) | 3/48 | 2/60 | 3(2)/69 |
| v6.4-C2 | 6 | 29 | 3.0 | 161 | 3 | 8/78 | 3/118 | 0/124 |
| v6.4-C3 | 1 | 3 | 0.0 | 0 | – | – | – | – |
| v6.4-C7 | 6 | 28 | 1.5 | 57 | 2 + Sne (3) | 1 + Rsph (2); Sgdp (2)/24 | 4/44 | 0/41 |
| CJ7-1 | 2 | 9 | 4.8 | 53 | 0 | Mei1 (5)/15 | 0/16 | 0/16 |
| CJ7-2 | 1 | 3 | 7.7 | 23 | 1 + Pde (5) | 0/4 | 0/16 | 1/18 |
| CT129-C | 7 | 35 | 6.0 | 265 | 1 + Twl (3) | 0/92 | 1/89 | 0/124 |
| CT129-D | 3 | 11 | 3.1 | 52 | 1 + Pfz (5) | 1/18 | 2/24 | 0/31 |
| Totals | 32 | 156 | 704 | 14 | 16/310 | 12/367 | 3/423 | |
The founder chimaeras are listed on the left, with the prefix designation reflecting the ES cells from which they were derived. ‘No. G2’ lists the total number of sons or daughters from all G1s that were backcrossed to a G1 parent. ‘Pups/mating’ is the total number of weaned offspring per mating period. A mating period consisted of at least one week in which a G1 and G2 were together in a cage before being separated. If two litters were produced in a mating, it was counted as two mating periods. G3 values indicate the number of pups weaned. All G3 animals were screened for visible mutations (‘vis.’); the number of variants not proven to be genetic is indicated and the names of proven mutations are indicated after the ‘+’, with the total number of animals exhibiting that mutation given in parentheses (‘ser’, a short-eared variant not discussed in the text). For the remaining phenotypic screens, the numerator is the number of variants identified and the denominator is the t otal number of animals examined. In those cases in which multiple animals in a family have the same heritable mutation, the number of such animals is given in parentheses, in some cases along with the mutant symbol. ‘Fert.’, infertile males; ‘hear.’, mice that failed both click box and ABR tests. The totals reflect the number of affected animals (in the numerator; underlined numbers indicate the adjusted number of independent phenotypes) over the total number of animals.
Fig. 2.
Phenotypes of mutants. a, Bone preparations of hindfeet from normal (+/+, left) and Pde (+/−) mutant mice. The length of the extra toe can vary, having either two phalanges, as is seen in the normal first digit, or three phalanges, as in digits two through five. In some cases, the extra toe arises as a bifurcation of a digit 1 metacarpal or phalange. Arrows highlight the variable manifestation of the poly-dactyly phenotype in hindfeet of one individual. L, left; R, right. b, Examples of sne phenotypic expression in hindfeet of homozygous mutants. c, Light microscopic and electron microscopic (EM; inset) images of sperm from the rsph mutant and associated sgdp phenotype. Normal sperm heads in mice are hook-shaped; an apparently normal shaped head is enveloped in the residual cytoplasmic sac in the EM photograph. Right, an example of an sgdp testis section in which relatively normal (albeit with apparently abnormal flagellar development in spermatids) seminiferous tubule segments are juxtaposed with agametic tubule segments. d, Skeletal preparations of the twl mouse. Arrows indicate the malformed fibulae in the mutant (−/−) compared with a normal animal (+/+). Also note the shortened and thickened tibiae in the affected mice. e, A pfz homozygote. f, Cross-section through the testes of a mei1 mutant and a normal animal (+/+). Note the accumulation of spermatocytes with condensed chromosomes in the most mature mutant tubules, and how others are either vacuolated or lined only by a thin spermatogonial layer. Presumed apoptotic cells appear as large, dark cells. Scale bars in c,f, 100 μM.
We screened 310 G3 males for fertility and identified 22 that failed to produce offspring. Of these, 11 had sperm or testis abnormalities. One male produced sperm in which all the heads were separated from the tails. Another had no epididymal sperm, apparently caused by defective flagellar formation. The remaining nine animals represent three mutations, called mei1 (meiosis defective 1), spermatogonial depletion (sgdp) and round sperm heads (rsph).
Mutant mei1 males are devoid of epididymal sperm and have small testes. Testis sections revealed a complete absence of post-meiotic cells (Fig. 2f). The most advanced seminiferous tubule section contained spermatocytes apparently arrested at the zygotene/pachytene stage of meiosis. Homozygous females are also sterile, having ovaries depleted or devoid of oocytes. Affected rsph males have sperm with rounded, rather than hooked, heads as visualized by light microscopy (Fig. 2c). Examination by scanning electron microscopy revealed that this is apparently due to a retention of cytoplasm around the sperm heads (Fig. 2c). Many sperm also have curled flagella (data not shown). The sgdp mutation causes some seminiferous tubule sections to become devoid of germ cells (Fig. 2c), reminiscent of Sertoli-cell-only syndrome in humans (MIM 305700). Other tubule regions remain relatively normal, although spermatid elongation appeared to be aberrant.
We screened 423 G3 mice from 28 families for deafness, identifying 12 animals with poor Preyer reflex. Of these, four had elevated auditory brainstem response (ABR) thresholds (Table 3): two were homozygous for a single recessive mutation originating from chimaera v6.4-C1 (Table 4), exhibiting high-frequency hearing loss; one carried an autosomal recessive mutation characterized by a subtle, unbalanced gait and elevated ABR threshold at low frequency (8 kHz); and the remaining mouse died before mating.
We failed to observe any retina or optic nerve abnormalities in 367 G3 mice. The lack of phenotypes such as retinal degeneration may reflect the young age (2–5 months) of the analysed animals. Several mice with other visible aberrations were born as single cases, and it is not yet known if their phenotypes are heritable. These include the following phenotypes (Table 4): curled tails, absent or small eyes, progressive ataxia/paralysis and unusually shaped heads or trunks. Many did not breed, and others are now being analysed.
Our finding that highly mutagenized ES cells retain germline competence provides unique options for the creation of mouse mutants. A wide range of mammalian-cell mutagens are available to achieve a desired mutational spectrum; this may be used to circumvent the mutational bias of ENU that confounds attempts to achieve saturation mutagenesis2, and to simplify detection of unselected mutations in genes of interest7 using frameshift mutagens such as ICR191. Most significant is the possibility of screening treated ES cells for mutations affecting basic cellular processes (such as DNA-damage repair) or following differentiation. Selection of recessive mutations could be achieved in deletion-bearing cells8,9. The ability to generate, screen and preserve clones at the cellular level affords enormous logistical benefit.
Methods
Treatment of cells
We maintained cultures of ES cells on primary embryonic fibroblasts under standard conditions. When they reached ~50% confluence, we applied EMS (methanesulphonic acid ethyl ester, 1.17 g/ml; Sigma) to the media for 16–20 h. In a typical experiment, we applied a range of EMS concentrations to a parallel series of ES cell cultures, each containing ~2×106 cells. After exposure, the cells were washed in PBS, trypsinized and counted. The bulk of cells were passaged onto fresh feeder plates and frozen for later use (to generate chimaeras). We seeded 1,000 of the remaining cells onto gelatin-coated 60 mm plates and counted the number of resulting colonies to determine the percentage survival of treated cells, following adjustments for plating efficiency (determined by plating untreated cells in parallel) and death during the treatment period (by comparing cell numbers following trypsinization of exposed and control cultures).
Mutation rate at Hprt
Following exposure to EMS, cells were grown for 10 d and passaged as needed to allow for dissipation of remaining Hprt activity. We then plated them at concentrations of 0.5–1×106 cells/150 mm gelatin-coated plate (no feeders) in media containing 6TG. Selection was removed after 3 d, and the number of colonies was counted 2 weeks after seeding. Untreated control cells yielded no 6TG-resistant colonies in v6.4 cells, and a maximum of one per plate with 129 cells. This background was subtracted in the reported frequency of EMS-induced mutations. The concentration of plated cells was critical; higher concentrations of cells plated caused the death of certain fractions of true Hprt-deficient cells, as determined in pilot experiments involving plating a constant number of Hprt-deficient ES cells with increasing numbers of wild-type cells.
Sequencing of Hprt mutations
We extracted total RNA from 6TG-resistant ES cell clones grown in individual wells of a 24-well culture dish, without feeders, using an RNeasy kit (Qiagen). Reverse transcription with Superscript (Gibco-BRL) was primed with random hexanucleotides. cDNA was amplified using the primers 5′–CGTCGTGATTAGCGAT GATG–3′ and 5′–TCTACCAGAGGGTAGGCTGG–3′, resulting in 852-bp amplimers. The PCR reaction was processed in a Qiaquick PCR cleanup columns (Qiagen), and sequenced from both ends using an ABI Model 377 automated sequencer.
Generation of chimaeric mice
We injected ES cells into C57BL/6J blasto-cysts by standard methods.
Fertility testing of males
We mated G3 males at ~2 months of age to a single (C57BL/6J×A/J) F1 female. The male was deemed infertile if no litters occurred over the following 2 months, and was sacrificed for examination of epididymal sperm. If the sperm appeared abnormal or were absent, then the testicles were fixed in Bouin’s, sectioned, and stained with haematoxylin and eosin.
Hearing
We first screened G3 mice with a ‘click box’, which emanates a high-frequency (18.6 Hz), pure-tone sound burst that is the optimum of mouse hearing sensitivity. This elicits a Preyer reflex (pinna twitch) in mice with normal hearing. An ABR test was performed on those that failed to display this reflex10. Mice with ABR thresholds above 55 (for click stimulus), 40 (for 8 kHz), 35 (for 16 kHz) or 60 (for 32 kHz) dB SPL were considered to have defective hearing10.
Clinical eye examinations
A slitlamp was used for evaluation of the anterior segment and an indirect opthalmoscope for assessment of the retina11. We found a variety of ocular abnormalities (including unilateral and bilateral mid-dilated pupils; oval, eccentric pupils; diffuse corneal opacity with a central clear area, and one mouse with persistent tunica vasculosa lentis and cataract), and we are investigating two to determine if the defects are genetic. The remainder were not pursued, as they had defects characteristic of C57BL/6J mice, the incidence and severity of which varies between colonies and due to environmental factors12.
Skeletal preparations
We carried out Alizarin red and Alcian blue staining of bone and cartilage as described12, except the skeletons were stored in a solution of 2:2:1 95% ethanol:glycerol:benzyl alcohol.
Testes histology
We fixed testes in Bouin’s for ~24 h, washed in 70% ethanol and embedded in paraffin. We stained sections (5 μM) with haematoxylin and eosin.
Acknowledgments
We thank T. Magnuson for CT129 ES cells; T. Gridley for CJ7 ES cells; A. Planchart for input on design of culture experiments; J. Sztein for assistance with sperm examination; C. O’Neill for animal care; and T. O’Brien, G. Cox, T. Magnuson and M.A. Handel for critical evaluation of the manuscript. This work was supported by NIH grant GM45415 to J.C.S. and a Cancer Center Grant CA34196 to The Jackson Laboratory V.L.B. was supported by an NIH training grant (HD07065) and fellowship NIH (HD08441). S.W.M.J. is an Assistant Investigator of The Howard Hughes Medical Institute. Hearing tests were performed under a contract (DC62108) to K. Johnson from the National Institute on Deafness and Other Communicative Disorders.
References
- 1.Bode VC. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the T region of mouse chromosome 17. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinchik EM, Carpenter DA. N-Ethyl-N-nitrosourea mutagenesis of a 6- to 11-cM subregion of the Fah-Hbb interval of mouse chromosome 7. Completed testing of 4557 gametes and deletion mapping and complementation analysis of 31 mutations. Genetics. 1999;152:373–383. doi: 10.1093/genetics/152.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sega GA. A review of the genetic effects of ethyl methanesulfonate. Mutat Res. 1984;134:113–142. doi: 10.1016/0165-1110(84)90007-1. [DOI] [PubMed] [Google Scholar]
- 4.Klungland A, Laake K, Hoff E, Seeberg E. Spectrum of mutations induced by methyl and ethyl methanesulfonate at the hprt locus of normal and tag expressing Chinese hamster fibroblasts. Carcinogenesis. 1995;16:1281–1285. doi: 10.1093/carcin/16.6.1281. [DOI] [PubMed] [Google Scholar]
- 5.Hsie AW, Brimer PA, Mitchell TJ, Gosslee DG. The dose-response relationship for ethyl methanesulfonate-induced mutations at the hypoxanthine-guanine phosphoribosyl transferase locus in Chinese hamster ovary cells. Somat Cell Genet. 1975;1:247–261. doi: 10.1007/BF01538449. [DOI] [PubMed] [Google Scholar]
- 6.Taft SA, Liber HL, Skopek TR. Mutational spectrum of ICR-191 at the hprt locus in human lymphoblastoid cells. Environ Mol Mutagen. 1994;23:96–100. doi: 10.1002/em.2850230204. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, et al. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nature Genet. 2000;24:314–317. doi: 10.1038/73557. [DOI] [PubMed] [Google Scholar]
- 8.You Y, et al. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nature Genet. 1997;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 10.Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.John SW, et al. Essential iris atrophy, pigment dispersion, and glaucoma in DBA/2J mice. Invest Ophthalmol Vis Sci. 1998;39:951–962. [PubMed] [Google Scholar]
- 12.Smith RS, Roderick TH, Sundberg JP. Microphthalmia and associated abnormalities in inbred black mice. Lab Anim Sci. 1994;44:551–560. [PubMed] [Google Scholar]
- 13.Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]