Abstract
The biological roles of the dual oxidases, DUOX1 and DUOX2, are dependent upon the tissue in which they are expressed. However, the mechanisms that control DUOX expression in these tissues are largely unexplored. Given the known role of DUOX for host defense in the gut and respiratory tract, we characterized potential mechanisms that control DUOX2 expression in response to interferon gamma (IFNγ) in respiratory tract epithelium. We discovered that IFNγ-mediated DUOX2 expression was regulated by a STAT-independent, JAK-independent pathway. These data provide insights into a novel IFNγ signaling pathway with potential importance for regulation of host defense responses.
Keywords: dual oxidase, DUOX, gamma interferon, JAK, STAT, signaling
Introduction
Growing evidence demonstrates that reactive oxygen species (ROS) such as superoxide and hydrogen peroxide (H2O2) are not just accidental byproducts of cellular respiration, but are intentionally generated to serve important cellular functions [1; 2; 3]. A key protein family responsible for the regulated generation of ROS in multiple cell types is the NOX/DUOX enzyme family [4]. Specifically, since the original discovery of DUOX enzymes [5; 6], there has been an exponentially growing body of literature characterizing the function of these enzymes in various aspects of biology and disease [7; 8; 9; 10; 11]
For example, Conner and others have demonstrated that DUOX-generated H2O2, in conjunction with lactoperoxidase and thiocyanate, kill bacteria in the respiratory tract [12; 13; 14; 15]. The predominant expression of DUOX proteins in epithelial cells of the respiratory and gastrointestinal tract [8; 16] supports the notion that these enzymes serve important host defense functions. We have previously shown that interferon gamma (IFNγ) and viral infection induce DUOX2 in respiratory epithelium [17]. These findings suggest that DUOX2 in the airway has antiviral host defense functions as well. However, the mechanisms of DUOX2 induction and its relationship to an innate viral response require further investigation.
Given the potential importance of DUOX2 for viral host defense, we first investigated the signaling mechanisms responsible for IFNγ–mediated DUOX2 mRNA expression. IFNγ has pleiotropic biological effects that primarily coordinate the immune response to intracellular pathogens [18; 19]. Most commonly, this biological effect is induced through binding of IFNγ to the heterodimerized IFNγ receptors (IFNγR1 and IFNγR2), which then activate Janus-family kinase (JAK) and signal-transducing activators of transcription 1 (STAT1) phosphorylation (reviewed in [20]).
For these studies, we utilized a respiratory tract epithelial cell line, HBE1, which we determined was capable of producing an IFNγ–mediated DUOX2 response similar to primary human respiratory tract epithelium. In parallel, we examined the response of CXCL10, a cytokine known to be increased by IFNγ through a JAK1-STAT1 pathway [21; 22], as a positive control for JAK-STAT signaling.
METHODS
Cell culture
For all our studies, we used a papilloma virus-immortalized human tracheobronchial epithelial cell line (HBE1) that was originally derived by Dr. J. Yankaskas [23] and generously donated to us by Dr. Reen Wu. Cells were maintained in submerged, confluent culture conditions with Ham’s F12/Hepes/DMEM supplemented with insulin (2 mg/ml), transferrin (2.5 mg/ml), epidermal growth factor (10 μg/ml), dexamethasone (0.05 mM), cholera toxin (10 μg/ml), bovine hypothalamus extract (1 ml/L), all-trans retinoic acid (30μM) and plasmocin (100 μl/L) [24]. Media was changed every 48 hours.
Cytokine and Inhibitor Treatments
Recombinant human IFNγ was purchased from R&D Systems, Inc. (Minneapolis, MN). Stock solutions were dissolved in sterile phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA) according to the manufacturer’s recommendations. Inhibitors of Janus Kinases (JAKs), PI3-Kinase (PI3-K), ERK1, ERK2, STAT3, and STAT5 were obtained from a single source (Calbiochem, La Jolla CA) and dissolved in dimethylsulfoxide (DMSO) according to the manufacturer’s instructions. All cytokine and inhibitor treatments were introduced as a complete media change to each well. Wells were pretreated with media containing DMSO alone or specific inhibitor (at IC50 based on manufacturer’s instructions) for three hours. IFNγ (100 ng/mL) was added to cells for two hours followed by a media change back to untreated conditions. This protocol was identical for all experiments.
RNA Isolation and Quantification
RNA was isolated using Trizol (Invitrogen, Carlsbad, CA) according to the extraction protocol supplied by the manufacturer. RNA purity and concentration was determined by spectroscopy (Nanodrop; Thermo Scientific). Three μg of total RNA for each sample was converted to cDNA using oligo-dT primers at 70°C for 10 min, followed by PCR amplification with MuMLV-reverse transcriptase (Promega, Inc., Madison WI) over 75 minutes in a 20 μl reaction volume. The cDNA stock was then diluted 1:5 in nuclease free water for real-time quantitative PCR (rt-QPCR). rt-QPCR was performed on an ABI PRISM© 7900HT Sequence Detection System (Applied Biosystems, Inc., Foster City, CA) using SYBR® GreenER™ qPCR Supermix (Invitrogen, Carlsbad, CA) and gene-specific, intron spanning primers for Duox2, and β-actin as described previously [17]. Gene-specific primers for CXCL10 were as follows: Forward primer = 5′-GCTGATGCAGGTACAGCGT-3′; Reverse primer = 5′-CACCATGAATCAAACTGCGA-3′. Reactions were carried out in triplicate for each sample. Mathematical calculations of fold induction relative to treatment controls (2−ΔΔCt) were performed as described previously [17].
Protein extraction and western blots
Protein extraction was carried out on ice in a 4°C cold room. Cells were lysed in RIPA buffer containing protease inhibitor (Thermo Fisher Scientific, Rockford Il), phosphatase inhibitors (PhosSTOP, Roche), and PMCF. Total protein concentration was determined using the Bradford protein assay (BioRad, Hercules, Ca). Protein electrophoreses was performed in 4-15% PAGE Ready Gels using standard buffers (BioRad, Hercules, Ca) and 60 μg of total protein per lane. Samples were denatured in a Laemelli loading buffer containing DTT at 90°C for 5 minutes before loading and subsequently transfered to PVDF for immunoblotting with STAT1 (1:500), αSTAT1Y701phos (1:500), αSTAT2 (1:500), or αSTAT2Y690phos (1:500) antibodies (AbCam; Cambridge, Ma). The nonphosphorylated STAT proteins served as controls for the phospho-proteins to ensure equal loading in each lane. Goat αRabbit-HRP IgG (Pierce) or donkey αMouse IgG (R&D Systems) were used at 1:1000 dilution for chemiluminescent detection (Pierce SuperSignal West Pico Substrate; Thermo Fisher Scientific).
Statistics
Data were expressed as mean ± standard deviation and statistical significance was determined using ANOVA with a Bonferroni correction for multiple comparisons using Prism software (Graph Pad, La Jolla Ca). At a minimum, all experiments were performed as three independent experiments with three replicates per experiment.
RESULTS
IFNγ induces the JAK-STAT1 canonical pathway in HBE1 cells
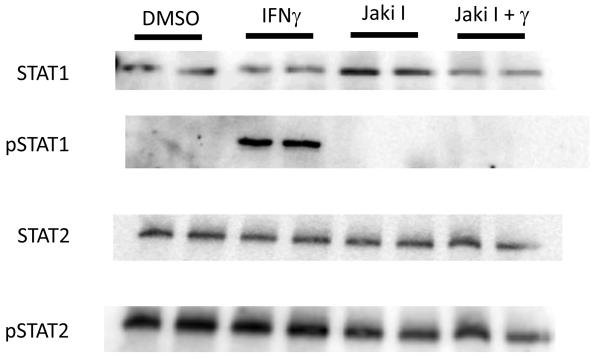
To determine the signaling mechanisms responsible for IFNγ-mediated DUOX2 expression, we selected the canonical JAK-STAT pathway as a logical starting point. HBE1 cultures were treated with either IFNγ (100 ng/mL) or vehicle control followed by protein harvest at 15 or 30 minutes after treatment. Western blot analyses demonstrated a clear increase in STAT1 phosphorylation at tyrosine residue 701 for both time points (Figure 1 and data not shown). To confirm that JAK signaling was responsible for this increase in STAT1 phosphorylation, we pretreated cells for three hours with a nonspecific JAK inhibitor (JAKi 1; 1μM) and repeated the IFNγ exposure. As expected, JAKi 1 abrogated IFNγ-mediated STAT1 phosphorylation (Figure1), which confirmed that IFNγ-induced JAK-STAT1 signaling occurred within 15 minutes of exposure in our cell culture system. In comparison, STAT2 phosphorylation did not change significantly with IFNγ treatment or JAK inhibition.
Figure 1. IFNγ induces JAK-mediated STAT1 phosphorylation in HBE1 cells.
HBE1 cells were pretreated with either vehicle control (DMSO) or JAK inhibitor I (Jaki I; 1 μM) for three hours. Cells were subsequent exposed to IFNγ (100 ng/mL) or media alone for 15 minutes and harvested for total protein. 60 μg of protein from each sample was loaded onto a 7% SDS PAGE gel and western blotting was performed using anti-STAT1 or anti-STAT2 antibodies. PVDF membranes were stripped and repeat western blotting was performed using unphosphorylated STAT protein as an internal control to ensure equal protein loading. Duplicate results are representative of three separate experiments. Fifteen or 30-minute IFNγ treatments produced similar results.
IFNγ-mediated DUOX2 induction does not require JAK signaling
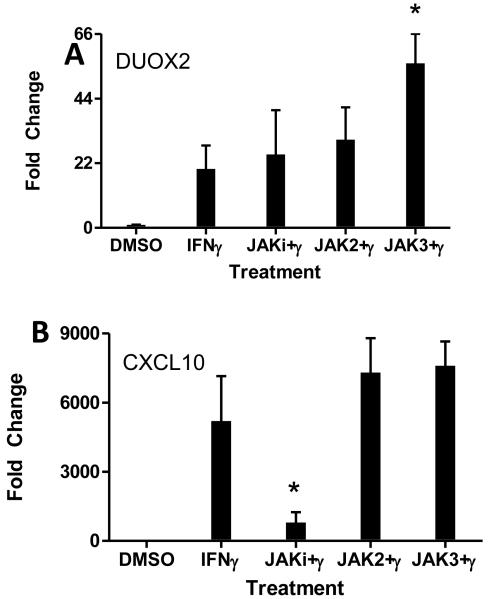
To establish that IFNγ induced DUOX2 though the JAK-STAT pathway, we measured DUOX2 mRNA levels in HBE1 cells by rt-QPCR before and after IFNγ treatment in the presence or absence of nonspecific JAK inhibitor (JAKi I; 1μM). Consistent with our previously published data, IFNγ induced a robust increase in DUOX2 mRNA at 24 hours (Figure 2A). Surprisingly, nonspecific JAK inhibition had no effect on DUOX2 mRNA levels. Similarly, chemical inhibition of JAK2 phosphorylation had no effect, whereas chemical inhibition of JAK3 resulted in a slight superinduction. Because IFNγ is known to increase CXCL10 through JAK1-STAT1 signaling [Neville], these unexpected results prompted us to analyze the same RNA samples for CXCL10 expression. In contrast to DUOX2 mRNA expression, nonspecific JAK inhibition reduced IFNγ-induced CXCL10 mRNA expression by 85% (Figure 2B), emphasizing an intact IFNγ-JAK1-STAT1 signaling pathway in our model.
Figure 2. Lack of DUOX2 mRNA inhibition by JAK inhibitors.
HBE1 cells were pretreated with various JAK inhibitors for 3 hours (JAKi1, 1 μM; Jak2i, 50 μM; Jak3i, 80 μM), followed by IFNγ (100 ng/mL) treatment for two hours. Cells were harvested for RNA extraction 24 hours after IFN-γ treatment. rt-QPCR analyses of mRNA expression are presented as fold-induction of DUOX2 (A) or CXCL10 (B) normalized to β-actin by defining 2−ΔΔCt value from the DMSO-treated sample as 1. Data were pooled from five separate experiments and represent mean ± SD. *; p < 0.05
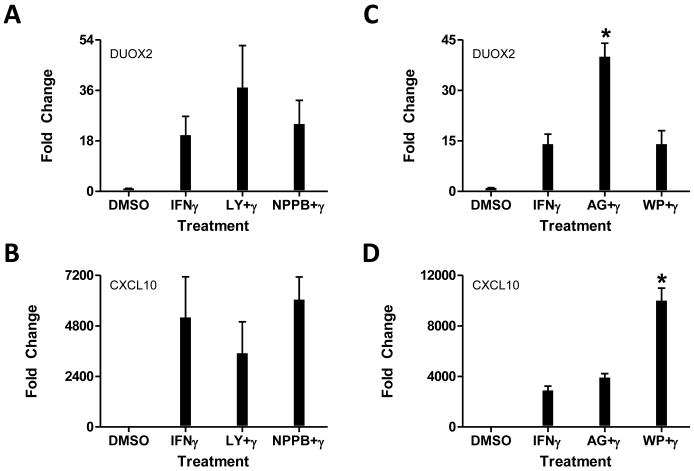
Similarly, inhibition of alternative JAK-mediated signaling pathways had no effect on DUOX2 expression (Figure 3). Using chemical inhibitors of PI3K (LY294002), ERK1/ERK2 (NPPB), STAT3 (WP1066), or STAT5 (AG490), we observed a nonsignificant superinduction or no change in IFNγ-mediated DUOX2 expression 24 hours after IFNγ treatment (Figure 3A and 3C). Of potential importance, CXCL10 expression inversely mirrored DUOX2 expression in response to these inhibitors (Figure 3B and 3D).
Figure 3. The effects of Erk and PI3K blockade on IFN-γ-mediated DUOX2 expression.
HBE1 cells were pretreated with inhibitors of PI3 kinase (LY; 50 μM), ERK (NPPB; 100 μM), STAT3 (WP; 10μM), or STAT5 (AG; 50μM) phosphorylation for 3 hours, followed by IFN-γ (100 ng/mL) treatment for two hours. Cells were harvested for RNA extraction 24 hours after IFN-γ treatment. rt-QPCR analyses of mRNA expression are presented as fold-induction of DUOX2 (A, C) or CXCL10 (B, D) normalized to β-actin by defining 2−ΔΔCt value from the DMSO-treated sample as1. Data were pooled from five separate experiments and represent mean ± SD.
IFNγ-mediated mRNA kinetics
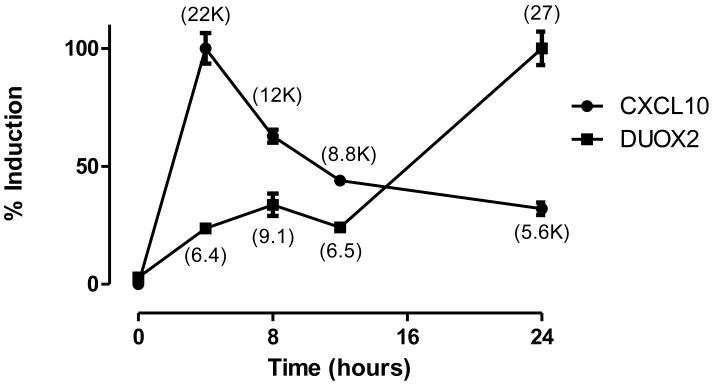
Together, these data suggested that IFNγ increases DUOX2 expression though a mechanism that is distinct from the IFNγ-JAK-STAT signaling cascade that is characteristic of CXCL10 and other IFNγ-inducible proteins. To better characterize these differences, HBE1 cells were treated with IFNγ for two hours and harvested for total RNA at multiple time points to determine DUOX2 or CXCL10 mRNA expression levels by rt-QPCR. CXCL10 mRNA increased robustly (22,000-fold) within four hours of IFNγ treatment and fell to approximately 25% of maximal induction at 24 hours. In stark contrast, DUOX2 mRNA levels modestly increased at the earlier time points, but reached maximal induction (27-fold) at 24 hours.
DISCUSSION
Our previous work and the work of others [14; 17; 25] has clearly indicated that IFNγ induces DUOX2 transcription in respiratory tract epithelium. To better understand the mechanisms responsible for this induction, we explored the early signaling events that were responsible for this expression. We hypothesized that canonical JAK1-STAT1 signaling and immediate transcriptional activation were primarily responsible, similar to the activation pathway for the closely related Nox protein gp91phox [26; 27; 28; 29]. However, our current data refute this notion and suggest that the regulatory mechanisms responsible for IFNγ-mediated DUOX2 expression are more complex than we initially postulated.
Although it is possible that the JAK1-STAT1 pathway was not robustly induced in our model system, our data suggest otherwise. We observed clear STAT1 phosphorylation by western blots within 15 or 30 minutes after IFNγ treatment, which was blocked by chemical inhibition of JAK activity (Figure 1 and data not shown). In addition, we performed parallel experiments with CXCL10, a gene known to be induced via an IFNγ-JAK-STAT1 pathway [22], and observed that IFNγ-mediated CXCL10 mRNA was substantially inhibited by JAK inhibitor at 24 hours. Also, because peak CXCL10 expression occurred at four hours (Figure 4), it is likely that the degree of JAK-mediated inhibition was significantly greater than we observed at the 24 hour time point (Figure 1). Together, these data support the notion that JAK1-STAT1 signaling was intact in our model system.
Figure 4. Time course comparison of DUOX2 and CXCL10 induction.
HBE1 cells were treated with IFNγ (100 ng/mL) for 2 hours and harvested for total RNA at various time points using the start of IFNγ exposure as t=0. rt-QPCR analyses of mRNA expression are presented as a percentage of fold-induction compared to maximum induction. Numbers in parentheses represent observed fold-induction for DUOX1 or CXCL10 normalized to β-actin by defining the average 2−ΔΔCt value of untreated samples at all time points as 1. K=1000-fold. Data are representative of four separate experiments and mean ± SD are shown.
Alternatively, IFNγ–mediated DUOX2 induction may occur through one of several previously established alternative pathways [30]. Several of these alternative pathways still require STAT1 (e.g. IRF family transcription factors), but multiple STAT-independent pathways have been described as well (e.g. STAT3, AP-1, or NF-κB). With the exception of PI3-K, however, these alternative pathways still utilize JAK. Based on our data, IFNγ–mediated DUOX2 expression appears to be independent of JAK altogether. Inhibition of PI3-K failed to decrease, or actually increased, DUOX2 transcription. Similarly, other mediators of alternative IFNγ signaling such as ERK1/2, STAT3, or STAT5, appear to play no role in DUOX2 regulation. This suggests a novel mechanism for IFNγ signaling that remains to be fully characterized.
Of potential importance, our data suggested that JAK3-STAT5 signaling suppresses DUOX2 expression (Figures 2 and 3). This highlights a possible cross-talk mechanism between IFNγ–mediated DUOX2 expression and JAK3/STAT5 signaling, where JAK3/STAT5 activation attenuates IFNγ-mediated DUOX2 expression or determines basal levels of DUOX2 expression. For example, we and others have previously shown that DUOX1 and DUOX2 in respiratory tract epithelium are differentially expressed both basally and in response to IFNγ or IL-4/IL-13 [17; 25]. Given these observed differences, it is possible that basal JAK3/STAT5 activation (i.e. low level IL-4/IL-13 signaling [31]) is responsible for ensuring higher basal DUOX1 and lower basal DUOX2 levels. These findings potentially give us insights to explain differential DUOX1 and DUOX2 expression in other tissue types as well.
The timing of DUOX2 induction versus CXCL10 induction similarly highlighted profound differences between the canonical JAK1-STAT1-CXCL10 pathway and IFNγ-mediated DUOX2 expression. After a two-hour pulse dose of IFNγ, CXCL10 mRNA rapidly increased to peak expression two hours later (four-hour timepoint) and decayed exponentially over the 24-hour observation period. Conversely, DUOX2 mRNA levels were less than 25% of peak levels at four hours, but reached maximal expression at 24 hours. These data confirm that the JAK1-STAT1 pathway is not responsible for transcriptional activation of DUOX2. If it were, we would expect DUOX2 gene expression to occur in parallel with CXCL10. Furthermore, these data suggest that IFNγ does not induce DUOX2 via immediate transcriptional activation. Although a modest increase in DUOX2 mRNA was seen at earlier timepoints, the predominant increase in DUOX2 is significantly delayed. We speculate that IFNγ induces transcription factors that secondarily activate the DUOX2 promoter, increases proteins that modulate DUOX2 RNA stability, or augments microRNA expression that impacts DUOX2 RNA transcription.
CONCLUSIONS
Our data indicate that, although IFNγ-JAK-STAT1 signaling occurs in our cell culture model, DUOX2 regulation does not occur through this signaling mechanism. Furthermore, IFNγ-mediated DUOX2 regulation did not utilize other common alternative pathways for IFNγ signaling. And, it is unlikely that IFNγ induces DUOX2 by immediate transcriptional activation. We anticipate that these unexpected findings will lead us to elucidate novel mechanisms of IFNγ-mediated gene regulation that will broaden our understanding of innate immunity.
ACKNOWLEGEMENTS
This work was supported by funds from the National Institutes of Health (HL085311).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–93. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- [2].Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–54. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- [3].Gabig TG, Babior BM. The O2(−) -forming oxidase responsible for the respiratory burst in human neutrophils. Properties of the solubilized enzyme. J Biol Chem. 1979;254:9070–4. [PubMed] [Google Scholar]
- [4].Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–9. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- [5].De Deken X, Wang D, Many MC, Costagliola S, Libert F, Vassart G, Dumont JE, Miot F. Cloning of two human thyroid cDNAs encoding new members of the NADPH oxidase family. J Biol Chem. 2000;275:23227–33. doi: 10.1074/jbc.M000916200. [DOI] [PubMed] [Google Scholar]
- [6].Dupuy C, Ohayon R, Valent A, Noel-Hudson MS, Deme D, Virion A. Purification of a novel flavoprotein involved in the thyroid NADPH oxidase. Cloning of the porcine and human cdnas. J Biol Chem. 1999;274:37265–9. doi: 10.1074/jbc.274.52.37265. [DOI] [PubMed] [Google Scholar]
- [7].van der Vliet A. NADPH oxidases in lung biology and pathology: host defense enzymes, and more. Free Radic Biol Med. 2008;44:938–55. doi: 10.1016/j.freeradbiomed.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fischer H. Mechanisms and function of DUOX in epithelia of the lung. Antioxid Redox Signal. 2009;11:2453–65. doi: 10.1089/ars.2009.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Grasberger H. Defects of thyroidal hydrogen peroxide generation in congenital hypothyroidism. Mol Cell Endocrinol. doi: 10.1016/j.mce.2010.01.029. [DOI] [PubMed] [Google Scholar]
- [10].Moreno JC, Bikker H, Kempers MJ, van Trotsenburg AS, Baas F, de Vijlder JJ, Vulsma T, Ris-Stalpers C. Inactivating mutations in the gene for thyroid oxidase 2 (THOX2) and congenital hypothyroidism. N Engl J Med. 2002;347:95–102. doi: 10.1056/NEJMoa012752. [DOI] [PubMed] [Google Scholar]
- [11].Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–50. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- [12].Forteza R, Salathe M, Miot F, Forteza R, Conner GE. Regulated hydrogen peroxide production by Duox in human airway epithelial cells. Am J Respir Cell Mol Biol. 2005;32:462–9. doi: 10.1165/rcmb.2004-0302OC. [DOI] [PubMed] [Google Scholar]
- [13].Salathe M, Holderby M, Forteza R, Abraham WM, Wanner A, Conner GE. Isolation and characterization of a peroxidase from the airway. Am J Respir Cell Mol Biol. 1997;17:97–105. doi: 10.1165/ajrcmb.17.1.2719. [DOI] [PubMed] [Google Scholar]
- [14].Gattas MV, Forteza R, Fragoso MA, Fregien N, Salas P, Salathe M, Conner GE. Oxidative epithelial host defense is regulated by infectious and inflammatory stimuli. Free Radic Biol Med. 2009;47:1450–8. doi: 10.1016/j.freeradbiomed.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Moskwa P, Lorentzen D, Excoffon KJ, Zabner J, McCray PB, Jr., Nauseef WM, Dupuy C, Banfi B. A novel host defense system of airways is defective in cystic fibrosis. Am J Respir Crit Care Med. 2007;175:174–83. doi: 10.1164/rccm.200607-1029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Donko A, Peterfi Z, Sum A, Leto T, Geiszt M. Dual oxidases. Philos Trans R Soc Lond B Biol Sci. 2005;360:2301–8. doi: 10.1098/rstb.2005.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Harper RW, Xu C, Eiserich JP, Chen Y, Kao CY, Thai P, Setiadi H, Wu R. Differential regulation of dual NADPH oxidases/peroxidases, Duox1 and Duox2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS Lett. 2005;579:4911–7. doi: 10.1016/j.febslet.2005.08.002. [DOI] [PubMed] [Google Scholar]
- [18].Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- [19].Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–91. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- [20].Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–95. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- [21].Luster AD, Unkeless JC, Ravetch JV. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315:672–6. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- [22].Neville LF, Mathiak G, Bagasra O. The immunobiology of interferon-gamma inducible protein 10 kD (IP-10): a novel, pleiotropic member of the C-X-C chemokine superfamily. Cytokine Growth Factor Rev. 1997;8:207–19. doi: 10.1016/s1359-6101(97)00015-4. [DOI] [PubMed] [Google Scholar]
- [23].Yankaskas JR, Haizlip JE, Conrad M, Koval D, Lazarowski E, Paradiso AM, Rinehart CA, Jr., Sarkadi B, Schlegel R, Boucher RC. Papilloma virus immortalized tracheal epithelial cells retain a well-differentiated phenotype. Am J Physiol. 1993;264:C1219–30. doi: 10.1152/ajpcell.1993.264.5.C1219. [DOI] [PubMed] [Google Scholar]
- [24].Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- [25].Rada B, Lekstrom K, Damian S, Dupuy C, Leto TL. The pseudomonas toxin pyocyanin inhibits the dual oxidase-based antimicrobial system as it imposes oxidative stress on airway epithelial cells. J Immunol. 2008;181:4883–93. doi: 10.4049/jimmunol.181.7.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cassatella MA, Bazzoni F, Flynn RM, Dusi S, Trinchieri G, Rossi F. Molecular basis of interferon-gamma and lipopolysaccharide enhancement of phagocyte respiratory burst capability. Studies on the gene expression of several NADPH oxidase components. J Biol Chem. 1990;265:20241–6. [PubMed] [Google Scholar]
- [27].Eklund EA, Skalnik DG. Characterization of a gp91-phox promoter element that is required for interferon gamma-induced transcription. J Biol Chem. 1995;270:8267–73. doi: 10.1074/jbc.270.14.8267. [DOI] [PubMed] [Google Scholar]
- [28].Foster N, Hulme SD, Barrow PA. Induction of antimicrobial pathways during early-phase immune response to Salmonella spp. in murine macrophages: gamma interferon (IFN-gamma) and upregulation of IFN-gamma receptor alpha expression are required for NADPH phagocytic oxidase gp91-stimulated oxidative burst and control of virulent Salmonella spp. Infect Immun. 2003;71:4733–41. doi: 10.1128/IAI.71.8.4733-4741.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mazzi P, Donini M, Margotto D, Wientjes F, Dusi S. IFN-gamma induces gp91phox expression in human monocytes via protein kinase C-dependent phosphorylation of PU.1. J Immunol. 2004;172:4941–7. doi: 10.4049/jimmunol.172.8.4941. [DOI] [PubMed] [Google Scholar]
- [30].Gough DJ, Levy DE, Johnstone RW, Clarke CJ. IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 2008;19:383–94. doi: 10.1016/j.cytogfr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- [31].Jiang H, Harris MB, Rothman P. IL-4/IL-13 signaling beyond JAK/STAT. J Allergy Clin Immunol. 2000;105:1063–70. doi: 10.1067/mai.2000.107604. [DOI] [PubMed] [Google Scholar]