Abstract
Several P450 enzymes localized in the endoplasmic reticulum and thought to be involved primarily in xenobiotic metabolism, including mouse and rat CYP1A1 and mouse CYP1A2, have also been found to translocate to mitochondria. We report here that the environmental toxin 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) induces enzymatically active CYP1A4/1A5, the avian orthologs of mammalian CYP1A1/1A2, in chick embryo liver mitochondria as well as in microsomes. P450 proteins and activity levels (CYP1A4-dependent 7-ethoxyresorufin-O-deethylase and CYP1A5-dependent arachidonic acid epoxygenation) in mitochondria were 23–40% of those in microsomes. DHET formation by mitochondria was twice that of microsomes and was attributable to a mitochondrial soluble epoxide hydrolase as confirmed by Western blotting with antiEPHX2, conversion by mitochondria of pure 11,12 and 14,15-EET to the corresponding DHETs and inhibition of DHET formation by the soluble epoxide hydrolase inhibitor, 12(-3-adamantan-1-yl-ureido)-dodecanoic acid (AUDA). TCDD also suppressed formation of mitochondrial and microsomal 20-HETE. The findings newly identify mitochondria as a site of P450-dependent arachidonic acid metabolism and as a potential target for TCDD effects. They also demonstrate that mitochondria contain soluble epoxide hydrolase and underscore a role for CYP1A in endobiotic metabolism.
Keywords: 2,3,7,8-tetrachlorodibenzo-p-dioxin; CYP1A4; CYP1A5; soluble epoxide hydrolase; arachidonic acid; EETs; DHETs; mitochondria; peroxisomes; chicken
INTRODUCTION
Activation of the aryl hydrocarbon receptor (AhR)1 by the environmental toxin 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and other AhR ligands leads to induction of enzymes in the cytochrome P450 1A family (mammalian CYPs 1A1 and 1A2 and the chick orthologs, 1A4 and 1A5). CYP1A enzymes have been considered to be catalysts of metabolism of foreign compounds, mainly toxic chemicals. However, there is increasing evidence that CYP1A enzymes can participate in metabolism of endogenous substrates, including estrogens, bilirubin, melatonin and arachidonic acid (reviewed in Rifkind, 2006 [1]). Like most P450s involved in xenobiotic metabolism, CYP1A enzymes are primarily located in microsomes (endoplasmic reticulum or ER) in contrast to P450 enzymes catalyzing steroid and bile acid synthesis and metabolism of Vitamin D which are selectively targeted to mitochondria [2]. It has become clear, largely based on work of Avadhani et al., that several P450s localized in the ER, including rat and mouse CYP1A1 in both liver and extrahepatic organs, can undergo processing with targeting to mitochondria [3–7]. It has recently been shown that TCDD-induced CYP1A2 is also found in mouse liver mitochondria [8].
Previous work from this laboratory showed that TCDD-induced CYP1A4 is entirely responsible for the increased 7-ethoxyresorufin deethylase (EROD) activity and CYP1A5 for the increased epoxygenation of the endogenous membrane fatty acid, arachidonic acid (aa), in chick embryo (CE) liver microsomes [9–11]. Further, TCDD treatment suppressed formation of another P450 aa metabolite, 20-HETE, in both mammalian and CE liver microsomes [12, 13].
P450 enzymes generate different aa metabolites from those produced by cyclooxygenases (prostaglandins) or lipoxygenases (leukotrienes). The P450 aa products include four regioisomeric aa epoxides (5,6–8,9–11,12–14,15- epoxyeicosatrienoic acids, EETs), each with 2 stereoisomers, mid-chain mono-hydroxylated products (HETEs) and monohydroxylated ω terminal alcohols (including 18-, 19- and 20-HETE) [14]. EETs can be converted to vic-dihydroxyeicosatrienoic acids (DHETs) by soluble epoxide hydrolase (EPHX2, sEH) or microsomal epoxide hydrolase (EPHX1) [15, 16]. TCDD-induced CYP1A5 [10] and human CYP1A2 [17, 18] generate mainly EETs as do some constitutive P450 aa epoxygenases (i.e. CYPs 2B, 2C and 2J) [1, 16, 19], while P450s in the CYP4 family are responsible for 20-HETE formation [19, 20].
P450 aa metabolites are biologically active compounds, whose effects have been most extensively investigated and recognized with respect to the cardiovascular system. EETs are considered to be generally vasodilatory and to provide protection in cardiac ischemia-reperfusion injury, while 20-HETE is considered to be mainly vasoconstrictive and to have a possible role in hypertension [16, 19]. The role of P450 aa metabolites in liver physiology and pathophysiology is not yet understood [21].
Activation of the AhR is required for TCDD to induce CYP1A, the major biochemical effect of TCDD, and for TCDD toxicity. The toxic effects include a wasting syndrome that appears to be associated with restricted nutrient supply and energy loss and may involve mitochondrial dysfunction [22–24]. The role of CYP1A products in TCDD toxicity has not been established.
In the research reported here we asked the following questions: Does TCDD induce CYP1A4 and 1A5 in CE liver mitochondria and, if induced, does mitochondrial CYP1A5 catalyze P450 dependent aa epoxygenation? Does TCDD affect 20-HETE formation in liver mitochondria as well as in microsomes?
The results show that in CE liver mitochondria: 1) TCDD induces CYP1A4 and CYP1A5, 2) the induced P450s are enzymatically active, as shown by EROD and aa epoxygenation, and 3) TCDD suppresses 20-HETE formation. The findings support a mitochondrial site of action for TCDD, demonstrate that liver mitochondria are a potential site of P450-dependent aa metabolism and reinforce a role for CYP1A in endobiotic metabolism. They also show that mitochondria contain soluble epoxide hydrolase resulting in greater DHET formation in mitochondria.
MATERIALS AND METHODS
Reagents
Chemicals were from Sigma-Aldrich (St. Louis, MO), unless otherwise indicated. Reagents used for high-pressure liquid chromatography (HPLC) were at HPLC-grade. Fertilized chicken eggs, White Leghorn strain, were obtained from Burr Farms (Hampton, CT). NCI Chemical Carcinogen Repository (Kansas City, MO) provided 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD).
Treatment of chick embryos (CE) and preparation of subcellular fractions from liver
Fertilized chicken eggs were incubated at 37° at high humidity. Embryos were treated at 16 or 17 days of gestation (hatching occurs at day 21), unless otherwise indicated, by injecting the compounds through a small hole in the shell into the fluids surrounding the embryo. TCDD treatment was at 1 nmol/egg (3.2 x 10−4 mg) in 0.005 ml of dioxane for 24 hr; controls received 0.005 ml of dioxane for the same time period.
To prepare mitochondria by differential centrifugation [25], liver homogenates (1 part tissue to 3 parts (w/v) 0.1 M KPO4, pH 7.4 or 300 mM sucrose in 5 mM HEPES, pH 7.4) were subjected to two centrifugations, each at 1,000g for 5 min, to remove nuclei and cell debris. The resulting supernatant was transferred to a new tube, centrifuged at 10,000g for 10 min, washed and recentrifuged. The 10,000g pellet containing mitochondria, was resuspended in 0.1 M KPO4 or 300 mM sucrose in 5 mM HEPES, pH 7.4. The remaining supernatant was centrifuged at 100,000g for 1 hr to pellet microsomes [26].
To prepare mitochondria using a Percoll density gradient, the procedure of Sims et al. [27] was followed with some modifications: CE liver (2 to 3 g, equivalent to about 10 livers) were homogenized in 1x Percoll isolation buffer (10 mM TRIS-HCl, 0.32 M sucrose, and 1 mM EDTA, pH 7.4) and centrifuged at 1,000g for 10 min (buffer to sample ratio of 14:1). The pellet was discarded and the supernatant was centrifuged again at 1,000g. The resulting supernatant was centrifuged at 12,000g for 15 min. That supernatant was removed and used to obtain microsomes as described above. The 12,000g pellet was resuspended in 14 ml Percoll isolation buffer and centrifuged again at 12,000g for 15 min. The supernatant was discarded and the pellet was resuspended in 9 ml of 15% Percoll in Percoll isolation buffer. Then, 3.5 ml of 40% Percoll, 3.5 ml of 23% Percoll and 3 ml of the sample in 15% Percoll were layered successively in a clear centrifuge tube (three tubes for each sample). The gradient was centrifuged at 31,000g for 5 min. The band at the 23/40% Percoll interface was collected from each tube as the mitochondrial fraction, using a blunt needle. The mitochondrial fractions from the three tubes were combined, diluted with 10 ml Percoll isolation buffer and centrifuged at 16,700g for 10 min. The supernatant was discarded; the pellet was resuspended in 10 ml Percoll isolation buffer and centrifuged at 6,900g for 10 min. The pellet was collected and used as “Percoll mitochondria”.
Peroxisomes were isolated using a Peroxisome Isolation kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, livers were weighed and homogenized in 1x peroxisome extraction buffer (diluted from 5x: 25 mM MOPS, pH 7.65 with 1.25 M sucrose, 5 mM EDTA and 0.5% ethanol provided with the kit), using 16 ml per 4 g liver. The homogenate was centrifuged at 1,000g for 10 min and the supernatant was transferred to a new tube and centrifuged at 2,000g for 10 min. The 2,000g pellet was washed twice in 300 mM sucrose in 5 mM HEPES, pH 7.4 (isolation buffer) and collected. This fraction was designated as “heavy mitochondria” in the manufacturer’s protocol and is referred to here as the “2,000g mitochondria”. The supernatant was centrifuged at 25,000g for 20 min. The resulting pellet (crude peroxisomal fraction), was diluted in 1x peroxisome extraction buffer to 1.2 ml, then 1.69 ml OptiPrep density gradient medium (60% iodixanol in water) and 1.61 ml of 1x OptiPrep dilution buffer (provided as 20x: 100 mM MOPS, pH 8.0 with 20 mM EDTA and 2% ethanol) were added to obtain 4.5 ml of a 22.5% OptiPrep solution. A gradient containing 3 layers was prepared in a clear ultracentrifuge tube; bottom, 2 ml of 27.5% OptiPrep solution in the OptiPrep dilution buffer; middle, 4.5 ml of the crude peroxisomal fraction in 22.5% OptiPrep; top, 2 ml of 20% OptiPrep solution. The gradient was centrifuged for 1.5 hr at 100,000g. The bottom layer containing the purified peroxisomes was removed using a blunt needle, resuspended in 5 ml of isolation buffer (described above), and centrifuged at 25,000g for 20 min. The supernatant was discarded and the pellet resuspended in isolation buffer. Subcellular fractions were stored at −80°.
P450-dependent arachidonic acid (aa) metabolism
Arachidonic acid metabolism was assayed by the method of Capdevila et al. [10, 28]. Reaction mixtures (0.25 ml total vol) contained 75 to 150 μg of protein as indicated in the figure legends, and 30 μM [1-14C] arachidonic acid (53 mCi/mmol) (Perkin Elmer, Torrance, CA). After preincubation for 2 min at 37°, reactions were started with 1 mM NADPH (samples without the addition of NADPH showed no activity), 10 mM isocitric acid, 0.2 U of isocitric dehydrogenase/ml, and 10 mM MgCl2 and incubated in a shaking water bath in air for 10 min at 37°. Addition of 0.1 ml of glacial acetic acid was followed by two extractions, each with 3 ml of ethyl acetate containing 0.005% butylated hydroxytoluene. The organic phases were pooled, dried under N2, and resuspended in 0.11 ml of 50% acetonitrile in water with 0.1% acetic acid. Products in 0.05 ml were resolved by reverse phase HPLC using a Vydac C18 column (Vydac, Hesperia, CA) (90 Å, 5 μm particle size, 4.6 x 250 mm) on a linear gradient from 50 to 100% acidified acetonitrile in water at 1 ml per min for 40 min. Radioactivity was measured using a Flo-One Beta Model S radioactivity flow detector (Packard Instrument Company, Downers Grove, IL). Products have been rigorously identified by derivatization and by reference to HPLC retention times of pure standards [10]. When different subcellular fractions were combined, they were added together to reaction mixtures before the initial pre-incubation step, keeping the total vol of the reaction mixture at 0.25 ml.
Effects of mitochondria on pure EETs were examined using [1-14C] 11,12 and 14, 15 EET synthesized and provided as gifts by Dr. Jorge Capdevila, Vanderbilt University, Nashville, TN, see legend to Fig. 5c for experimental details.
7-Ethoxyresorufin deethylase (EROD)
Reaction mixtures contained 1.25 mM EDTA, 1 mg/ml BSA, 1 mM NADPH, 10 μM dicumarol, 4 μM 7-ER, and 20 μg liver protein in 0.039 M Tris buffer, pH 8.3 (0.24 ml total vol) [10]. After preincubation at 37° for 1 min without NADPH, reactions were started with NADPH, incubated for 5 min in a shaking water bath in air at 37°, and stopped with 0.25 ml of ice-cold acetone. Incubation times and tissue concentrations were within the linear range for product formation. Resorufin formation was measured by fluorescence (excitation wavelength 558 nm, emission wavelength 590 nm) and compared to a resorufin standard curve. Background resorufin levels in non-incubated samples were subtracted; each determination was made in triplicate.
SDS-Polyacrylamide Gel Electrophoresis (PAGE)/Western Blotting
SDS-PAGE [29] was performed on slab gels (14.5 cm x 14.5 cm x 1.0 mm) using 3% and 10% acrylamide for the stacking and running gels, respectively. Samples were prepared in 2x sample buffer (125 mM Tris/HCl, pH 6.8, 4% SDS, 16% glycerol, 10% β-mercaptoethanol, 0.002% bromophenol blue), (1:1, v/v), and kept at 100° for 2 min. Protein migrated into the stacking gel for 2 hr at 15 mA and through the separating gel at 30 mA for 3–4 hr. For Western blotting, proteins separated on SDS/PAGE gels were transferred to nitrocellulose at 90 mA overnight at 4°. Completeness of transfer was monitored by post-transfer staining with Coomassie Blue. Membranes were washed with T-PBS (0.1% Triton-X 100, 0.1% SDS in 1x PBS), blocked for 1 hr with 5% non-fat dry milk in T-PBS and then incubated with the following primary antibodies at the dilutions given, in 3% non-fat dry milk in T-PBS: immunopurified antisera to CYP1A4 or CYP1A5 (1:2000) [10]; calnexin (1:7500), porin/VDAC (1:5000), ferredoxin reductase (1:4000), ALA synthase (1:500) (all from ABCAM, Cambridge, MA); rat P450 reductase (1:4000) (rabbit antibody to purified rat reductase [26], Pocono Rabbit Farms, Canadensis, PA); PMP70 (1:10,000) (Sigma, Saint Louis, MO); soluble epoxide hydrolase (EPHX2/sEH) (1:250) (Protein Tech Group, Chicago, IL); CYP4 (1:4000) (rabbit antibody to purified chick CYP4 (Rifkind, AB et al., unpublished), Pocono Rabbit Farms). Preliminary experiments established optimal antibody concentrations for recognition of the chicken proteins. Primary antibodies were incubated for 1 to 5 hr at room temperature. After 3 five min washes with T-PBS, blots were incubated with horseradish peroxidase-linked goat anti-rabbit IgG (1:1000–1:7500) for 1 hr at room temperature in 3% non-fat dry milk in T-PBS. Detection was by chemiluminescence using “Western blotting detection reagent” (Amersham, Buckinghamshire, UK). Densitometry was performed using AlphaEaseFC software (Alpha Innotech, San Leandro, CA).
Immunofluorescence
CYP1A4 cDNA [11] was inserted in the sense orientation in a pCXIZ construct [30] (pCXIZ-CYP1A4) (gift from Dr. Takashi Mikawa, U.C.S.F., San Francisco, CA) after digestion with the restriction enzyme XbaI and used to stably transfect D17 dog fibroblasts (CCL-183, ATCC, Manassas, VA). The transfected cells were incubated in DMEM medium (Cellgro) containing 10 % FBS (Cellgro) with 250 nM Mitotracker 580 (Invitrogen-Molecular Probes) (mitochondrial marker) for 15 min and then fixed in acetone/methanol (1:1, v/v) for 10 min at −20°. The protocol followed was from BD Biosciences with the following modifications: For CYP1A4 detection, cells were incubated with rabbit antiCYP1A4 IgG as primary antibody (1:200 dilution) [10] and donkey anti-rabbit Alexa Fluor 488 as secondary antibody (1:200 dilution). Nuclei were stained with DAPI (Sigma). An Axiovert 35 Zeiss fluorescence microscope was used for visualization.
Other Procedures
Protein concentrations were measured by the method of Lowry et al. [31]. Differences between group means were compared using unpaired t tests (GraphPad Prism 4 software, GraphPad, San Diego, CA). p values <0.05 were accepted as statistically significant. Cleavage and phosphorylation sites of N-terminal sequences of selected proteins were predicted using SignalP 3.0 and NetPhosK 1.0 servers (link of ExPASy) (Expert Protein Analysis System program, Swiss Institute of Bioinformatics, Geneva) [32, 33]. A high probability for putative PKC phosphorylation was assigned for threonine or serine residues with a phosphorylation probability of 50% or more. Alignment of amino acid sequences was performed using DNASTAR MegAlign program (DNASTAR Inc., Madison, WI).
RESULTS
Fig. 1 shows an alignment of the N-terminal amino acid sequences for chicken CYPs 1A4 and 1A5, mouse and rat CYP1A1 and mouse CYP1A2, the CYP1A P450s that have been reported to translocate to mitochondria [3–8], and human CYP1A1. CYPs 1A4 and 1A5 both contain the highly conserved positively charged arginine and lysine residues aligning with amino acids 34, 39 and 42 in rat and mouse CYP1A1 and identified as mitochondrial targeting signals [2, 3]. The chick CYP1A enzymes also contain the trypsin-like cleavage site (underlined) also in rat and mouse CYP1A1 and mouse 1A2 found in P450s that translocate to mitochondria [3, 6], as well as putative PKC phosphorylation sites (circled), also suggested to be involved in mitochondrial translocation [7]. Human CYP1A1 contains the conserved positive amino acids, cleavage site and a putative serine phosphorylation site. The presence of putative mitochondrial targeting signals and cleavage sites in chick CYPs 1A4 and 1A5 illustrates the conservation of these signals among species and provides a structural basis for both chick P450s to be targeted to mitochondria.
Figure 1. Comparison of CYP1A N-terminal amino acid sequences: conserved mitochondrial targeting signals.
N-terminal amino acid sequences of chicken CYP1A4 (accession #: NP_990478) and CYP1A5 (accession #: NP_990477), mouse CYP1A1 (accession #: NP_034122) and CYP1A2 (accession #: NP_034123), human CYP1A1 (accession #: NP_000490) and rat CYP1A1 (accession #: NP_036672) were aligned using DNASTAR software (ClustalW method). The three conserved positively charged amino acids indicated by the ‘+’ preceding the proline-rich hinge region that constitute the mitochondrial targeting signal are shown. Circles indicate putative phosphorylation sites; predicted cleavage sites are underlined.
Initial experiments (data not shown) performed on liver mitochondria prepared from solvent or TCDD-treated CE by a standard differential centrifugation method (see “Materials and Methods”) showed that the mitochondria (10,000g pellet) contained CYP1A4 and 1A5 as shown by Western blotting and were active in CYP1A4-dependent EROD and CYP1A5-dependent aa metabolism at levels close to those in microsomes. However, the mitochondria were contaminated with microsomes to the extent of 40 to 50% as evidenced by Western blotting with microsomal markers P450 reductase or calnexin. (Microsomes were free of mitochondrial contamination as indicated by use of the mitochondrial markers, ALA synthase and porin.) Notably, similar degrees of microsomal contamination have been reported for mammalian mitochondria prepared by differential centrifugation [34–36].
Numerous attempts to diminish microsomal contamination, using multiple washes, centrifugation over a sucrose cushion or sucrose gradient [8] and osmotic swelling followed by shrinkage [37, 38] were unsuccessful. Addition of a re-homogenization step prior to osmotic shock, however, reduced microsomal contamination to 27% suggesting that the microsomes were closely attached to mitochondrial fragments but could be dislodged by physical disruption.
Isolation of mitochondria using a Percoll density gradient [27] substantially reduced microsomal contamination to 6.5 ± 1.1% and 10.6 ± 0.9% (SE) (n = 6) using calnexin or P450 reductase, respectively, as microsomal markers. A representative blot is shown in Fig. 2a. No differences in contamination were seen in preparations for control or TCDD-treated CE. Western blots in the top and middle panels in Fig. 3a also illustrate the mitochondrial enrichment and the reduction in microsomal contamination for mitochondria prepared by Percoll gradient as compared to differential centrifugation (10,000g pellet).
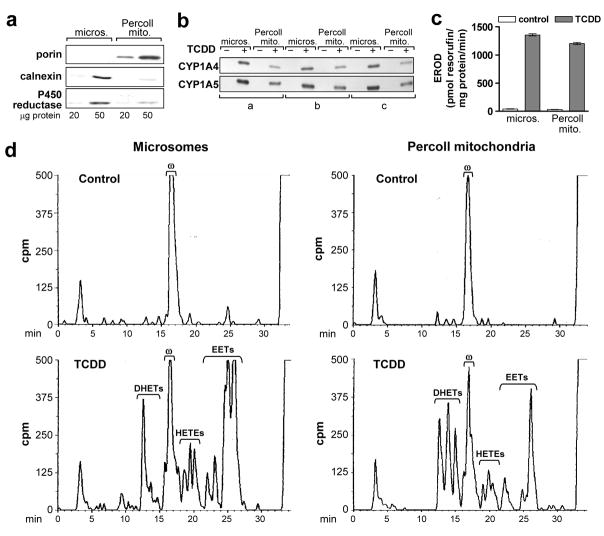
Figure 2. TCDD induced CYP1A4 and 1A5 and associated enzyme activities in mitochondria isolated by Percoll density gradient centrifugation.
Microsomes (micros.) and mitochondria (Percoll mito.) were prepared from livers of 17 day old CE 24 hr after treatment with TCDD at 1 nmol/egg or the solvent dioxane (control) (3 groups each for control and TCDD treated CE, 8 to 10 eggs per group). Mitochondria were isolated using a Percoll density gradient; microsomes were prepared from the same livers, concurrently, as described in “Materials and Methods”. a. Mitochondrial purity. Representative Western blot of Percoll mitochondria and microsomes from one of six groups using antibodies against porin (mitochondrial marker, 31 kDa), calnexin (ER marker, 78 kDa) and P450 reductase (ER marker, 78 kDa), at 20 and 50 μg protein per lane; samples from control or TCDD treated CE produced essentially the same results. b. CYP1A4 and 1A5. Western blotting of mitochondria and microsomes with antisera immunoselective for CYP1A4 or 1A5 (55 and 55.5 kDa, respectively); 10 μg of protein per lane. See Fig. 4c for specificity of antibodies. c. CYP1A4 mediated EROD activity. Mean values ± SE are shown (n = 3 for control and TCDD treated groups). d. P450-dependent arachidonic acid (aa) metabolism. Representative reverse phase HPLC chromatograms for P450-dependent aa metabolism by liver mitochondria and microsomes assayed as described in “Materials and Methods”. Retention times for EETs and DHETs (min) are as follows: 25.5–27.0 (5, 6 EET); 24.8–25.5 (8, 9 EET); 24.0–24.8 (11, 12 EET); 22.8–23.8 (14, 15 EET); 14.5–15.8 (8, 9 DHET); 13.5–14.5 (11, 12 DHET) and 12–13.5 (14, 15 DHET). Retention time for 20-HETE (ω): 16.0–17.5 min, for aa: 32.5 – 35 min. Total counts (cpm) for each chromatogram (left to right): 170,924; 168,865; 173,800 and 177,203; 75 μg protein per reaction mixture. Peaks eluting before 10 min are polar non-enzymatic breakdown products present also in the blanks (aa metabolism reaction mixtures without tissue or NADPH). Results for the two other control and TCDD treated groups were essentially identical.
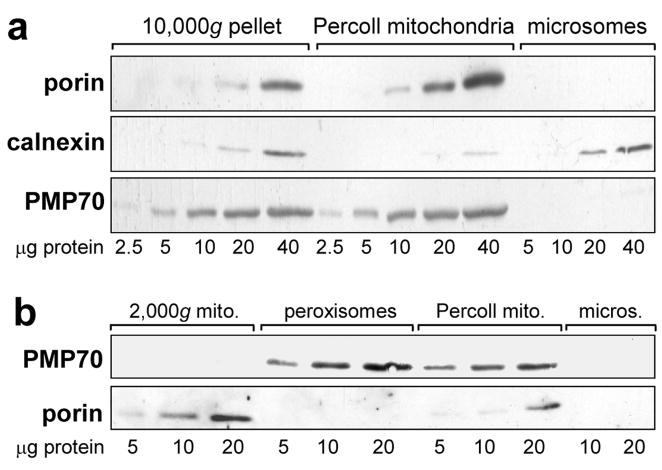
Figure 3. Peroxisomal contamination of mitochondrial fractions and isolation of peroxisomes.
Samples shown are representative of livers from three independent groups of control and TCDD-treated CE. No differences were observed in the results using samples from control or TCDD treated CE. a. Western blots showing peroxisomal contamination of mitochondria prepared by Percoll-gradient or differential centrifugation. Mitochondria isolated by differential centrifugation (10,000g pellet), or utilizing a Percoll density gradient (Percoll mitochondria) and microsomes, immunoblotted using antibodies for porin (mitochondrial marker, 31 kDa), calnexin (ER marker, 78 kDa) and PMP70 (peroxisomal marker, 70 kDa). b. Isolation of peroxisomes free of mitochondria and a mitochondrial fraction (2,000g mitochondria) free of peroxisomes. Western blots of peroxisomes isolated using a Peroxisome Isolation Kit (Sigma-Aldrich), the 2000g mitochondria produced during that procedure, Percoll mitochondria and microsomes, using antibodies against PMP70 and porin.
CYP1A4 and 1A5 were present in the Percoll mitochondria at 49 ± 4% and 62.3 ± 2.6 % of their levels in microsomal fractions, respectively, as determined by densitometry of Western blots (Fig. 2b) (n = 3). The Percoll mitochondria from livers of TCDD-treated CE had 88 ± 4% of the microsomal levels for EROD (Fig. 2c) and 60% ± 0.6% (SE) for aa epoxygenase activity (EETs plus DHETs) (Fig. 2d) (n = 3 independent control and TCDD-treated groups). The substantial P450 activity of mitochondria with reduced microsomal contamination supported the localization of CYP1A in mitochondria.
20-HETE (ω), essentially the sole P450 aa product in livers of control CE, was suppressed by TCDD treatment in mitochondria, as previously observed in microsomes [12, 13], here by means of 27.5 ± 2% in microsomes and 25 ± 8% in mitochondria (p = 0.001 and 0.045, respectively; n = 3).
The aa metabolite pattern differed in mitochondria and microsomes from TCDD treated livers. Although mitochondria had less total aa epoxygenase activity compared to microsomes, DHET production in mitochondria was twice that of microsomes (Fig. 2d). The lower DHET formation in microsomes made it unlikely that microsomal contamination of mitochondria accounted for conversion of EETs to DHETs. In other experiments microsomes from TCDD-treated CE liver when coincubated with control CE liver mitochondria generated 6.1-fold more DHETs than when coincubated with control microsomes, confirming that the epoxide hydrolase activity was associated with the mitochondria (data not shown).
As peroxisomes and mitochondria have similar sedimentation characteristics [39], and peroxisomes had been reported to be present in some mitochondrial preparations [35, 36], it was necessary to exclude peroxisomal contamination as a source of epoxide hydrolase in the mitochondrial fraction. Western blotting with an antibody to PMP70, a peroxisomal marker (Fig. 3a, bottom panel), showed that the Percoll mitochondria were, in fact, contaminated with peroxisomes (while microsomes from the same livers were free of peroxisomes). This peroxisomal contamination was not a consequence of the Percoll density gradient approach per se as mitochondria prepared by differential centrifugation were also contaminated with peroxisomes (Fig. 3a, 10,000g pellet). In other experiments (not shown) mitochondria from mouse livers prepared by differential centrifugation were just as contaminated with peroxisomes as the 10,000g CE mitochondria shown in Fig. 3; thus the peroxisomal contamination of mitochondria was not a particular characteristic of CE liver.
Peroxisomes were purified from CE liver (see “Materials and Methods) to use in assessing peroxisomal soluble epoxide hydrolase activity. The peroxisomal fraction obtained was shown to be free of mitochondria by Western blotting using an antibody against porin (Fig. 3b).
During the isolation of peroxisomes, a fraction designated “heavy mitochondria” in the manufacturer’s instructions (henceforth “2,000g mitochondria”) was obtained (see “Materials and Methods”). This fraction proved to be enriched in mitochondria and lack peroxisomal contamination when compared to Percoll mitochondria as shown by immunoblotting with porin and PMP70, respectively (Fig. 3b). It was also virtually free of microsomal contamination as shown by immunoblotting for calnexin (Fig. 4a) (0.45 ± 0.29% (SE), by densitometry; n = 6). Immunoblotting with P450 reductase (Fig. 4b) confirmed the low microsomal contamination of the 2,000g mitochondria. On the other hand, ferredoxin reductase, which, when coupled with ferredoxin, transfers electrons from NADPH to mitochondrial P450 [2], was present in mitochondria but not in microsomes (Fig. 4b).
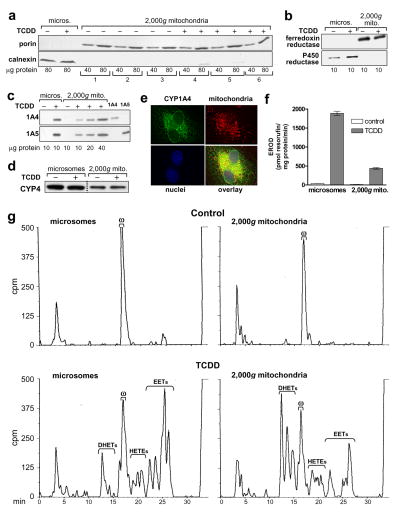
Figure 4. P450 levels and activities in 2,000g mitochondrial preparations; CYP1A4 mitochondrial localization by immunofluorescence.
2,000g mitochondria and microsomes were prepared as described in “Materials and Methods” from CE liver 24 hr after treatment with TCDD at 1 nmol/egg or dioxane, the solvent control, (8–10 eggs per group). a. Mitochondrial purity. Western blots of six groups of 2,000g mitochondria (from 3 control and 3 TCDD-treated groups of CE) using antibodies for porin (mitochondrial marker, 31 kDa) and calnexin (ER marker, 78 kDa); 40 and 80 μg of protein/lane. Microsomes from livers of control and TCDD treated CE are shown for comparison, at 80 μg of protein/lane. b. Western blotting for P450 reductase and ferredoxin reductase. Representative Western blots for microsomes and 2,000g mitochondria from control and TCDD treated CE, with antibodies to ferredoxin reductase or P450 reductase (48 and 78 kDa, respectively); all samples at 10 μg of protein/lane. c. Western blotting for CYP1A4 and CYP1A5. Representative Western blots for microsomes (lanes 1 and 2) at 10 μg per lane and 2,000g mitochondria (lanes 3–6), at 10 to 40 μg per lane, from control and TCDD treated CE livers, using immunospecific antibodies for CYP1A4 or CYP1A5 (55 and 55.5 kDa, respectively); last two lanes: purified CYP1A4 and CYP1A5 at 1.0 pmol/lane to demonstrate specificity of the antibodies. d. Western blotting for CYP4. Representative immunoblot for microsomes and 2,000g mitochondria from control and TCDD treated CE livers with an antibody to chick CYP4 (53 kDa) (see “Materials and Methods”); 25 μg of protein per lane. Microsomes and 2,000g mitochondria from the same membrane are shown (dotted line indicates where samples not relevant to these data were deleted from the picture shown). e. Immunofluorescence evidence for colocalization of CYP1A4 and mitochondria. D17 fibroblasts were stably transfected with a pCXIZ-CYP1A4 sense construct (see “Materials and Methods”). Cells were incubated with primary antibody to CYP1A4, using donkey anti-rabbit Alexa Fluor 488 as the secondary antibody (upper left panel), Mitotracker 580 to stain mitochondria (upper right panel) and DAPI (Sigma-Aldrich) to stain nuclei (lower left panel). Overlay (lower right panel) shows colocalization of mitochondria and CYP1A4 (yellow signal). An Axiovert 35, Zeiss fluorescence microscope was used for visualization (images shown were acquired at 63x). f. CYP1A4 mediated EROD activity. Values (mean ± SE) for EROD activity in microsomes and 2,000g mitochondria (n = 3 for each group). g . P450-dependent arachidonic acid (aa) metabolism. Representative HPLC chromatograms for P450-dependent aa metabolism by microsomes and 2,000g mitochondria from livers of control and TCDD treated CE, assayed at 75 and 150 μg protein per reaction mixture, for microsomes and mitochondria, respectively. Retention times for EETs, DHETs, 20-HETE and aa (min) as in Fig. 2d. Total counts (cpm) clockwise from top left to bottom left were 168,742; 186,617; 184,784 and 174,994.
The availability of a mitochondrial preparation essentially free of peroxisomal and microsomal contamination offered an opportunity to assess accurately microsomal and mitochondrial CYP1A4 and 1A5 activities and the contribution of mitochondria to DHET formation.2 Fig. 4c shows that CYP1A4 and 1A5 were absent from 2,000g mitochondria and microsomes from control CE livers but were present in the corresponding fractions from livers of TCDD-treated CE, at 32.1 ± 2.9% and 36.6 ± 7.8% (SE) of the levels in microsomes, respectively, as shown by densitometry of Western blots (n = 3).
2,000g mitochondria from control CE liver, like microsomes, generated only 20-HETE (Fig. 4g, upper panels) at mean levels about 31 ± 2% (SE) of those for microsomes (p < 0.0001; n = 3). TCDD decreased 20-HETE formation compared to the respective controls by 39 ± 9% in microsomes and 23 ± 3% in mitochondria (p< 0.005 and < 0.01, respectively; n = 3 for each; Table 1). CYP4 was present in mitochondria from livers of control and TCDD treated CE at 29 ± 1.3% of the levels in microsomes (p < 0.0001; n = 6) as shown by Western blotting using an antibody against a CYP4 purified from CE liver (Fig. 4d). The CYP4 was found in separate studies to catalyze 20-HETE formation in microsomes (manuscript in preparation). TCDD did not appear to affect CYP4 protein levels.
Table 1.
TCDD effects on P450 arachidonic acid metabolism in CE microsomes and 2,000g mitochondria.
| Arachidonic acid epoxygenase products | 20-HETE | ||||
|---|---|---|---|---|---|
| EETs | DHETs | DHETs as % of EETs+DHETs ± SE | TCDD as % of control ± SE | ||
| pmol/min/mg protein ± SE | pmol/min/mg protein ± SE | ||||
| Microsomes | |||||
| Control | 9 ± 2 | ND | 246 ± 8 | ||
| TCDD | 523 ± 49 a | 84 ± 15 a | 13 ± 2 | 150 ± 14 a | 61 ± 6 |
| Mitochondria | |||||
| Control | ND | 2 ± 2 | 80 ± 3 | ||
| TCDD | 97 ± 13 a,b | 160 ± 22 a,b | 62 ± 8 b | 62 ± 2 a,b | 77 ± 2 |
Microsomes and 2,000g mitochondria were prepared as described in “Materials and Methods” from livers of 17 day old CE treated for 24 hr with 1 nmol/egg TCDD, 8–10 eggs per group. Mean values ± SE are shown for n = 3 independent preparations.
significantly different from respective control;
significantly different from TCDD microsomes. ND, not detectable.
Fig. 4e presents immunofluorescence evidence showing that CYP1A4, when overexpressed in fibroblasts, co-localized with mitochondria. These fibroblasts, when transfected with CYP1A4 cDNA had been found in previous experiments (not shown) to transcribe and translate CYP1A4 into enzymatically active protein. The immunofluorescence data provide independent confirmation that CYP1A4 can be localized to mitochondria and further that this can occur in cells from a mammalian species as well as chick and is not restricted to hepatocytes.
CYP1A4 mediated EROD (Fig. 4f) and CYP1A5 mediated aa epoxygenase activities (Fig. 4g) were present in microsome- and peroxisome-free mitochondria at 23.2 % ± 0.8% and 42 ± 6.0% of the levels in microsomes, respectively (see also Table 1; n = 3 for both assays).
Although total epoxygenase activity (EETs plus DHETs) in 2,000g mitochondria was 42% of that in microsomes from TCDD-treated CE liver, DHET formation in the mitochondria was about twice as high as in microsomes per mg protein. Furthermore, the formation of DHETs as a percentage of the total epoxygenase products was 4.8-fold higher for mitochondria than microsomes (Table 1).
Some differences were observed for the regioisomeric distribution of the epoxygenase products in mitochondria and microsomes. The 5,6 regioisomers were a slightly lower percentage of the total EETs and DHETs in mitochondria (mean percentages ± SE: 36.3% ± 1.7 for microsomes and 30.7% ± 0.9% for mitochondria (p<0.04, n = 3)), while the 11, 12 regioisomers were a greater percentage of the total in the mitochondria (26.3 ± 1.8 % in the mitochondria and 18.3% ± 0.9 in the microsomes (p<0.015, n = 3)). No significant differences were found for the 8,9 and 14,15 regioisomers in mitochondria and microsomes (mean distributions were 20.3 and 20.7% for mitochondria and microsomes for the 8,9- and 23.3 % and 25% for the 14,15-regioisomer respectively).
The evidence that mitochondria (which contained only ferredoxin reductase, Fig. 4b) supported CYP1A4 and 1A5 catalysis demonstrated that ferredoxin reductase could mediate electron transfer from NADPH to the mitochondrial forms of those P450s. We further investigated whether P450 reductase could support mitochondrial as well as microsomal CYP1A activities. Purified P450 reductase added to reaction mixtures containing microsomes or 2,000g mitochondria from TCDD-treated CE liver, at an excess of reductase to P450, increased both microsomal and mitochondrial CYP1A5 aa epoxygenase activities. The P450 reductase increased both mitochondrial and microsomal aa epoxygenation but increased the microsomal activity 90% more than the mitochondrial. Supplementation of microsomes or 2,000g mitochondria from livers of TCDD treated CE with 2,000g mitochondria from control CE liver (as a source of ferredoxin reductase) increased both microsomal and mitochondrial EROD activities but increased the mitochondrial activity 30% more than the microsomal activity.
The evidence in Fig. 4 that DHETs were generated by mitochondria essentially free of peroxisomes suggested that the mitochondrial DHET formation was attributable to a soluble epoxide hydrolase (sEH) intrinsic to mitochondria, rather than to peroxisomal contamination. Use of an antibody against sEH (EPHX2) (Fig. 5a) showed that the mitochondria and peroxisomes both contained sEH. Microsomes, in contrast, were essentially free of both sEH and peroxisomes indicating that the microsomal DHET formation (Fig. 4g) was attributable to a microsomal epoxide hydrolase distinct from the mitochondrial or peroxisomal sEH. TCDD did not increase sEH levels in 2,000g mitochondria (not shown).
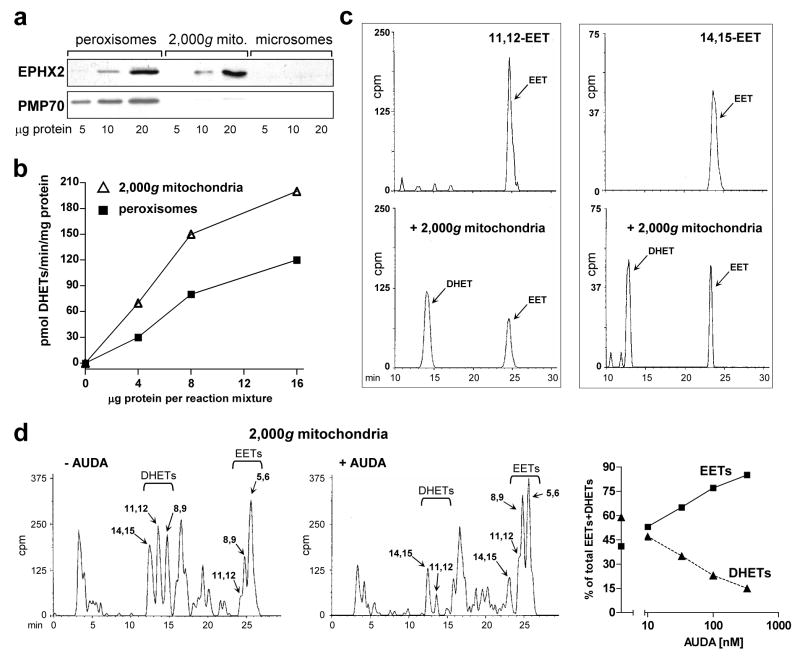
Figure 5. Mitochondrial soluble epoxide hydrolase.
a. Presence of soluble epoxide hydrolase (sEH) in CE liver mitochondria and peroxisomes. Western blots of peroxisomes, 2,000g mitochondria and microsomes prepared as described in “Materials and Methods”, with antibodies to soluble epoxide hydrolase (EPHX2/sEH, 63 kDa) and PMP70 (peroxisomal marker, 70 kDa). The blot shows representative samples from control livers. b. DHET formation by peroxisomes and 2,000g mitochondria. Four, 8 and 16 μg of protein from peroxisomes (squares) and mitochondria (triangles) from control CE livers were added to standard reaction mixtures for assaying microsomal aa metabolism, using microsomes from TCDD-treated CE liver (75 μg microsomal protein per reaction mixture) to generate EETs, as described in “Materials and Methods”. The production of EETs and DHETs after 10 min of incubation was measured. DHET formation, reflecting the sum of 8,9–11,12- and 14,15-DHETs (5,6-DHET was absent), is plotted in the graph. DHETs formed by microsomes alone (without addition of peroxisomes or 2,000g mitochondria; see Fig. 4g for example) were subtracted. c. Conversion by mitochondria of [1-14C] 11,12 and 14,15 EETs to the corresponding DHETs. [1-14C] 11,12 or 14,15 EET were incubated alone (top panels) or with 2,000g mitochondria from control (untreated) CE liver (bottom panels), at 50 μg of mitochondrial protein, for 20 min in a shaking water bath at 37°. The reaction was stopped and products were extracted and resolved on HPLC as described for aa metabolism in “Materials and Methods”. The EETs and corresponding DHETs exhibited the expected retention times (see legend to Fig. 2d). Total cpm in the chromatogram pairs were comparable: 1288 and 1505 for 11,12 EET alone or with mitochondria respectively, and 414 and 467 for 14,15 EET alone or with mitochondria. d. AUDA inhibits DHET formation by mitochondria. Reverse phase HPLC chromatograms for aa metabolism by 2,000g mitochondria (150 μg per reaction mixture) from livers of CE treated with TCDD for 24 hr in the absence (left panel) or presence (right panel) of 330 nM of AUDA are shown. Total cpm were 154,179 and 145,361 for the left and right panels respectively. Arachidonic acid metabolism was assayed as described in “Materials and Methods” except that mitochondria were first preincubated with AUDA for 10 min at 30° before adding the other components of the reaction mixture. The right panel shows the dose response relationships for suppression of DHET formation by AUDA at 10 to 330 nM (squares, EETs; triangles, DHETs). The distribution of EETs and DHETs in the absence of AUDA (43.3 ± 1.2 % and 56.7 ± 1.2 %, respectively) are plotted on the y axis; the SE is included in the symbol. Control values for mitochondrial EET and DHET formation in the absence of AUDA (pmol/mg/min ± SE) were 103 ± 5.6 for EETs, and 133 ± 3.8 for DHETs, n = 3.
The ability of the sEH in peroxisomes and 2,000g mitochondria to convert EETs to DHETs was compared by coincubating peroxisomes or 2,000g mitochondria from control CE liver with liver microsomes from TCDD-treated CE (as a source of EETs, see “Materials and Methods”). The baseline microsomal DHET formation (see Fig. 4g) was subtracted. Mean DHET formation by the 2,000g mitochondria was 30 ± 2.5% (SE) greater than by peroxisomes (Fig. 5b) strongly supporting the conversion of EETs to DHETs by a mitochondrial sEH.
To confirm the presence of sEH in mitochondria we incubated pure [1-14C] labeled 11, 12 and 14, 15 EET with 2,000g mitochondria from control (untreated) CE liver. The results (Fig. 5c) show that under the conditions used, the mitochondria converted 66% of the 11,12 EET and 58% of the 14,15 EET to the corresponding DHETs.
Further confirmation that DHET formation by mitochondria was attributable to mitochondrial sEH was obtained in experiments in which mitochondria from livers of TCDD treated CE were incubated with the sEH inhibitor, AUDA (Fig. 5d). Dose-dependent inhibition of DHET formation was observed, ranging from 17% inhibition at 10 nM AUDA, the lowest concentration tested, to 74% inhibition at the highest concentration, 330 nM. The latter findings are consistent with the reported IC50 for the isolated recombinant chicken sEH of 13.7 nM [40].
Together, the data in Fig. 5 show that DHETs in the microsome- and peroxisome-free 2,000g mitochondria were generated by a mitochondrial sEH, supporting the inference that mitochondria contain sEH, which in the presence of TCDD-induced CYP1A can convert EETs to DHETs.
DISCUSSION
Several P450 enzymes regarded as being localized to the ER have also been found in liver, brain and lung mitochondria, including mouse and rat CYPs 1A1, 1A2, 2A5, 2B1 2E1, 3A4, 4 [3–8]. We show here that TCDD induced enzymatically active CYPs 1A4 and 1A5, the avian orthologs of mammalian CYP1A1 and 1A2, in CE liver mitochondria in substantial amounts within 24 hr of TCDD treatment. Those findings and the evidence that CYP1A is induced in mitochondria in diverse species suggest that mitochondrial CYP1A induction is likely to have physiologic or pathophysiologic significance.
Mitochondrial CYP1A enzymes in CE liver were similar to the rat and mouse orthologs with respect to (a) relative amounts and activity levels as compared to those in microsomes (mitochondrial CYP1A was present in CE livers at 20 to 40% of the microsomal levels and in rat and mouse livers at 20 to 30% of the microsomal levels [3, 8]), (b) strict requirement for NADPH for catalytic activity as for their microsomal counterparts, and (c) ability to use either ferredoxin reductase or P450 reductase for electron transfer from NADPH but with greater effectiveness of ferredoxin reductase for the mitochondrial P450s [2, 7].
It has been suggested that a portion of CYP1A1 nascent chains escape transport to the ER and are imported into the mitochondria after endoprotease cleavage [2, 7]. Interestingly, 25 to 33% of rat CYP1A1 nascent chains resisted ER insertion [3], which is remarkably consistent with the amounts of mitochondrial CYP1A found in mammalian and CE liver. CYP1A4 and 1A5 both have the conserved positively charged amino acids and endoprotease cleavage sites in the N-terminal region (see Fig. 1) associated with signaling for mitochondrial import and found in rat and mouse CYP1A1 and in mouse CYP1A2. However, their migration on SDS-gels differed less from the microsomal or purified P450s than would be expected, based on cleavage of amino acids at the sites shown in Fig. 1 (~ 4 kDa), although differences of 0.5 kDa in MW for CYPs1A4 and 1A5 can be seen on the 10% polyacrylamide gels used here.
PKC-dependent phosphorylation sites have also been identified as potential signals for translocation of mouse CYP1A1 to mitochondria [7]. Conserved threonines in CYP1A4 and 1A5, as well as in mammalian CYP1A enzymes (see Fig. 1), identified as high-probability PKC dependent phosphorylation sites, suggest that a phosphorylation dependent mechanism might be involved for mitochondrial transfer of CYP1A4 and 1A5. Reported activation of PKC by TCDD [41] is consistent with this hypothesis.
Although the liver is a well recognized site of P450 aa metabolism [21, 42] and P450 aa metabolism was first identified in liver microsomes [43], effects of P450 aa metabolites in liver physiology and pathophysiology have not yet been delineated. Effects of EETs on liver cells or liver microsomes include increased intracellular Ca2+, vasopressin stimulated glycogenolysis, activation of phosphorylase and ADP ribosylation (reviewed in Spector and Norris [16]). Moreover, putative targets for cardiovascular actions of EETs (i.e KATP and BKCa channels, and signaling pathways regulating inflammatory responses, angiogenesis and cell cycling (via mitogen activated protein kinase (MAPK), Akt and Src-dependent pathways)) are present and active in liver [44, 45].
We show here for the first time that mitochondria are a site of P450 dependent aa metabolism, making it possible that the P450 products have mitochondria-specific effects. For example, it has been hypothesized that some mitochondrial ion channels can be affected by EETs [46]. Also, EETs and 20-HETE are associated with increased production of reactive oxygen species [19, 47], and could contribute to mitochondrial ROS production. Further, P450 aa products can activate PPARα, a regulator of mitochondrial fatty acid oxidation [48, 49]. Induction of CYP1A dependent aa metabolism in mitochondria provides a new venue for P450 aa metabolism, one that has significant implications for affecting mitochondrial function. Further, the regioisomeric differences in epoxygenase products for mitochondria and microsomes could result in different biologic effects in microsomes and mitochondria.
The increased DHET production in CE liver mitochondria was the major factor distinguishing the mitochondrial from the microsomal epoxygenase products. DHETs, conversion products of EETs, are generated principally by a soluble epoxide hydrolase (sEH; EPHX2) and to a lesser extent by a microsomal epoxide hydrolase (EPHX1) [15]. Although sEH is primarily localized in cytosol [50] its presence in mitochondria has been controversial. Initially sEH was thought to be localized to mitochondria as well as cytosol, but it was subsequently assigned to peroxisomes in addition to cytosol [35, 36, 51, 52]. Our results indicate that the majority of the sEH in liver of the CE, as in other species, is in cytosol (about 75%; data not shown). We show here that sEH is also localized not only in peroxisomes but in mitochondria as well. The results show further that the mitochondrial sEH is responsible for the mitochondrial DHET formation.
The chick sEH was recognized by an antibody to mouse sEH, and the rank order for conversion of EETs to DHETs by CE liver sEH observed in these experiments was the same as reported for the mammalian enzyme: 14,15-EET > 11,12-EET = 8,9-EET, with negligible conversion of 5,6-EET [16]. The findings are consistent with conservation of structure and function of this enzyme among species.
Conversion of EETs to DHETs generally reduces EET activities, and DHETs are thought to have an adverse effect on cardiac function by impairing responses to ischemia-reperfusion injury and increasing hypertension, perhaps by counteracting the vasodilating effects of EETs, [16, 19]. Accordingly, sEH inhibitors have been proposed for use as antihypertensive agents or cotherapies for enhancing anti-inflammatory effects of drugs [53]. On the other hand, DHETs have some of the effects of EETs: they can activate BKCa channels [54] and have been reported to activate PPARα in rat liver, COS and HepG2 cells [48, 55]. Moreover, a R287Q mutation of human sEH resulting in decreased enzymatic activity has been associated with increased atherosclerosis and insulin resistance, suggesting a possible protective role for DHETs [56].
Although human CYP1A2 and avian CYP1A5 (more than other mammalian CYP1A2 enzymes [12, 57]) have been demonstrated to have epoxygenase activities, and CYP1A induction was discovered to enhance aa epoxygenation early in the history of P450 mediated aa metabolism [43], attention to CYP1A epoxygenase activity has been eclipsed by interest in constitutive aa epoxygenases such as CYP2C and 2J, P450s, present in vascular endothelial and smooth muscle cells or heart [19]. Several factors should serve to increase attention to CYP1A2/1A5 mediated aa metabolism: (1) CYP1A2 is induced not only in liver, but in kidney, a well recognized site of action for P450-dependent aa metabolites [19, 58], (2) CYP1A enzymes are induced not only by toxic chemicals but also by many aryl hydrocarbon receptor ligands such as tryptophan derivatives and other substances in food, as well as some endogenous CYP1A substrates, i.e. bilirubin and melatonin [1, 59, 60], and (3) CYP1A2, and to a lesser extent CYP1A1, catalyzes metabolism of those and other endogenous substrates (reviewed in Rifkind [1]). Thus, CYP1A induction and associated effects may be expected to have much wider implications than has been appreciated.
These results also have potential implications for TCDD toxicity. TCDD produces a diverse toxicity syndrome with effects including tumor promotion, immune system dysfunction and a wasting syndrome characterized by suppressed gluconeogenesis and reliance on fatty acid metabolism as a source of energy, and cardiac contractile dysfunction [22, 24, 61]. The mechanisms by which aryl hydrocarbon receptor activation leads to TCDD toxicity and the role of the induced CYP1A in TCDD toxicity are not yet understood. Consistent with the picture of energy failure suggested by TCDD effects, TCDD has been reported to reduce ATP levels in mouse liver [23]. Mitochondria control energy supply via generation of ATP and fatty acid metabolism. The discovery that TCDD induces CYP1A enzymes in liver mitochondria demonstrates that mitochondria are a direct target for TCDD action.
TCDD induces mitochondrial CYP1A4, the avian ortholog of mammalian CYP1A1, to levels comparable to CYP1A5. CYP1A4 is inactive in aa metabolism, but CYP1A1 has been implicated in generation of reactive oxygen intermediates which have been considered as possible mediators of some of the adverse effects of TCDD [62]. Moreover, CYP1A1 knockout mice exhibit reduced TCDD toxicity [63].
Mitochondria from CE liver, like mouse liver [8], also contained a CYP4 enzyme which generates 20-HETE. TCDD has been found to suppress formation of 20-HETE in mammalian and avian liver microsomes [12, 13] and is shown here also to suppress 20-HETE in mitochondria. Notwithstanding the dominant role of 20-HETE as virtually the only constitutive P450 product in CE liver microsomes and mitochondria, hepatic effects of 20-HETE are unknown. In the cardiovascular system, 20-HETE has mainly been found to be vasoconstrictive and is considered a possible factor in hypertension [16, 19]. 20-HETE, however, is also an activator of PPARα [48] and therefore could be involved in alterations of fatty acid metabolism by TCDD. The consequences of CYP1A4, 1A5 and CYP4 expression in liver mitochondria and of TCDD effects on CYP aa product formation warrant further investigation.
Finally, we offer a cautionary comment. We found high peroxisomal contamination as shown by Western blotting in CE and mouse liver mitochondria prepared by standard differential centrifugation and Percoll gradient techniques. Although peroxisomal contamination of mitochondrial preparations measured by peroxisome-specific enzyme assays had been well recognized [36] a search of more recent literature revealed that studies examining mitochondrial function rarely looked for peroxisomal contamination. Peroxisomes are the locus of catalase, a major determinant of reactive oxygen species production [64], and they share mitochondrial functions including β-oxidation of fatty acids [65]. As peroxisomal contamination is potentially a confounding factor in studies of mitochondrial function, it should be excluded routinely.
Acknowledgments
This work was supported by NIH grants RO1-ES03606 (A.B.R) and T32-CA062948 (S.D-M); 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) was provided by the National Cancer Institute’s Chemical Carcinogen Reference Standards Repository operated under contract by Midwest Research Institute, NO. NO2-CB-66600. We thank Dr. Jorge Capdevila, Vanderbilt University, Nashville, TN for providing radiolabeled EETs.
Footnotes
Abbreviations: aa, arachidonic acid; AhR, aryl hydrocarbon receptor; AUDA, 12(-3-adamantan-1-yl-ureido)-dodecanoic acid; CE, chick embryo; EET, epoxyeicosatrienoic acid; DHET, vic-dihydroxyeicosatrienoic acid; sEH, EPHX2, soluble epoxide hydrolase; ER, endoplasmic reticulum; EROD, 7-ethoxyresorufin deethylase; P450, cytochrome P450; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin;ω, 20-HETE, 20-hydroxyeicosatetraenoic acid.
We noted that for mouse liver, the 2,000g pellet exhibited reduced but not absent peroxisomal and microsomal contamination when compared to the 10,000g pellet (reductions of 39% and 65%, respectively).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rifkind AB. CYP1A in TCDD toxicity and in physiology-with particular reference to CYP dependent arachidonic acid metabolism and other endogenous substrates. Drug Metab Rev. 2006;38:291–335. doi: 10.1080/03602530600570107. [DOI] [PubMed] [Google Scholar]
- 2.Omura T. Mitochondrial P450s. Chem Biol Interact. 2006;163:86–93. doi: 10.1016/j.cbi.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Addya S, Anandatheerthavarada HK, Biswas G, Bhagwat SV, Mullick J, Avadhani NG. Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. J Cell Biol. 1997;139:589–599. doi: 10.1083/jcb.139.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anandatheerthavarada HK, Vijayasarathy C, Bhagwat SV, Biswas G, Mullick J, Avadhani NG. Physiological role of the N-terminal processed P4501A1 targeted to mitochondria in erythromycin metabolism and reversal of erythromycin-mediated inhibition of mitochondrial protein synthesis. J Biol Chem. 1999;274:6617–6625. doi: 10.1074/jbc.274.10.6617. [DOI] [PubMed] [Google Scholar]
- 5.Bhagwat SV, Mullick J, Raza H, Avadhani NG. Constitutive and inducible cytochromes P450 in rat lung mitochondria: xenobiotic induction, relative abundance, and catalytic properties. Toxicol Appl Pharmacol. 1999;156:231–240. doi: 10.1006/taap.1999.8646. [DOI] [PubMed] [Google Scholar]
- 6.Bhagwat SV, Biswas G, Anandatheerthavarada HK, Addya S, Pandak W, Avadhani NG. Dual targeting property of the N-terminal signal sequence of P4501A1. Targeting of heterologous proteins to endoplasmic reticulum and mitochondria. J Biol Chem. 1999;274:24014–24022. doi: 10.1074/jbc.274.34.24014. [DOI] [PubMed] [Google Scholar]
- 7.Dasari VR, Anandatheerthavarada HK, Robin MA, Boopathi E, Biswas G, Fang JK, Nebert DW, Avadhani NG. Role of protein kinase C-mediated protein phosphorylation in mitochondrial translocation of mouse CYP1A1, which contains a non-canonical targeting signal. J Biol Chem. 2006;281:30834–30847. doi: 10.1074/jbc.M510725200. [DOI] [PubMed] [Google Scholar]
- 8.Genter MB, Clay CD, Dalton TP, Dong H, Nebert DW, Shertzer HG. Comparison of mouse hepatic mitochondrial versus microsomal cytochromes P450 following TCDD treatment. Biochem Biophys Res Commun. 2006;342:1375–1381. doi: 10.1016/j.bbrc.2006.02.121. [DOI] [PubMed] [Google Scholar]
- 9.Kanetoshi A, Ward AM, May BK, Rifkind AB. Immunochemical identity of the 2,3,7,8-tetrachlorodibenzo-p-dioxin- and beta-naphthoflavone-induced cytochrome P-450 arachidonic acid epoxygenases in chick embryo liver: distinction from the omega-hydroxylase and the phenobarbital-induced epoxygenase. Mol Pharmacol. 1992;42:1020–1026. [PubMed] [Google Scholar]
- 10.Rifkind AB, Kanetoshi A, Orlinick J, Capdevila JH, Lee C. Purification and biochemical characterization of two major cytochrome P-450 isoforms induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in chick embryo liver. J Biol Chem. 1994;269:3387–3396. [PubMed] [Google Scholar]
- 11.Gilday D, Gannon M, Yutzey K, Bader D, Rifkind AB. Molecular cloning and expression of two novel avian cytochrome P450 1A enzymes induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1996;271:33054–33059. doi: 10.1074/jbc.271.51.33054. [DOI] [PubMed] [Google Scholar]
- 12.Lee CA, Lawrence BP, Kerkvliet NI, Rifkind AB. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induction of cytochrome P450-dependent arachidonic acid metabolism in mouse liver microsomes: evidence for species-specific differences in responses. Toxicol Appl Pharmacol. 1998;153:1–11. doi: 10.1006/taap.1998.8468. [DOI] [PubMed] [Google Scholar]
- 13.Diani-Moore S, Papachristou F, Labitzke E, Rifkind AB. Induction of CYP1A and CYP2-mediated arachidonic acid epoxygenation and suppression of 20-hydroxyeicosatetraenoic acid by imidazole derivatives including the aromatase inhibitor vorozole. Drug Metab Dispos. 2006;34:1376–1385. doi: 10.1124/dmd.106.009498. [DOI] [PubMed] [Google Scholar]
- 14.Capdevila JH, Falck JR, Harris RC. Cytochrome P450 and arachidonic acid bioactivation. Molecular and functional properties of the arachidonate monooxygenase. J Lipid Res. 2000;41:163–181. [PubMed] [Google Scholar]
- 15.Zeldin DC, Kobayashi J, Falck JR, Winder BS, Hammock BD, Snapper JR, Capdevila JH. Regio- and enantiofacial selectivity of epoxyeicosatrienoic acid hydration by cytosolic epoxide hydrolase. J Biol Chem. 1993;268:6402–6407. [PubMed] [Google Scholar]
- 16.Spector AA, Norris AW. Action of epoxyeicosatrienoic acids on cellular function. Am J Physiol Cell Physiol. 2007;292:C996–1012. doi: 10.1152/ajpcell.00402.2006. [DOI] [PubMed] [Google Scholar]
- 17.Rifkind AB, Lee C, Chang TK, Waxman DJ. Arachidonic acid metabolism by human cytochrome P450s 2C8, 2C9, 2E1, and 1A2: regioselective oxygenation and evidence for a role for CYP2C enzymes in arachidonic acid epoxygenation in human liver microsomes. Arch Biochem Biophys. 1995;320:380–389. doi: 10.1016/0003-9861(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 18.Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. Metabolism of retinoids and arachidonic acid by human and mouse cytochrome P450 1b1. Drug Metab Dispos. 2004;32:840–847. doi: 10.1124/dmd.32.8.840. [DOI] [PubMed] [Google Scholar]
- 19.Roman RJ. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 20.Kroetz DL, Xu F. Regulation and inhibition of arachidonic acid omega-hydroxylases and 20-HETE formation. Annu Rev Pharmacol Toxicol. 2005;45:413–438. doi: 10.1146/annurev.pharmtox.45.120403.100045. [DOI] [PubMed] [Google Scholar]
- 21.Sacerdoti D, Gatta A, McGiff JC. Role of cytochrome P450-dependent arachidonic acid metabolites in liver physiology and pathophysiology. Prostaglandins Other Lipid Mediat. 2003;72:51–71. doi: 10.1016/s1098-8823(03)00077-7. [DOI] [PubMed] [Google Scholar]
- 22.Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 23.Shertzer HG, Genter MB, Shen D, Nebert DW, Chen Y, Dalton TP. TCDD decreases ATP levels and increases reactive oxygen production through changes in mitochondrial F(0)F(1)-ATP synthase and ubiquinone. Toxicol Appl Pharmacol. 2006;217:363–374. doi: 10.1016/j.taap.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lentnek M, Griffith OW, Rifkind AB. 2,3,7,8-Tetrachlorodibenzo-p-dioxin increases reliance on fats as a fuel source independently of diet: evidence that diminished carbohydrate supply contributes to dioxin lethality. Biochem Biophys Res Commun. 1991;174:1267–1271. doi: 10.1016/0006-291x(91)91558-t. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Vizarra E, Lopez-Perez MJ, Enriquez JA. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods. 2002;26:292–297. doi: 10.1016/S1046-2023(02)00034-8. [DOI] [PubMed] [Google Scholar]
- 26.Nakai K, Ward AM, Gannon M, Rifkind AB. Beta-naphthoflavone induction of a cytochrome P-450 arachidonic acid epoxygenase in chick embryo liver distinct from the aryl hydrocarbon hydroxylase and from phenobarbital-induced arachidonate epoxygenase. J Biol Chem. 1992;267:19503–19512. [PubMed] [Google Scholar]
- 27.Sims NR. Rapid isolation of metabolically active mitochondria from rat brain and subregions using Percoll density gradient centrifugation. J Neurochem. 1990;55:698–707. doi: 10.1111/j.1471-4159.1990.tb04189.x. [DOI] [PubMed] [Google Scholar]
- 28.Capdevila JH, Falck JR, Dishman E, Karara A. Cytochrome P-450 arachidonate oxygenase. Methods Enzymol. 1990;187:385–394. doi: 10.1016/0076-6879(90)87045-5. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Mikawa T. Retroviral targeting of FGF and FGFR in cardiomyocytes and coronary vascular cells during heart development. Ann N Y Acad Sci. 1995;752:506–516. doi: 10.1111/j.1749-6632.1995.tb17459.x. [DOI] [PubMed] [Google Scholar]
- 31.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 32.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 33.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 34.Eliasson M, Brock S, Bengtsson Ahlberg M. Evidence for mitochondrial metabolism of 7,12-dimethylbenz(a)anthracene in porcine ovaries: comparison with microsomal metabolism. Toxicology. 1997;122:11–21. doi: 10.1016/s0300-483x(97)00074-7. [DOI] [PubMed] [Google Scholar]
- 35.Gill SS, Hammock BD. Epoxide hydrolase activity in the mitochondrial and submitochondrial fractions of mouse liver. Biochem Pharmacol. 1981;30:2111–2120. doi: 10.1016/0006-2952(81)90230-6. [DOI] [PubMed] [Google Scholar]
- 36.Meijer J, Lundqvist G, DePierre JW. Comparison of the sex and subcellular distributions, catalytic and immunochemical reactivities of hepatic epoxide hydrolases in seven mammalian species. Eur J Biochem. 1987;167:269–279. doi: 10.1111/j.1432-1033.1987.tb13333.x. [DOI] [PubMed] [Google Scholar]
- 37.Krasnikov BF, Kim SY, McConoughey SJ, Ryu H, Xu H, Stavrovskaya I, Iismaa SE, Mearns BM, Ratan RR, Blass JP, Gibson GE, Cooper AJ. Transglutaminase activity is present in highly purified nonsynaptosomal mouse brain and liver mitochondria. Biochemistry. 2005;44:7830–7843. doi: 10.1021/bi0500877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hovius R, Lambrechts H, Nicolay K, de Kruijff B. Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim Biophys Acta. 1990;1021:217–226. doi: 10.1016/0005-2736(90)90036-n. [DOI] [PubMed] [Google Scholar]
- 39.Pallotti F, Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2001;65:1–35. doi: 10.1016/s0091-679x(01)65002-7. [DOI] [PubMed] [Google Scholar]
- 40.Harris TR, Morisseau C, Walzem RL, Ma SJ, Hammock BD. The cloning and characterization of a soluble epoxide hydrolase in chicken. Poult Sci. 2006;85:278–287. doi: 10.1093/ps/85.2.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machemer DE, Tukey RH. The role of protein kinase C in regulation of TCDD-mediated CYP1A1 gene expression. Toxicol Sci. 2005;87:27–37. doi: 10.1093/toxsci/kfi220. [DOI] [PubMed] [Google Scholar]
- 42.Qu W, Rippe RA, Ma J, Scarborough P, Biagini C, Fiedorek FT, Travlos GS, Parker C, Zeldin DC. Nutritional status modulates rat liver cytochrome P450 arachidonic acid metabolism. Mol Pharmacol. 1998;54:504–513. doi: 10.1124/mol.54.3.504. [DOI] [PubMed] [Google Scholar]
- 43.Capdevila J, Chacos N, Werringloer J, Prough RA, Estabrook RW. Liver microsomal cytochrome P-450 and the oxidative metabolism of arachidonic acid. Proc Natl Acad Sci U S A. 1981;78:5362–5366. doi: 10.1073/pnas.78.9.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takano M, Noma A. The ATP-sensitive K+ channel. Prog Neurobiol. 1993;41:21–30. doi: 10.1016/0301-0082(93)90039-u. [DOI] [PubMed] [Google Scholar]
- 45.Barfod ET, Moore AL, Roe MW, Lidofsky SD. Ca2+-activated IK1 channels associate with lipid rafts upon cell swelling and mediate volume recovery. J Biol Chem. 2007;282:8984–8993. doi: 10.1074/jbc.M607730200. [DOI] [PubMed] [Google Scholar]
- 46.Seubert JM, Zeldin DC, Nithipatikom K, Gross GJ. Role of epoxyeicosatrienoic acids in protecting the myocardium following ischemia/reperfusion injury. Prostaglandins Other Lipid Mediat. 2007;82:50–59. doi: 10.1016/j.prostaglandins.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward NC, Rivera J, Hodgson J, Puddey IB, Beilin LJ, Falck JR, Croft KD. Urinary 20-hydroxyeicosatetraenoic acid is associated with endothelial dysfunction in humans. Circulation. 2004;110:438–443. doi: 10.1161/01.CIR.0000136808.72912.D9. [DOI] [PubMed] [Google Scholar]
- 48.Cowart LA, Wei S, Hsu MH, Johnson EF, Krishna MU, Falck JR, Capdevila JH. The CYP4A isoforms hydroxylate epoxyeicosatrienoic acids to form high affinity peroxisome proliferator-activated receptor ligands. J Biol Chem. 2002;277:35105–35112. doi: 10.1074/jbc.M201575200. [DOI] [PubMed] [Google Scholar]
- 49.Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290:G852–858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- 50.Eriksson AM, Zetterqvist MA, Lundgren B, Andersson K, Beije B, DePierre JW. Studies on the intracellular distributions of soluble epoxide hydrolase and of catalase by digitonin-permeabilization of hepatocytes isolated from control and clofibrate-treated mice. Eur J Biochem. 1991;198:471–476. doi: 10.1111/j.1432-1033.1991.tb16037.x. [DOI] [PubMed] [Google Scholar]
- 51.Waechter F, Bentley P, Bieri F, Staubli W, Volkl A, Fahimi HD. Epoxide hydrolase activity in isolated peroxisomes of mouse liver. FEBS Lett. 1983;158:225–228. doi: 10.1016/0014-5793(83)80583-3. [DOI] [PubMed] [Google Scholar]
- 52.Enayetallah AE, French RA, Barber M, Grant DF. Cell-specific subcellular localization of soluble epoxide hydrolase in human tissues. J Histochem Cytochem. 2006;54:329–335. doi: 10.1369/jhc.5A6808.2005. [DOI] [PubMed] [Google Scholar]
- 53.Inceoglu B, Schmelzer KR, Morisseau C, Jinks SL, Hammock BD. Soluble epoxide hydrolase inhibition reveals novel biological functions of epoxyeicosatrienoic acids (EETs) Prostaglandins Other Lipid Mediat. 2007;82:42–49. doi: 10.1016/j.prostaglandins.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu T, Katakam PV, VanRollins M, Weintraub NL, Spector AA, Lee HC. Dihydroxyeicosatrienoic acids are potent activators of Ca(2+)-activated K(+) channels in isolated rat coronary arterial myocytes. J Physiol. 2001;534:651–667. doi: 10.1111/j.1469-7793.2001.t01-1-00651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang X, Hu S, Xu B, Snyder GD, Harmon S, Yao J, Liu Y, Sangras B, Falck JR, Weintraub NL, Spector AA. 14,15-Dihydroxyeicosatrienoic acid activates peroxisome proliferator-activated receptor-alpha. Am J Physiol Heart Circ Physiol. 2006;290:H55–63. doi: 10.1152/ajpheart.00427.2005. [DOI] [PubMed] [Google Scholar]
- 56.Larsen BT, Campbell WB, Gutterman DD. Beyond vasodilatation: non-vasomotor roles of epoxyeicosatrienoic acids in the cardiovascular system. Trends Pharmacol Sci. 2007;28:32–38. doi: 10.1016/j.tips.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Capdevila JH, Karara A, Waxman DJ, Martin MV, Falck JR, Guenguerich FP. Cytochrome P-450 enzyme-specific control of the regio- and enantiofacial selectivity of the microsomal arachidonic acid epoxygenase. J Biol Chem. 1990;265:10865–10871. [PubMed] [Google Scholar]
- 58.Zhao X, Imig JD. Kidney CYP450 enzymes: biological actions beyond drug metabolism. Curr Drug Metab. 2003;4:73–84. doi: 10.2174/1389200033336892. [DOI] [PubMed] [Google Scholar]
- 59.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 60.Diani-Moore S, Labitzke E, Brown R, Garvin A, Wong L, Rifkind AB. Sunlight generates multiple tryptophan photoproducts eliciting high efficacy CYP1A induction in chick hepatocytes and in vivo. Toxicol Sci. 2006;90:96–110. doi: 10.1093/toxsci/kfj065. [DOI] [PubMed] [Google Scholar]
- 61.Canga L, Paroli L, Blanck TJ, Silver RB, Rifkind AB. 2,3,7,8-tetrachlorodibenzo-p-dioxin increases cardiac myocyte intracellular calcium and progressively impairs ventricular contractile responses to isoproterenol and to calcium in chick embryo hearts. Mol Pharmacol. 1993;44:1142–1151. [PubMed] [Google Scholar]
- 62.Shertzer HG, Nebert DW, Puga A, Ary M, Sonntag D, Dixon K, Robinson LJ, Cianciolo E, Dalton TP. Dioxin causes a sustained oxidative stress response in the mouse. Biochem Biophys Res Commun. 1998;253:44–48. doi: 10.1006/bbrc.1998.9753. [DOI] [PubMed] [Google Scholar]
- 63.Uno S, Dalton TP, Sinclair PR, Gorman N, Wang B, Smith AG, Miller ML, Shertzer HG, Nebert DW. Cyp1a1(−/−) male mice: protection against high-dose TCDD-induced lethality and wasting syndrome, and resistance to intrahepatocyte lipid accumulation and uroporphyria. Toxicol Appl Pharmacol. 2004;196:410–421. doi: 10.1016/j.taap.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 64.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 65.Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK. Peroxisomal beta-oxidation--a metabolic pathway with multiple functions. Biochim Biophys Acta. 2006;1763:1413–1426. doi: 10.1016/j.bbamcr.2006.08.034. [DOI] [PubMed] [Google Scholar]