Abstract
The β-amyloid precursor protein (APP) plays a major role in Alzheimer’s disease. The APP intracellular domain (AICD), together with Fe65 and Tip60, localizes to spherical nuclear AFT complexes that might represent sites of transcription. We now show that endogenous AICD is targeted to similar nuclear spots. AFT complexes were closely associated with Cajal and PML bodies but did not localize to nucleoli or splicing speckles. Live imaging revealed that AFT complexes were highly mobile within nuclei. Following pharmacological inhibition of transcription AFT complexes merged into a few large assemblies. We have previously shown that AICD regulates the expression of its own precursor APP. Transfection of APP promoter plasmids as substrates resulted in cytosolic AFT complex formation at the labeled APP promoter plasmids. In addition, identification of chromosomal APP or KAI1 gene loci by fluorescence in situ hybridization showed their close association with nuclear AFT complexes. The transcriptional activator Notch intracellular domain (NICD) localized to the same nuclear spots as occupied by AFT complexes, suggesting that these nuclear compartments correspond to transcription factories. Fe65 and Tip60 also co-localized with APP in the neurites of primary neurons. Pre-assembled AFT complexes may serve to assist fast nuclear signaling upon endoproteolytic APP cleavage.
Keywords: AICD, NICD, transcription, nuclear signaling, life imaging, Tip60, Fe65, nuclear bodies, FISH
Introduction
The transmembrane β-amyloid precursor protein APP is sequentially cleaved by membrane-anchored secretases (12). The first proteolytic cut by α- or β-cleavage releases its extracellular domain, followed by intramembraneous γ/ε-cleavage generating the APP intracellular domain (AICD) (45). In the case of initial β-cleavage proteolytic processing of APP generates the amyloid β (Aβ) peptide, which oligomerizes to create toxic species that cause behavioural deficits and neurodegeneration, characteristics of Alzheimer’s disease (AD) (52). The transmembrane Notch receptor is similarly cleaved by α- and γ-secretases and the liberated Notch intracellular domain (NICD) translocates to the nucleus to regulate transcription (13, 51). In addition, an Aβ-like peptide can be cleaved from Notch by γ secretase and secreted (41). The analogy of APP and Notch processing is suggestive of a possible function for AICD in nuclear signaling.
Experiments involving the fusion of the yeast Gal4 DNA-binding domain to AICD revealed transcriptional activity in luciferase assays (7). Transactivation was enhanced by the AICD-binding protein Fe65 and the histone acetylase Tip60. Fe65 was shown to bind to APP and stabilize AICD, which together localized to the nuclear compartment (22, 34, 58). In the nucleus Fe65 and AICD associate with Tip60 and localize to multiple spherical nuclear compartments (57). Different APP-interacting proteins have been shown to differentially regulate APP processing and AICD subcellular localization. MINT-1/X11α was reported to stabilize APP (5), exert an inhibitory effect on AICD-mediated transactivation in the Gal4 system (4) and prevent nuclear translocation of AICD (57). The jun-interacting protein (Jip1) is another APP-adaptor protein that stabilizes APP (31, 54), connects APP to Kinesin (26), shows transactivation activity in the Gal4 system (Scheinfeld et al., 2003) and can target AICD to the nucleus, where it localizes to speckle structures (57).
Several genes regulated by APP have been identified, among them KAI1 (2), APP, BACE (57), neprilysin (44), p53 (1), α-actinin and transgelin (36) and the EGF receptor that is negatively regulated by AICD (61). Nevertheless some studies questioned the nuclear function of AICD (6) and the impact on gene transcription (17).
The nucleus possesses a variety of compartments and even interphase chromosomes are not spread throughout the nucleus but are confined to much smaller chromosome territories (33). Actively transcribed DNA can loop out of these territories to come into contact with transcription factories (11, 30, 35). The nucleolus is the most prominent structure and is dedicated to the transcription and modification of ribosomal RNAs. Many smaller structures, bodies and speckles have been identified that harbor proteins involved in executing nuclear functions, such as transcription, splicing or snRNP biogenesis (24). The formation of numerous distinct nuclear spots by AICD, Fe65 and Tip60 (AFT complexes) prompted us to analyze the nature of these nuclear bodies. We present evidence that they represent the sites of transcription of AICD-regulated genes. Furthermore, we show that NICD localizes to the same structures, pointing to a possible cross-talk between APP and Notch in nuclear signaling.
Materials and Methods
Cell culture
Human embryonic kidney cells (HEK 293, DSMZ, ACC 305) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen #52100-039) supplemented with 10% fetal calf serum (FCS) and Penicillin/Streptomycin (PenStrep, Invitrogen #10378-016) at 37°C, 5% CO2, 95% humidity.
Primary astrocytes were prepared from postnatal day 4 to 6 C57Bl/6 and APP knockout mice (27). Cerebelli were removed into CSS buffer (120 mM NaCl, 5.4 mM KCl, 0.8 mM MgCl2, 25 mM Tris-HCl, 15 mM D-Glucose) containing 0.05% Trypsin (Invitrogen, # 25300-062) and incubated for 10 minutes at room temperature. Cerebelli were triturated in DMEM/10% FCS with Pasteur pipettes of decreasing diameter, centrifuged and triturated again in A-DMEM (DMEM (Invitrogen-Gibco, #52100-021) with the addition of 1 mM sodium pyruvate, 44 mM sodium hydrogen carbonate, 1% BSA, 60 ng/ml EGF (Roche, #855731), 10 μg/ml Insulin (Sigma, #I-4011), 66 μg/ml Transferrin (Roche, #652 202) and 1% PenStrep). Cells were cultured on poly-D-Lysine coated (100ug/ml) coverslips at 37°C, 7% CO2 in A-DMEM until 80% confluency and the medium changed to DMEM/10% FCS for differentiation of the astrocytes.
Primary neurons were prepared from E14-16 C57Bl/6 mice. Meninges were removed and cortices were dissected in ice-cold CSS. The cortices of 5-7 embryos were treated with 7.2 U Dispase II (Roche, #10295825001) for 10 minutes at room temperature and triturated in DMEM with 10% horse serum (HS), 1 mM sodium pyruvate, 44 mM sodium hydrogen carbonate, 1% BSA and PenStrep. After dissociation the cells were plated at a density of 9000 cells/mm2 on astrocyte monolayers. Transfections were preformed after 4-6 days in vitro. Primary neurons from the cortex of E18 Wistar rats were similarly prepared and 50000 cells plated onto polyornithine-coated coverslips. Neurons were grown for 7 days in Neurobasal medium supplemented with B27 and L-Glutamine.
For DNA transfection in HEK 293 or primary cells, Lipofectamine 2000 (Invitrogen, #11668-019) was used according to the manufacturer’s protocol. Transfection medium was replaced after 2 h with fresh medium. Cells were fixed or imaged 18 to 22 hours later. A clonal HEK 293 cell line with inducible expression of AICD has been previously described (57).
Drugs
Actinomycin D (Sigma-Aldrich, #A1410) was used at a final concentration of 2 μM. Leptomycin B (LMB) (Calbiochem, #431050) was used at a final concentration of 5 to 20 ng/ml. DAPT (Calbiochem, #565770) was used at a final concentration of 1 μM.
DNA constructs
The following expression constructs have been described: APP-Citrine, HA-Fe65, Myc-Fe65 and CFP-Tip60 (57). The improved Cyan Fluorescent protein (CFP) Cerulean was derived from pECFPnuc (Clontech-Invitrogen) by site-directed mutagenesis according to the published sequence (48). Full-length NICD was amplified from a plasmid kindly provided by Raphael Kopan (13). Two PCR reaction products (Forward primer 1: 5′-CCGGAGTGCTGCTGTCCCGCAAGCGC; reverse primer 1: 5′-AGTTTAAACTTATTTAAATGCCTCTGGAATGTGGG; forward primer 2: 5′-AGTGCTGCTGTCCCGCAAGCGC; reverse primer 2: 5′-CTAGAGTTTAAACTTATTTAAATGCCTCTGGAATGTGGG) were mixed, heated and re-annealed to generate BspEI and XbaI overhangs, and ligated to the C-terminus of Cerulean. APP promoter plasmids containing 1.2 kbp (-1141/+105, +1 transcription start site preceding the ATG (at +148) of APP (23) were nick-labeled with Digoxigenin according to the manufacturers protocol (DIG-Nick Translation, Roche).
Immunocytochemistry
Cells were first washed with PBS and fixed for 20 min with 4% paraformaldehyde (PFA) at room temperature or for 10 min with Methanol at -20°C. They were then washed with TBS containing 0.05% Triton X-100 and blocked with TBS containing 0.2% Triton X-100, 5% horse serum and 5% goat serum. Primary antibodies were applied in blocking solution over night at 4°C. Antibodies were mouse anti-Myc (1:100; Roche, #11667149001), rat anti-HA (1:100; Roche, #1867423), sheep anti-Digoxigenin (Roche, #11333089001), C23/nucleolin (1:50; Santa Cruz, sc-8031), PML (1:50; Santa Cruz, sc-966), Coilin (1:200; Sigma, C-1662), SC-35 (1:200; Sigma, S-4045), APP C-terminus (55, 56), Tip60 (Abcam, ab23886). Cy3- or Cy5-conjugated secondary antibodies (Jackson Labs) were applied at 1:250 and after subsequent washing the cells were embedded in Mowiol with the addition of 2.5% 1.4 Diazobicyclo (2.2.2.) octane (DABCO, Sigma-Aldrich, #D-2522). DRAQ5 (Biostatus, Leicestershire, UK) or DAPI (Sigma-Aldrich, #D-9564) were used to counterstain nuclei.
Fluorescence in situ hybridization
Bacterial artificial chromosomes harboring genomic sequences of APP (RP11-66H5 and RP11-75H10) or KAI1 (RP58K22) were obtained from the German Resource Center for Genome Research (RZPD) and amplified using Large-Construct Kits (Qiagen # 12462). BACs were labeled by nick translation with a kit from Vysis (Abbott #7J001) using SpectrumOrange-dUTP (#6J9415) according to the manufacturer’s protocol. Clonal HEK293 were induced to express AICD and transfected with HA-Fe65 and CFP-Tip60. Cells were fixed in methanol/acetic acid, washed briefly in PBS and incubated for 30 minutes with 0.04 mg/ml RNase A. After washing in 2 × SSC the cells were digested for 3 minutes with 0.1 mg/ml pepsin in 10 mM HCl and washed in 2 × SSC. Cells were dehydrated in a rising alcohol series and transferred to prehybridization solution containing 50% formamide, 2 × SSC, 10% dextrane sulfate with freshly added salmon sperm (210 μg/ml) and human Cot-1 DNA (42 μg/ml). Labeled BAC clones were added to the prehybridization mix and denatured for 5 minutes at 80°C. Addition of hybridization mix to cells was followed by 5 minute incubation at 80°C and 16 hours hybridization at 37°C. Stringency washes were done in 2 × SSC, 50% formamide at 42°C. After postfixation with Formalin the cells were immunostained with antibodies detecting the HA tag of Fe65. In some cases the immunostaining preceded the hybridization, which resulted in better staining of nuclear spots.
Pulse Chase
Clonal HEK 293 cells were grown for 48 hours in the absence or presence of tebufenozide, which induces AICD expression. Cells were incubated in medium without methionine and cysteine for 30 minutes followed by pulsing with Easy Tag 35S Label Mix (200 μCi per plate) for 20 minutes. Cells were washed and chased for various times in regular DMEM/FCS. Cells were lysed in 1% Triton X100, 40 mM TrisHCl, pH 7.4, 150 mM KCl, 5% glycerol, 5 mM β-mercaptoethanol, Protease inhibitor cocktail (Roche, #1 697 498), homogenized with a G22 syringe and centrifuged for 10 minutes at 500 g. The supernatant was immunoprecipitated using an APP C-terminal antibody coupled to Protein A Sepharose. The precipitates were separated with 10-20% Tricine precast gradient gels (Invitrogen, #EC66252). Gels were dried and exposed to Biomax MR films for 4 days to 3 weeks.
Confocal Microscopy
Images were acquired on a Leica TCS/SP2 confocal microscope (Leica, Wetzlar, Germany) with a 63x water immersion objective. The Argon Laser line of 458 nm was used to excite CFP (PMT window: 465 - 485 nm) and the 514 nm line to excite Citrine (PMT window: 525-545 nm). A 543 nm HeNe laser was used to excite Cy3 (PMT window: 553 - 600 nm). A 633 nm HeNe laser was used to excite Cy5 (PMT window: 655 - 710 nm). Between 5 and 15 sections in the z-axis (xy-mode) were acquired, and maximum projections of the sections encompassing the nucleus were performed. Antibody staining with Cy3 is always colour-coded in red and Cy5-staining in blue.
Live imaging
Cells grown on coverslips were transferred to DMEM with HEPES and lacking phenolred (Invitrogen-Gibco), supplemented with 10% FCS (Invitrogen-Gibco). The TCS/SP2 confocal microscope was equipped with a Plexiglas chamber from Life Imaging Systems, heating the setup to 37°C. Citrine and CFP Fluorescence was imaged at 400 Hz with 512 × 512 resolution and an averaging of two sequential scans. A stack of 4 to 6 sections in the z-axis was acquired every 15 to 45 seconds. Leica software was used to perform maximum projections of stacks and a blur filter of 3 was applied to the images.
Results
Fe65 dominates over Jip1 and X11/MINT in binding AICD and targeting it to nuclear spots
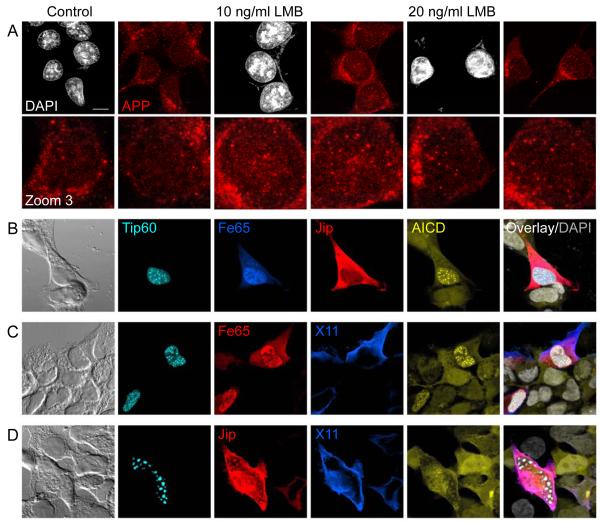
We have reported that AICD fused to a fluorescent protein (Citrine) can be detected via the Citrine fluorescence, together with Fe65 and Tip60, in nuclear spots (AFT complexes) (57). In contrast, using fusion proteins with an HA-tag connecting the AICD and Citrine sequences, HA-tag antibody staining detected APP throughout the cell, co-localizing with Citrine fluorescence, but failed to label nuclear AFT complexes (supplementary figure 1). It is likely that standard paraformaldehyde-fixation shields nuclear AICD and the fused HA-tag, which is embedded in a multiprotein complex, from antibody detection. Using Methanol fixation we were able to stain endogenous nuclear AICD with antibodies. We could detect AICD in nuclear spots as seen for AFT complexes in transfected cells. Inhibition of nuclear export by Leptomycin B (LMB) resulted in dramatic increases in nuclear AICD spots (Figure 1A). Thus, endogenous AICD, as shown for transfected Citrine-AICD (57), undergoes nuclear cycling and is targeted to nuclear spots.
Figure 1.
AICD is targeted to nuclear spots with Fe65 dominating this localization in competition with other APP-binding proteins. (A) HEK 293 cells were treated with Leptomycin B (LMB, 10 or 20 ng/ml) for 24 hours to block nuclear export. Staining of endogenous APP with a C-terminal antibody reveals the accumulation of AICD in nuclear spots. (B-D) A clonal HEK 293 cell line with inducible expression of Citrine-AICD was co-transfected with CFP-Tip60 and a combination of HA-Fe65 and FLAG-Jip1b (B) or Myc-Fe65 and HA-X11α/MINT1 (C) or FLAG-Jip1b and HA-X11α (D). Confocal microscopy can clearly discriminate CFP, Citrine, Cy3 (red), Cy5 (Blue) and DAPI (grey) emissions. Targeting of AICD to nuclear spots was seen in all cells with Fe65 expression regardless of the co-expressed proteins. Bar, 10 μm.
Different cytosolic APP-binding proteins have different effects on the processing of APP and the cellular distribution of AICD. We have previously shown that Fe65 and Jip1b localize AICD to morphologically different nuclear structures whereas X11 retains it in the cytosol. To analyze the competition between different APP-binding proteins we co-transfected various combinations of Fe65, Jip1b and X11α together with Tip60 into a clonal cell line with inducible AICD expression. In cells with strong co-expression of Fe65 and Jip we observed the formation of nuclear AFT complexes, whereas Jip1b was restricted to the cytosol and never localized to nuclear spots (Figure 1B, n = 18 cells). When the expression of Fe65 was very low it localized to Tip60 speckles and did not form spots. In this case there was a co-localization of Jip1b with nuclear speckles (n = 6 cells). X11α can bind AICD and prevent it entering the nucleus but it did not prevent the formation of AFT complexes when co-transfected with Fe65 (Figure 1C, n = 9 cells). The nuclear translocation of AICD to speckles formed by Jip1b and Tip60 was reduced by X11α that retained AICD in the cytosol (Figure 1D, n = 8 cells). In every combination Fe65 turned out to dominate AICD localization, targeting it to spherical nuclear AFT complexes.
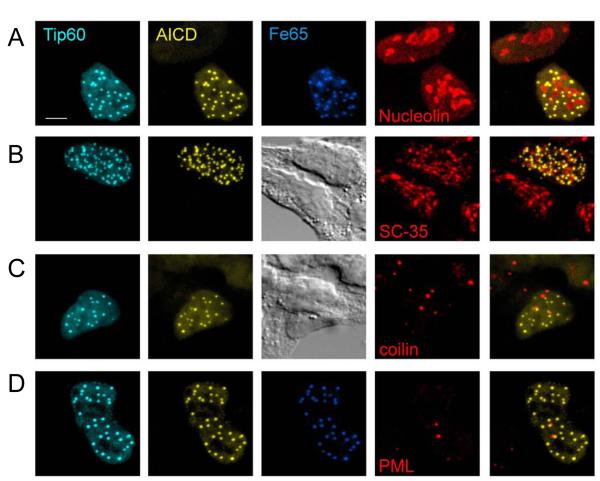
AFT complexes are closely associated with Cajal and PML bodies
Compartmentalization of the nucleus has been reported for various processes related to nuclear function such as transcription, splicing or snRNA synthesis and modification. To analyze the relationship of AFT complexes to described nuclear compartments we performed co-staining of cells transfected to generate AFT complexes. Nucleoli, detected by an antibody against nucleolin/C23, have a fragmented appearance in HEK 293 cells. The majority of nuclear AFT complexes did not localize to nucleoli (Fig 2A), which is also evident as a sparing of nucleolar territorium in further figures. Only one to three AFT complexes per cell could be seen to co-localize with the nucleolar territorium. We next stained splicing speckles with an antibody against SC-35. We did not detect any overlap of AFT complexes with splicing speckles (Figure 2B), which contrasts to a previous report using an antibody against the phosphorylated T668 of APP (38). In our hands this antibody stains neither nuclear AFT complexes nor any other nuclear structure in HEK 293 cells. In a rat neuronal progenitor cell line transfected with human APP the T668-APP antibody labels nuclear speckle structures in all cells, irrespective of the over-expression of APP (data not shown). When Tip60 is expressed alone it localizes to irregular speckle structures reminiscent of splicing speckles. These structures showed no overlap with nucleoli and also did not stain with SC-35 (supplementary figure 2).
Figure 2.
AFT complexes associate with Cajal and PML bodies but are distinct from splicing speckles. Clonal AICD-expressing cells were transfected with CFP-Tip60 and HA-Fe65 to generate AFT complexes. Antibodies against marker proteins of nuclear compartments were used to identify the nature of AFT complexes. No co-localization was seen with Nucleoli (A) or splicing speckles (B). The few cases where AFT complexes are seen inside the nucleolar territorium are probably due to spots lying over the nucleolus. Staining of Cajal bodies (C) and PML bodies (D), both of which have a spherical structure shows that the majority of these compartments are closely associated with an AFT spot. Bar, 5 μm.
The spherical geometry of AFT complexes prompted us to analyze co-localization with Cajal bodies (CBs) and PML bodies, both of which show a similar morphology. Staining of CBs with an antibody against p80 coilin revealed one to seven strongly stained spherical CBs per nucleus (Average = 2.8, n = 31 cells), sometimes accompanied by a weak diffuse staining throughout the nucleoplasm (Figure 2C). No direct overlap was detected between AFT complexes and CBs but 87% of the CBs are in close proximity of AFT complexes. A similar picture was seen when staining for PML to reveal one to three prominent PML bodies (Average = 2.1, n = 9 cells) and far weaker diffuse staining of the nucleoplasm (Figure 2D). Again, there was no overlap but a close association of 79% of the PML bodies with AFT complexes.
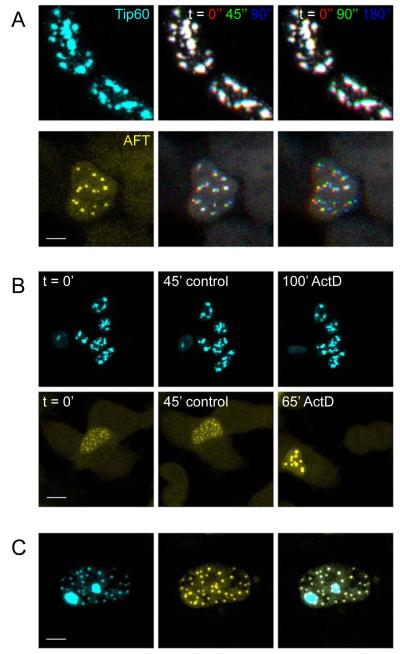
AFT complexes are highly mobile and fuse upon inhibition of transcription
Proteins in the nucleus have been shown to possess a high mobility and bigger structures, for example CBs, have been reported to move within the nucleoplasm. We performed live imaging at 37°C to analyze the mobility of Tip60 speckles and AFT complexes via the fused Cyan and Yellow Fluorescent proteins respectively. Hek293 cells are mobile in culture and changes of cell position and shape are accompanied with movement of the nuclei. Despite these movements Tip60 speckles are relatively stable structures. AFT complexes, in contrast, show a high mobility. The first lane in figure 3A shows a single stack of confocal images of a living cell. Consecutive stacks were imaged 45 seconds apart. Maximum projections of the first three recorded stacks were color-coded in RGB and overlaid (Figure 3A, second lane. Stack corresponding to lane 1 in red, 45seconds later in green and 90 seconds later in blue). In the last lane the RGB-coded time series is spaced 90 seconds apart, so that red has the identical position and green corresponds to the blue position in lane 2. Tip60 speckles appearing white in the last lane have not changed position during 3 minutes. The lower cell has moved upward as seen by the lower red and the upper green/blue staining of speckles. AFT complexes, in contrast to Tip60 speckles, show a high mobility as can be seen in the RGB images showing different extents of separation in various directions (see also supplementary movies).
Figure 3.
Live cell imaging shows that AFT complexes reside in highly mobile nuclear compartments that fuse upon inhibition of transcription. Live cell imaging was performed with the confocal microsope heated to 37°C. (A) Tip60 speckles imaged via the fused CFP were relatively stable, whereas AFT complexes visualized by the fused Citrine are highly mobile. The first lane shows a single confocal stack. In the second lane three consecutive stacks imaged 45 seconds apart are color-coded in RGB. In the third lane the consecutive stacks are imaged 90 seconds apart. HEK cells are mobile in culture, resulting in concomitant movement of the nucleus, which can be seen by red coloring of Tip60 speckles at the bottom and the blue at the top. On top of this movement AFT complexes are highly mobile with different velocities (see also supplementary movies). (B) Inhibition of transcription by ActD had no effect on Tip60 speckles but caused a rapidly initiated fusion of AFT complexes. (C) The different behavior of Tip60 speckle and AFT spot compartments is also mirrored in the fact that these two compartments do not co-localize in cells where not all of the Tip60 has been relocated to AFT complexes. Bar, 7.5 μm in A upper, 5 μm in A lower, 20 μm in B upper, 10 μm in B lower, 5 μm in C.
We next blocked transcription with Actinomycin D (ActD). No major effects on speckle morphology were visible after 100 minutes of ActD treatment were visible (n = 24 cells in 7 experiments, Fig 3B, upper and supplementary movie 1). Inhibition of transcription had dramatic effects on AFT complexes however, leading to fusion into larger structures becoming evident a few minutes after the addition of ActD (Figure 3B lower and supplementary movie 2). We imaged a total of 16 cells (13 experiments) with AFT complexes and all showed fusion of spots starting soon after inhibition of transcription. We imaged one cell with AFT complexes for 3 hours before adding ActD to analyze the effects of long-term imaging on AFT spot dynamics. Constantly high mobility of spots was seen throughout the recording period with fusion again beginning shortly after the addition of ActD (supplementary movie 3). The different behavior of Tip60 speckles and AFT complexes points to different nuclear compartments highlighted by these structures. In many of the Fe65-transfected AICD-expressing cells all of the Tip60 is redistributed to spots. In cells where some of the Tip60 is still found in speckles AFT complexes never co-localize with Tip60 speckles but are found at the periphery of the speckle compartment (Figure 3C).
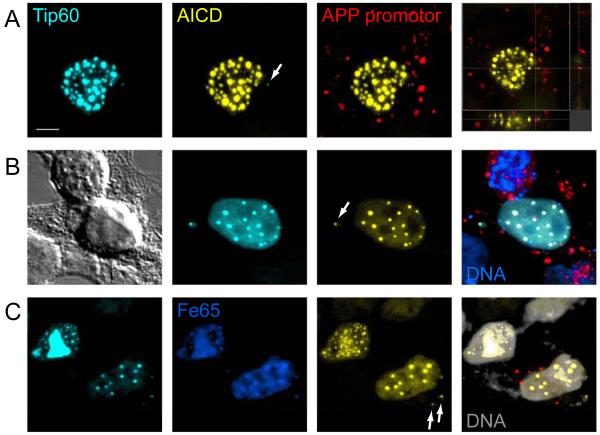
The APP promoter is a substrate for AFT spot formation
We have acquired over 300 confocal images of cells with AFT complexes and in every case these spots were confined to the nucleus. We have recently shown that APP is a target gene for AICD-regulated transcription (57). Assuming that AFT complexes are sites of transcription, the APP promoter represents a substrate that should induce AFT spot formation. We transfected digoxigenin-labeled plasmids, containing APP promoter fragments, together with Tip60 and Fe65, into AICD-expressing cells. During transient transfection plasmids are endocytosed and subsequently localized to the cytoplasm. Retrograde transport leads to accumulation of plasmids in the perinuclear space from where they can enter the nucleus when the nuclear membrane breaks down during mitosis (62). Thus, 20 hours after transfection many APP promoter plasmids are still localized to the cytoplasm and this resulted in the formation of extranuclear AFT complexes in the vicinity of nuclei (Figure 4A). Staining of the APP promoter plasmids revealed both cytoplasmic and extracellular plasmid cDNA. In many cases we observed plasmid labeling in the close vicinity of extranuclear complexes, less than 1 μm away. Binding of AFT complexes and additional unidentified proteins to the APP promoter sequence probably shields the Dig-labeled DNA from detection by anti-Dig antibodies. Only the backbone vector sequences will be detected, which would be approximately 700 nm away with a 2000 base pair stretch of DNA (base pair stacking distance 0.34 nm). Transfected plasmids are retrogradely transported to the vicinity of the nucleus in 24 hours. To generate extranuclear AFT complexes in a larger distance from the nucleus we first transfected Fe65 and Tip60 into AICD-expressing cells, which leads to the formation of nuclear spots. Three hours before fixing the cells were additionally transfected with the APP promoter plasmid. Extranuclear AFT complexes could now be observed at greater distances from the nucleus (Figure 4B). We imaged 30 cells with extranuclear AFT complexes and detected a digoxigenin-labeled promoter plasmid in the vicinity of 9 of these spots. Extensive washing resulted in better removal of digoxigenin-labeled DNA, not taken up by cells, clearly showing that cells transfected with APP promoter sequences also show extranuclear AFT complexes (Figure 4C).
Figure 4.
APP promoter sequences can induce the formation of extranuclear AFT complexes. (A) Co-transfection of Dig-labeled (red) promoter plasmids into cells transfected to generate AFT complexes induced the formation of extranuclear AFT complexes close to sites of labeled promoter plasmid (arrow). (B) Fe65 and Tip60 were transfected to generate AFT complexes and APP promoter plasmids were subsequently transfected 3 hours prior to fixation. Extranuclear AFT complexes can now be observed at greater distances from the nucleus (arrow) with nearby labeling of promoter DNA. (C) Extensive washing removes much of the extracellular promoter plasmid DNA, while extranuclear AFT complexes are still found to be associated with APP promoters. Bar, 5 μm.
Nuclear AFT complexes localize to the genomic loci of AICD-regulated genes
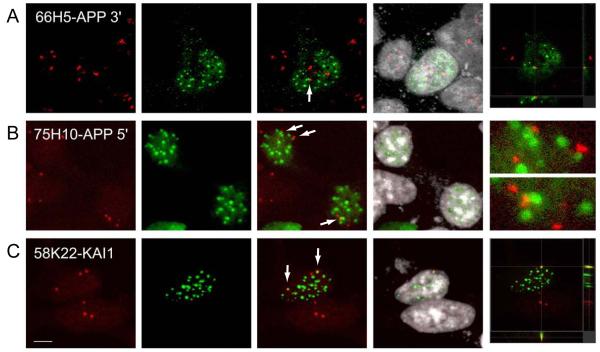
To analyze if AFT complexes localize to AICD-regulated genes we labeled the endogenous DNA loci of APP and KAI1 by fluorescence in situ hybridization (FISH). We used two different Bacterial artificial chromosomes (BACs) hybridizing with the 3′ end of the APP locus (RP11-66H5) and the 5′ end/promoter region (RP11-75H10). On average we observed FISH signals in 3.4 loci (range 2-5), which could be caused by aneuploidy of HEK293 cells. Alternatively, the additional hybridization signals could also stem from binding of BAC sequences to unspecific chromosomal loci. AICD-expressing cells were transfected with Fe65 and Tip60 to generate AFT complexes. FISH destroys the fluorescence of Citrine and CFP, therefore a staining of the myc tag of Fe65 was used to identify AFT complexes. 54% of the 66H5 FISH signals (n = 48 cells) and 65% of the 75H10 signals (n = 36 cells) were tightly associated with AFT complexes (Figure 5). Performing FISH to detect the chromosomal KAI1 locus (RP58K22) we observed on average 2.8 hybridization signals (range 2-4, n = 37 cells) with 45% of them being tightly associated with AFT complexes. Thus, AFT complexes are in contact with the chromosomal loci of two AICD-regulated genes.
Figure 5.
AFT complexes are closely associated with the chromosomal loci of AICD-regulated genes. Fluorescence in situ hybridization was performed on clonal cells transfected to generate AFT complexes. (A) Hybridization with a BAC containing sequences of the 3′ end of the APP gene. Close association of the FISH signal with an AFT spot is marked by an arrow. (B) FISH signals from a BAC containing sequences from the APP promoter are similarly associated with AFT complexes (arrows). (C) Hybridization with a BAC containing sequences of the KAI1 gene. Two of the labeled genomic loci are associated with AFT complexes. Bar, 7.5 μm.
AICD expression leads to increased turnover of APP
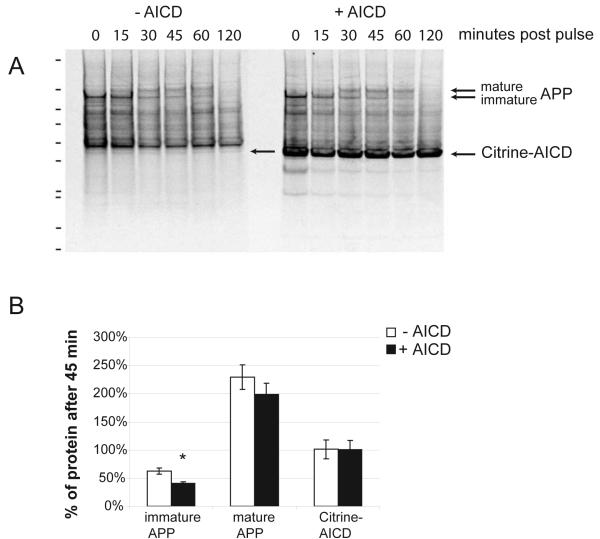
We have previously shown that AICD increases the levels of APP mRNA and protein. Whereas the increase in full-length protein was not significant, there was a significant increase in the levels of α- and β-stubs, suggesting that in addition to APP synthesis the cleavage and turnover is enhanced. To analyze the half-life of APP we pulse chased clonal cells with or without the induction of Citrine-AICD expression. APP was immunoprecipitated with an antibody against the C-terminus that also detects Citrine-AICD. The autoradiograms of SDS-PAGE separated precipitates show the rapid conversion of immature APP to mature glycosylated APP and further degradation with a half-life of approximately 45 minutes (Figure 6A). We repeated experiments at 45 minutes when immature and glycosylated APP are clearly detectable (n = 5). Quantification of APP levels showed that the induction of AICD expression significantly reduced the half-life of immature APP and also reduced mature APP (Figure 6B). Citrine-AICD levels in contrast stayed constant during 45 minutes in both induced and non-induced (leakage expression) conditions (Figures 6A and B).
Figure 6.
AICD expression increases the turnover of APP. (A) Autoradiograph of SDS PAGE from clonal AICD-expressing cell lines. Cells without or with induction of AICD expression were pulsed with Easy Tag 35S Label Mix and chased for various times. Immunoprecipitation with an antibody against the C-terminus of APP retrieves endogenous APP and the induced Citrine-AICD (arrows). Full-length APP quickly matures and is thereafter degraded. (B) Quantification at 45 minutes post pulse revealed a reduced half-life when Citrine AICD was expressed, which was significant for immature APP (n = 5). No changes in the levels of Citrine-AICD, either due to leakage expression or induction, where seen in 45 minutes.
The Notch intracellular domain co-localizes with nuclear AFT complexes
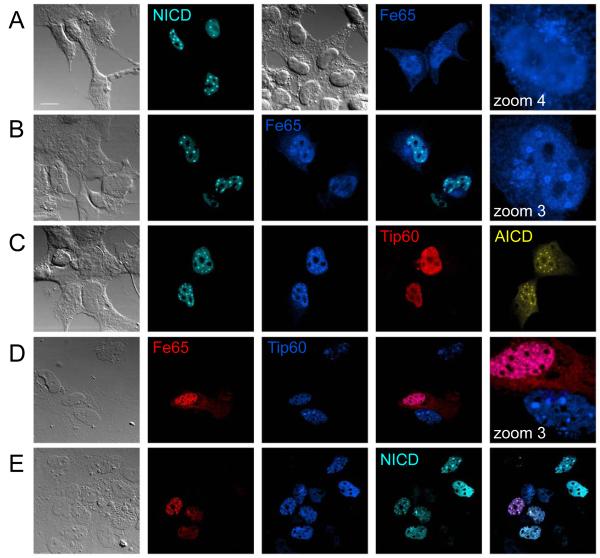
As shown for APP, ectodomain shedding of the Notch receptor is followed by γ-secretase cleavage. The released Notch intracellular domain (NICD) has been shown to activate the transcription of several NICD-target genes. To analyze the nuclear localization of NICD, we tagged it at the N-terminus with the enhanced Cyan fluorescent protein Cerulean (Cer). Expression of Cer-NICD in HEK 293 cells resulted in nuclear localization to spherical spots even in the absence of expressing additional proteins (Figure 7A). Co-expression of Fe65 that on its own distributes throughout the cell in HEK 293 (Figure 7A) (57), showed that it co-localizes with NICD in nuclear spots (Figure 7B). In cells expressing AICD, Fe65 and Tip60, the co-expression of NICD revealed that NICD localizes to identical nuclear structures as highlighted by AFT complexes (Figure 7C). To analyze if endogenous AICD is involved in the formation of nuclear NICD spots we transfected primary astrocytes derived from APP knockout mice. Fe65 can bind to Tip60 and localize it to nuclear spots in the absence of AICD (Figure 7D). Similarly NICD can localize to nuclear spots together with FE65 and Tip60, in the absence of AICD (Figure 7E).
Figure 7.
NICD localizes to the same nuclear structures as AICD. (A) Transfection of Cerulean-NICD into HEK 293 showed that it localizes to nuclear complexes, whereas HA-Fe65 is distributed throughout the cells with diffuse accumulation in the nucleus. (B) Co-transfection of NICD and Fe65 targeted Fe65 to the same nuclear complexes. (C) Clonal Citrine-AICD expressing cells were transfected with Myc-Tip60 and HA-Fe65 to generate AFT complexes. Co-transfected NICD localizes to all nuclear AFT complexes. (D) APP knockout fibroblasts were transfected with Fe65 and Tip60. Together they localize in nuclear complexes, whereas Tip60 alone is seen in speckles. (E) NICD transfected together with Tip60 and Fe65 in APP knockout fibroblasts localizes to identical nuclear complexes. Bar, 15 μm.
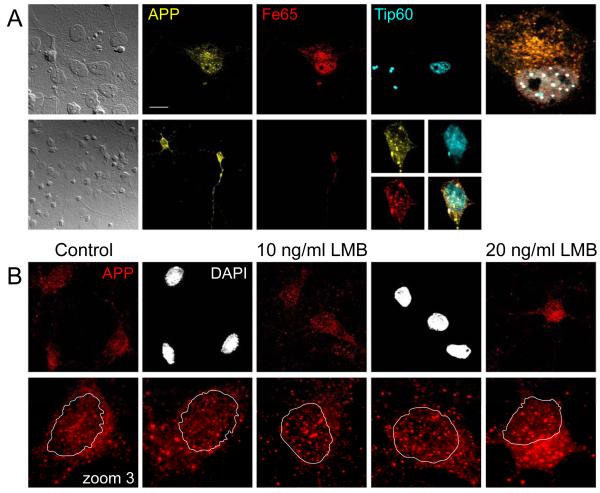
AICD cycles to nuclear spots in neurons
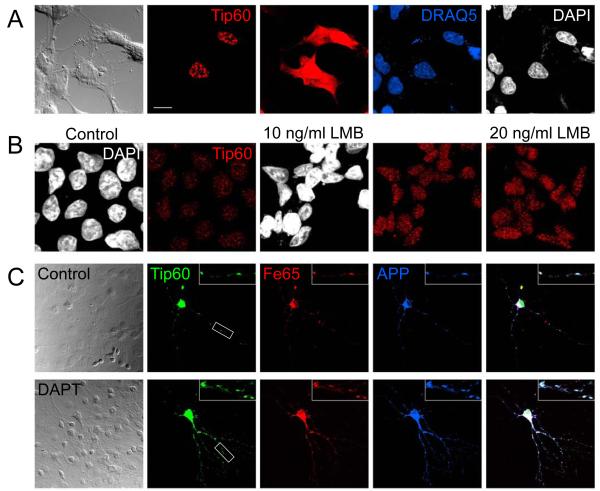
We have observed the formation of AFT complexes in various cell lines (HEK 293, SH-SY5Y, COS-7, rat progenitor cells and fibroblasts). Transfection of APP-Citrine together with CFP-Tip60 and HA-Fe65 into mouse primary astrocytes and neurons similarly lead to the formation of nuclear AFT complexes (Figure 8A). Thus, nuclear signaling by APP is conserved in primary brain cells. To analyze whether endogenous AICD similarly cycles between the cytosol and the nucleus in primary cells we inhibited nuclear export with LMB in rat neurons. Antibody staining detected APP in vesicular structures throughout the processes. We outlined the DAPI-stained nuclei to better identify nuclear AICD in the zoomed pictures of individual neurons (Figure 8B). AICD can already be detected in nuclear spots under control conditions. Inhibition of nuclear export with LMB caused a clear accumulation of AICD in nuclear spots.
Figure 8.
AICD localizes to nuclear AFT complexes in neurons and astrocytes. (A) Primary astrocytes (upper row) and neurons (lower row) were co-transfected with APP-Citrine, HA-Fe65 and CFP-Tip60. Spherical nuclear complexes are formed in both cell types. (B) Staining for endogenous APP shows that AICD is localized to nuclear spots. Inhibition of nuclear export with LMB leads to accumulation of AICD in spots. The lower row shows zoomed neuronal somata with the outline of the DAPI stained nucleus. Bar, 15 μm in A upper, 30 μm and 7.5 μm in A lower, 10 μm in B.
Tip60 co-localizes with APP and Fe65 in neuronal processes
We transfected HEK293 cells with a Tip60 construct harboring an N-terminal myc tag and detected nuclear speckles with myc antibody staining. In addition, with increased photomultiplier gain, we detected the presence of Tip60 in the cytosol (Figure 9A). Although some Tip60 would be expected to be found in the cytosol, where nuclear proteins are synthesized, Tip60 staining was widespread even entering the processes. This was not seen when staining other nuclear proteins (i.e. RNA polymerase II, not shown). To analyze if Tip60 can cycle from the nucleus to the cytosol we blocked nuclear export with LMB in HEK 293 cells. After 24 hours we saw a strong increase in the staining of endogenous Tip60 in nuclear speckles (Figure 9B). Thus, in addition to AICD and Fe65, Tip60 is actively exported from the nucleus.
Figure 9.
Tip60 undergoes nucleo-cytoplasmic cycling and co-localizes with APP and Fe65 throughout the neurites. (A) Myc-tagged Tip60 is localized to nuclear speckles in HEK 293. Increased photomultiplier gain revealed that Tip60 is also present throughout the cytosol. Cells were counterstained with two different DNA-binding dyes (DAPI and DRAQ5) that both stain nuclei. (B) Endogenous Tip60 localizes to nuclear speckles in HEK 293 cells. Inhibition of nuclear export with LMB for 24 hours leads to a strong accumulation of Tip60 in nuclear speckles as can be seen in confocal images acquired with identical photomultiplier settings. (C) Co-transfection of Myc-Tip60, RFP-Fe65 and APP-3HA into primary neurons cultured on astrocytes. Cells were fixed 20 hours after transfection and viewed in the confocal microscope. AFT complexes were formed throughout the neurites in control cells. Inhibition of γ-secretase with DAPT leads to a strong accumulation of AFT complexes in the neurites. Boxed insert is zoomed threefold. Bar, 15 μm in A, 10 μm in B 30 μm in C and 10 μm for the enlarged insert.
We transfected primary neurons with Myc-Tip60, APP-HA and RFP-Fe65 and again nuclear AICD was not detectable by HA-tag staining. The staining of Tip60 showed the strongest signal in nuclear speckles and in addition there was a punctate staining throughout the neurites, co-localizing with Fe65 and APP (Figure 9C). Inhibition of gamma-secretase with DAPT for 24 hours prevented the cleavage of APP and the nuclear translocation of AICD. Under these conditions we observed a strong increase in the amounts of APP, Fe65 and Tip60 localized to neuritic punctae.
Discussion
Regulated intramembrane proteolysis (RIP) by the γ-secretase complex releases the intracellular domains of various type I transmembrane proteins, which are then able to signal to the nucleus. Here we show that AICD liberated from APP localizes to nuclear AFT complexes that represent sites of transcription of AICD-target genes.
APP has a high turnover but the generated AICD is rapidly degraded by the endosomal/lysosomal system resulting in barely detectable cellular levels in Western blots (55). In addition we have reported that AICD in nuclear multiprotein complexes is not accessible by antibodies. Nevertheless, we were able to detect endogenous AICD in nuclear spots after blocking nuclear export. AICD, together with Fe65, constantly cycles between the nucleus and the cytosol (57). Thus, after inhibition of export, AICD accumulates and is seen to localize to nuclear spots similar to AFT complexes detected after transfection with fluorescent-protein tagged constructs.
We have shown that Fe65 effectively competes with other APP-binding proteins, MINT1/X11α and Jip1, and targets AICD to nuclear spots in the presence of Tip60. These spots do not co-localize with splicing speckles or nucleoli but associate closely with Cajal and PML bodies. Both of these nuclear structures can function in transcription and they have been shown to co-localize with an snRNA gene locus, showing a tight association, but no overlap, as seen with AFT complexes (25, 53). The interphase nucleus is a dynamic structure with chromatin remodeling accompanying the transcription of genes. Proteins are highly mobile in the nucleus (8, 46) and even bigger structures, such as PML bodies or Cajal bodies, show a variety of partly energy-dependent movements (37, 47). We observed high mobility of AFT complexes, whereas Tip60 speckles are stationary structures. The histone acetylase Tip60 is involved in many nuclear functions requiring chromatin remodeling, such as transcription and DNA repair. Nuclear proteins are known to recycle between different compartments. We have previously shown that the NKSY to NASA mutation in Tip60, which was reported to abolish the interaction with Fe65 (7), prevents localization of Tip60 to spherical spots containing AFT complexes (57). Tip60 speckles are not identical to splicing speckles and could represent storage or recycling compartments. The fluctuations of Tip60 fluorescence in stationary speckles could resemble the flux of Tip60 to different compartments including AFT complexes, where it supports nuclear functions by acetylating histones and modifying chromatin structure.
Blocking transcription did not influence Tip60 speckles but led to a dramatic fusion of AFT complexes. This is similar to the behavior of RNA polymerase II, which is dispersed in actively transcribing cells and concentrates in speckle-like domains after inhibition of transcription (60). The redistribution of AFT complexes points to a function in transcription. We have localized AFT complexes to two different AICD-regulated genes where they might directly be involved in regulating transcription. We further showed that NICD, another known transcriptional regulator (13, 51), localizes to identical nuclear spots as AFT complexes. These nuclear structures are therefore possibly identical to transcription factories, each of which can associate with a variety of actively transcribed genes from different chromosomal loci (11, 42). The co-localization of AICD and NICD in the same transcription factories suggests that they might regulate the transcription of similar genes. Cross-talk between different signaling pathways is a common phenomenon. APP and Notch signaling pathways interact at various points, including direct interactions of Notch and APP (15, 40) or competition for binding proteins such as Fe65, Jip1 or Numb, which can physically associate with AICD and NICD (16, 19, 49). A recent report showed that NICD can repress AFT-mediated transcription by direct interaction (21). We have shown that NICD can localize Fe65 to nuclear spots and therefore the co-localization of AICD and NICD in nuclear spots is most likely to mutually influence transcriptional activity. The AICD-regulated gene KAI1 has also been shown to be regulated by NICD (17) and other transcription factors such as NF-κB (2) or the estrogen receptor α (3). Many proteins signal to the nucleus and have overlapping sets of target genes. Therefore, it is a daunting task to unequivocally identify genes regulated by AICD (10, 43).
Further support that AFT complexes localize to promoters of transcribed genes was seen when APP promoter plasmid transfection led to the formation of AFT complexes at plasmids localized in the cytosol. In addition, these experiments strongly suggest that transient transfection results in cytosolic localization of the plasmids (62). Most studies using luciferase reporters of AICD-mediated transcription assay 24 hours after transient transfection, using a reporter construct lacking a nuclear targeting sequence. These plasmids will enter the nucleus only after breakdown of the nuclear membrane during mitosis. Nevertheless, as all nuclear proteins involved in transcription are translated in the cytosol the plasmids can compete with nuclear import factors and enable the detection of AICD transcriptional activity (7). A recent report showing reduced transcriptional activity by co-expressing Tip60 (59) could be explained by the trapping of AICD-Fe65 complexes in the nucleus as we observed in the confocal microscope. Overexpression of Tip60 creates nuclear docking sites that can sequestrate AICD-Fe65 complexes that constantly cycle between cytosolic and nuclear compartments and can also be induced to accumulate in spots after inhibition of nuclear export in the absence of Tip60 over-expression (57). Nuclear trapping of AICD should reduce luciferase expression when the reporter plasmids are localized to the cytosol. Trapping of cycling transcription factors, which are subject to nuclear export, by nuclear binding partners has also been shown for β-catenin. Overexpression of the β-catenin binding transcription factor TCF localizes β-catenin to the nucleus (18).
We had previously reported that AICD can regulate the expression of APP. The mRNA levels of APP were increased, accompanied by a rise in full-length APP and processed C-terminal stubs. We have now confirmed by pulse chase experiments that AICD not only increases the expression of APP mRNA but also leads to enhanced processing of the protein, leading to a reduced half-life of full-length APP. Thus, AICD regulates the turnover of APP, possibly via the transcription of other proteins involved in APP metabolism, such as BACE (57).
Nuclear AFT complexes are formed in all cell types analyzed so far, including primary neurons and astrocytes. In neurons we also observed APP-Fe65-Tip60 complexes in neurites, showing an irregular geometry in contrast to the spherical spots seen in the nucleus. These complexes probably lack components that are found in nuclear AFT complexes since antibodies directed against the tag fused to AICD can detect AFT complexes in neurites, but fail to identify nuclear AICD, which can only be detected via the fused fluorescent protein. Preassembled complexes of APP, Fe65 and Tip60 would generate AFT complexes for nuclear signaling directly after γ-secretase cleavage of APP. This is in line with our observation that endogenous Tip60 is subject to nuclear export and thus shows the same cycling between nuclear and cytosolic compartments as AICD and Fe65. The ErbB4 receptor is another example of RIP followed by nuclear signaling. Two proteins that interact with the ErbB4-ICD are TAB2 and N-CoR, components of nuclear co-repressor complexes. They translocate only after γ-secretase cleavage of ErbB4 into the nucleus to regulate transcription (39, 50). Other histone modifying proteins have also been described to cycle across the nuclear membrane. For example, histone deacetylases (HDAC4 and 5) are phosphorylated by neuronal activity-regulated CamKIIα, leading to their nuclear export (9, 28, 32).
The APP holoprotein has a very short half-life due to constant proteolytic processing by α-, β- and γ-secretases. We have provided evidence that the liberated intracellular domain cycles into the nucleus, where it localizes to spherical AFT complexes that are involved in the transcription of AICD-regulated genes. Other transcriptional activators such as NICD co-localize to identical transcription factories, pointing to a cross-talk between different nuclear signaling pathways. Notably, Fe65 and Tip60 are already bound to APP in neuronal processes. APP cleavage by γ-secretase releases AFT complexes from the membrane that are translocated to nuclear transcription factories forming a direct pathway for membrane to nucleus signaling.
Supplementary Material
Supplementary figure 1. AICD in AFT complexes is not accessible by antibodies. HEK 293 were transfected with CFP-Tip60, Myc-Fe65 and APP with a C-terminal fusion of two HA tags followed by the Citrine fluorescent protein (APP-2HA-Citrine). Staining for the HA tags shows a total overlap with Citrine throughout the cell except the nucleus. Nuclear AFT complexes are detected by Citrine fluorescence, but antibody access to the HA tags connecting AICD and Citrine is prevented by the big macromolecular complex of AFT complexes. Lower row of pictures were imaged in x-z mode of the confocal microscope. Bar, 10 μm.
Supplementary figure 2. Tip60 speckles do not co-localize with nucleoli or splicing speckles. HEK 293 cells were transfected with CFP-Tip60 and counterstained for nucleolin (A) or SC-35 (B). Lower row in (B) is a x-z confocal image. Bar, 5 μm.
Supplementary movie 1. Live imaging of CFP-Tip60 speckles in HEK cells. Movie corresponds to cells shown in figure 3B. Z-stacks were recorded every 30 seconds and the movie is played with 5 pictures per second (150-fold speed). Because of the mobility of HEK cells the recording was sometimes interrupted to readjust the imaging window in x-y or z directions. The last stack of the control period is intensity-enhanced to identify the time point of ActD addition. 45 minutes control (15′ + 15′ + 15′) followed by 100 minutes ActD (10′ + 15′ + 3′ + 13′ + 45′ + 14′).
Supplementary movie 2. Live imaging of Citrine-AICD in HEK cells transfected with Fe65 and Tip60 to generate AFT complexes. Blocking transcription with ActD results in fusion of AFT complexes into larger structures. Movie corresponds to cell shown in figure 3B. Z-stacks were recorded every 45 seconds and the movie is played with 3 pictures per second (135-fold speed). Because of the mobility of HEK cells the recording was sometimes interrupted to readjust the imaging window in x-y or z directions. The last stack of the control period is intensity-enhanced to identify the time point of ActD addition. 45 minutes control followed by 65 minutes ActD (45′ + 20′).
Supplementary movie 3. Live imaging of Citrine-AICD in HEK cells transfected with Fe65 and Tip60 to generate AFT complexes. An extended control measurement was performed to exclude that the ActD-induced fusion of AFT complexes is not simply caused by the duration of the live imaging. Z-stacks were recorded every 45 seconds and the movie is played with 5 pictures per second (225-fold speed). Because of the mobility of HEK cells the recording was sometimes interrupted to readjust the imaging window in x-y or z directions. The last stack of the control period is intensity-enhanced to identify the time point of ActD addition. 172 minutes control (24′ + 31′ + 31′ + 51′ + 35′) followed by 72 minutes ActD (22′ + 50′).
Figure 10.
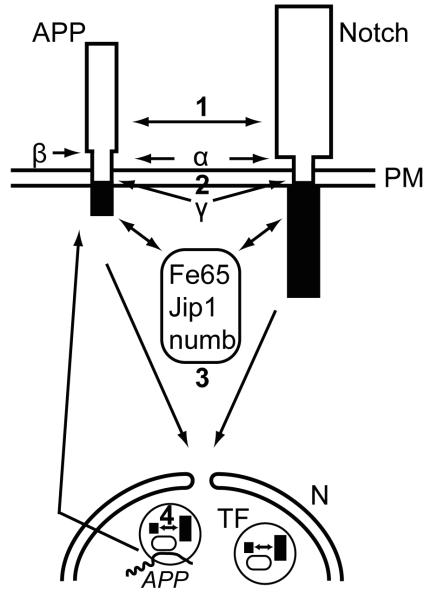
APP and Notch interact at different levels of their nuclear signaling pathways and their intracellular domains co-localize in transcription factories. Notch and APP can directly interact (1) at the plasma membrane (PM) (15, 40). Both proteins undergo ectodomain shedding by the same α-secretases and regulated intramembrane proteolysis by γ-secretase complexes (13, 29) leading to competition for these proteolytic cleavages (2) that are necessary for the translocation of the intracellular domains to the nucleus (N). Several adaptor proteins such as Fe65, Jip1 or numb can physically associate with AICD or NICD (3), thereby influencing their capacity of nuclear translocation and transcriptional activity (16, 19, 49). AICD is transported to the nucleus by Fe65 and together with Tip60 forms spherical nuclear AFT complexes (57) that represent transcription factories (TF). AICD and NICD co-localize in nuclear transcription factories, where they both can interact with Fe65 (4) and mutually influence transcriptional activity (17, 21). Transcription factories containing AFT complexes associate with the genomic loci of regulated genes, i.e. APP and KAI1. Increased generation of AICD enhances the expression of APP and the β-secretase BACE1 (57), leading to enhanced APP turnover due to this positive feedback loop. NICD can associate with Fe65 and AICD in transcription factories and negatively (21) or positively (17) modulate the expression of AICD-regulated genes.
Acknowledgements
We are grateful to Michelle Meier for excellent technical assistance. We would like to thank Raphael Kopan for supplying us with a construct of NICD. An antibodiy against the APP C-terminus was kindly provided by Nicolas Sergeant. This work was supported by the Transregio SFB (6027) on Structure and Function of Membrane Proteins, the SNF NCCR on Neural Plasticity and Repair and the EU grant LSHM-CT-2003-503330 (APOPIS). DKL was supported by grants from Alzheimer’s Association (Zenith Award) and NIA/National Institutes of Health.
References
- 1.Alves da Costa C, Sunyach C, Pardossi-Piquard R, Sevalle J, Vincent B, Boyer N, Kawarai T, Girardot N, St George-Hyslop P, Checler F. Presenilin-dependent gamma-secretase-mediated control of p53-associated cell death in Alzheimer’s disease. J Neurosci. 2006;26:6377–85. doi: 10.1523/JNEUROSCI.0651-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 3.Bao J, Cao C, Zhang X, Jiang F, Nicosia SV, Bai W. Suppression of APP signaling into nucleus by estrogens mediated through the complex formation between the estrogen receptor and Fe65. Mol Cell Biol. 2006 doi: 10.1128/MCB.01280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biederer T, Cao X, Sudhof TC, Liu X. Regulation of APP-dependent transcription complexes by Mint/X11s: differential functions of Mint isoforms. J Neurosci. 2002;22:7340–51. doi: 10.1523/JNEUROSCI.22-17-07340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borg JP, Yang Y, De Taddeo-Borg M, Margolis B, Turner RS. The X11alpha protein slows cellular amyloid precursor protein processing and reduces Abeta40 and Abeta42 secretion. J Biol Chem. 1998;273:14761–6. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- 6.Cao X, Sudhof TC. Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem. 2004;279:24601–11. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–20. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 8.Carmo-Fonseca M, Platani M, Swedlow JR. Macromolecular mobility inside the cell nucleus. Trends Cell Biol. 2002;12:491–5. doi: 10.1016/s0962-8924(02)02387-5. [DOI] [PubMed] [Google Scholar]
- 9.Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–9. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen AC, Selkoe DJ. Response to: Pardossi-Piquard et al., “Presenilin-Dependent Transcriptional Control of the Abeta-Degrading Enzyme Neprilysin by Intracellular Domains of betaAPP and APLP.”. Neuron. 2007;46:541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]; Neuron. 53:479–83. [Google Scholar]
- 11.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–5. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 12.De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113(Pt 11):1857–70. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- 13.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–22. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 14.Eisele YS, Baumann M, Klebl B, Nordhammer C, Jucker M, Kilger E. Gleevec Increases Levels of the Amyloid Precursor Protein Intracellular Domain (AICD) and of the A{beta}-degrading Enzyme Neprilysin. Mol Biol Cell. 2007 doi: 10.1091/mbc.E07-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fassa A, Mehta P, Efthimiopoulos S. Notch 1 interacts with the amyloid precursor protein in a Numb-independent manner. J Neurosci Res. 2005;82:214–24. doi: 10.1002/jnr.20642. [DOI] [PubMed] [Google Scholar]
- 16.Fischer DF, van Dijk R, Sluijs JA, Nair SM, Racchi M, Levelt CN, van Leeuwen FW, Hol EM. Activation of the Notch pathway in Down syndrome: cross-talk of Notch and APP. Faseb J. 2005;19:1451–8. doi: 10.1096/fj.04-3395.com. [DOI] [PubMed] [Google Scholar]
- 17.Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Muller U, De Strooper B. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7:739–45. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 19.Kim JW, Kim MJ, Kim KJ, Yun HJ, Chae JS, Hwang SG, Chang TS, Park HS, Lee KW, Han PL, Cho SG, Kim TW, Choi EJ. Notch interferes with the scaffold function of JNK-interacting protein 1 to inhibit the JNK signaling pathway. Proc Natl Acad Sci U S A. 2005;102:14308–13. doi: 10.1073/pnas.0501600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MY, Ann EJ, Kim JY, Mo JS, Park JH, Kim SY, Seo MS, Park HS. Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol Cell Biol. 2007 doi: 10.1128/MCB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SY, Kim MY, Mo JS, Park HS. Notch1 intracellular domain suppresses APP intracellular domain-Tip60-Fe65 complex mediated signaling through physical interaction. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamcr.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001;276:40288–92. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- 23.Lahiri DK, Robakis NK. The promoter activity of the gene encoding Alzheimer beta-amyloid precursor protein (APP) is regulated by two blocks of upstream sequences. Brain Res Mol Brain Res. 1991;9:253–7. doi: 10.1016/0169-328x(91)90009-m. [DOI] [PubMed] [Google Scholar]
- 24.Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–53. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 25.LaMorte VJ, Dyck JA, Ochs RL, Evans RM. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci U S A. 1998;95:4991–6. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazarov O, Morfini GA, Lee EB, Farah MH, Szodorai A, DeBoer SR, Koliatsos VE, Kins S, Lee VM, Wong PC, Price DL, Brady ST, Sisodia SS. Axonal transport, amyloid precursor protein, kinesin-1, and the processing apparatus: revisited. J Neurosci. 2005;25:2386–95. doi: 10.1523/JNEUROSCI.3089-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li ZW, Stark G, Gotz J, Rulicke T, Gschwind M, Huber G, Muller U, Weissmann C. Generation of mice with a 200-kb amyloid precursor protein gene deletion by Cre recombinase-mediated site-specific recombination in embryonic stem cells. Proc Natl Acad Sci U S A. 1996;93:6158–62. doi: 10.1073/pnas.93.12.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linseman DA, Bartley CM, Le SS, Laessig TA, Bouchard RJ, Meintzer MK, Li M, Heidenreich KA. Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca(2+) //calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J Biol Chem. 2003;278:41472–81. doi: 10.1074/jbc.M307245200. [DOI] [PubMed] [Google Scholar]
- 29.Lleo A, Berezovska O, Ramdya P, Fukumoto H, Raju S, Shah T, Hyman BT. Notch1 competes with the amyloid precursor protein for gamma-secretase and down-regulates presenilin-1 gene expression. J Biol Chem. 2003;278:47370–5. doi: 10.1074/jbc.M308480200. [DOI] [PubMed] [Google Scholar]
- 30.Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002;157:579–89. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda S, Yasukawa T, Homma Y, Ito Y, Niikura T, Hiraki T, Hirai S, Ohno S, Kita Y, Kawasumi M, Kouyama K, Yamamoto T, Kyriakis JM, Nishimoto I. c-Jun N-terminal kinase (JNK)-interacting protein-1b/islet-brain-1 scaffolds Alzheimer’s amyloid precursor protein with JNK. J Neurosci. 2001;21:6597–607. doi: 10.1523/JNEUROSCI.21-17-06597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenzie GJ, Stevenson P, Ward G, Papadia S, Bading H, Chawla S, Privalsky M, Hardingham GE. Nuclear Ca2+ and CaM kinase IV specify hormonal- and Notch-responsiveness. J Neurochem. 2005;93:171–85. doi: 10.1111/j.1471-4159.2005.03010.x. [DOI] [PubMed] [Google Scholar]
- 33.Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- 34.Minopoli G, de Candia P, Bonetti A, Faraonio R, Zambrano N, Russo T. The beta-amyloid precursor protein functions as a cytosolic anchoring site that prevents Fe65 nuclear translocation. J Biol Chem. 2001;276:6545–50. doi: 10.1074/jbc.M007340200. [DOI] [PubMed] [Google Scholar]
- 35.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Muller T, Concannon CG, Ward MW, Walsh CM, Tirniceriu AL, Tribl F, Kogel D, Prehn JH, Egensperger R. Modulation of Gene Expression and Cytoskeletal Dynamics by the APP Intracellular Domain (AICD) Mol Biol Cell. 2006 doi: 10.1091/mbc.E06-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muratani M, Gerlich D, Janicki SM, Gebhard M, Eils R, Spector DL. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat Cell Biol. 2002;4:106–10. doi: 10.1038/ncb740. [DOI] [PubMed] [Google Scholar]
- 38.Muresan Z, Muresan V. A phosphorylated, carboxy-terminal fragment of beta-amyloid precursor protein localizes to the splicing factor compartment. Hum Mol Genet. 2004;13:475–88. doi: 10.1093/hmg/ddh054. [DOI] [PubMed] [Google Scholar]
- 39.Ni CY, Murphy MP, Golde TE, Carpenter G. gamma - Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–81. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 40.Oh SY, Ellenstein A, Chen CD, Hinman JD, Berg EA, Costello CE, Yamin R, Neve RL, Abraham CR. Amyloid precursor protein interacts with notch receptors. J Neurosci Res. 2005;82:32–42. doi: 10.1002/jnr.20625. [DOI] [PubMed] [Google Scholar]
- 41.Okochi M, Fukumori A, Jiang J, Itoh N, Kimura R, Steiner H, Haass C, Tagami S, Takeda M. Secretion of the Notch-1 Abeta-like peptide during Notch signaling. J Biol Chem. 2006;281:7890–8. doi: 10.1074/jbc.M513250200. [DOI] [PubMed] [Google Scholar]
- 42.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 43.Pardossi-Piquard R, Dunys J, Kawarai T, Sunyach C, Alves da Costa C, Vincent B, Sevalle J, Pimplikar S, St George-Hyslop P, Checler F. Response to Correspondence: Pardossi-Piquard et al., “Presenilin-Dependent Transcriptional Control of the Abeta-Degrading Enzyme Neprilysin by Intracellular Domains of betaAPP and APLP.”. Neuron. 2007;46:541–554. doi: 10.1016/j.neuron.2007.01.024. [DOI] [PubMed] [Google Scholar]; Neuron. 53:483–6. [Google Scholar]
- 44.Pardossi-Piquard R, Petit A, Kawarai T, Sunyach C, Alves da Costa C, Vincent B, Ring S, D’Adamio L, Shen J, Muller U, St George Hyslop P, Checler F. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005;46:541–54. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Passer B, Pellegrini L, Russo C, Siegel RM, Lenardo MJ, Schettini G, Bachmann M, Tabaton M, D’Adamio L. Generation of an Apoptotic Intracellular Peptide by gamma-Secretase Cleavage of Alzheimer’s Amyloid beta Protein Precursor. J Alzheimers Dis. 2000;2:289–301. doi: 10.3233/jad-2000-23-408. [DOI] [PubMed] [Google Scholar]
- 46.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–9. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 47.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–74. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–9. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 49.Roncarati R, Sestan N, Scheinfeld MH, Berechid BE, Lopez PA, Meucci O, McGlade JC, Rakic P, D’Adamio L. The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci U S A. 2002;99:7102–7. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-Dependent ErbB4 Nuclear Signaling Regulates the Timing of Astrogenesis in the Developing Brain. Cell. 2006;127:185–97. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 51.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–6. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 52.Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol. 2004;6:1054–61. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 53.Sun J, Xu H, Subramony SH, Hebert MD. Interactions between coilin and PIASy partially link Cajal bodies to PML bodies. J Cell Sci. 2005;118:4995–5003. doi: 10.1242/jcs.02613. [DOI] [PubMed] [Google Scholar]
- 54.Taru H, Kirino Y, Suzuki T. Differential roles of JIP scaffold proteins in the modulation of amyloid precursor protein metabolism. J Biol Chem. 2002;277:27567–74. doi: 10.1074/jbc.M203713200. [DOI] [PubMed] [Google Scholar]
- 55.Vingtdeux V, Hamdane M, Begard S, Loyens A, Delacourte A, Beauvillain JC, Buee L, Marambaud P, Sergeant N. Intracellular pH regulates amyloid precursor protein intracellular domain accumulation. Neurobiol Dis. 2007;25:686–96. doi: 10.1016/j.nbd.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 56.Vingtdeux V, Hamdane M, Gompel M, Begard S, Drobecq H, Ghestem A, Grosjean ME, Kostanjevecki V, Grognet P, Vanmechelen E, Buee L, Delacourte A, Sergeant N. Phosphorylation of amyloid precursor carboxy-terminal fragments enhances their processing by a gamma-secretase-dependent mechanism. Neurobiol Dis. 2005;20:625–37. doi: 10.1016/j.nbd.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 57.von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci. 2004;117:4435–48. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- 58.Walsh DM, Fadeeva JV, LaVoie MJ, Paliga K, Eggert S, Kimberly WT, Wasco W, Selkoe DJ. gamma-Secretase cleavage and binding to FE65 regulate the nuclear translocation of the intracellular C-terminal domain (ICD) of the APP family of proteins. Biochemistry. 2003;42:6664–73. doi: 10.1021/bi027375c. [DOI] [PubMed] [Google Scholar]
- 59.Yang Z, Cool BH, Martin GM, Hu Q. A dominant role for FE65 (APBB1) in nuclear signaling. J Biol Chem. 2006;281:4207–14. doi: 10.1074/jbc.M508445200. [DOI] [PubMed] [Google Scholar]
- 60.Zeng C, Kim E, Warren SL, Berget SM. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. Embo J. 1997;16:1401–12. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang YW, Wang R, Liu Q, Zhang H, Liao FF, Xu H. Presenilin/{gamma}-secretase-dependent processing of beta-amyloid precursor protein regulates EGF receptor expression. Proc Natl Acad Sci U S A. 2007;104:10613–8. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou R, Geiger RC, Dean DA. Intracellular trafficking of nucleic acids. Expert Opin Drug Deliv. 2004;1:127–40. doi: 10.1517/17425247.1.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. AICD in AFT complexes is not accessible by antibodies. HEK 293 were transfected with CFP-Tip60, Myc-Fe65 and APP with a C-terminal fusion of two HA tags followed by the Citrine fluorescent protein (APP-2HA-Citrine). Staining for the HA tags shows a total overlap with Citrine throughout the cell except the nucleus. Nuclear AFT complexes are detected by Citrine fluorescence, but antibody access to the HA tags connecting AICD and Citrine is prevented by the big macromolecular complex of AFT complexes. Lower row of pictures were imaged in x-z mode of the confocal microscope. Bar, 10 μm.
Supplementary figure 2. Tip60 speckles do not co-localize with nucleoli or splicing speckles. HEK 293 cells were transfected with CFP-Tip60 and counterstained for nucleolin (A) or SC-35 (B). Lower row in (B) is a x-z confocal image. Bar, 5 μm.
Supplementary movie 1. Live imaging of CFP-Tip60 speckles in HEK cells. Movie corresponds to cells shown in figure 3B. Z-stacks were recorded every 30 seconds and the movie is played with 5 pictures per second (150-fold speed). Because of the mobility of HEK cells the recording was sometimes interrupted to readjust the imaging window in x-y or z directions. The last stack of the control period is intensity-enhanced to identify the time point of ActD addition. 45 minutes control (15′ + 15′ + 15′) followed by 100 minutes ActD (10′ + 15′ + 3′ + 13′ + 45′ + 14′).
Supplementary movie 2. Live imaging of Citrine-AICD in HEK cells transfected with Fe65 and Tip60 to generate AFT complexes. Blocking transcription with ActD results in fusion of AFT complexes into larger structures. Movie corresponds to cell shown in figure 3B. Z-stacks were recorded every 45 seconds and the movie is played with 3 pictures per second (135-fold speed). Because of the mobility of HEK cells the recording was sometimes interrupted to readjust the imaging window in x-y or z directions. The last stack of the control period is intensity-enhanced to identify the time point of ActD addition. 45 minutes control followed by 65 minutes ActD (45′ + 20′).
Supplementary movie 3. Live imaging of Citrine-AICD in HEK cells transfected with Fe65 and Tip60 to generate AFT complexes. An extended control measurement was performed to exclude that the ActD-induced fusion of AFT complexes is not simply caused by the duration of the live imaging. Z-stacks were recorded every 45 seconds and the movie is played with 5 pictures per second (225-fold speed). Because of the mobility of HEK cells the recording was sometimes interrupted to readjust the imaging window in x-y or z directions. The last stack of the control period is intensity-enhanced to identify the time point of ActD addition. 172 minutes control (24′ + 31′ + 31′ + 51′ + 35′) followed by 72 minutes ActD (22′ + 50′).