Abstract
The lipid extract of the marine sponge Mycale sp. inhibited the activation of hypoxiainducible factor-1 (HIF-1) in a human breast tumor T47D cell-based reporter assay. Bioassay-guided isolation and structure elucidation yielded 18 new lipophilic 2,5-disubstituted pyrroles, and eight structurally related known compounds. The active compounds inhibited hypoxia-induced HIF activation with moderate potency (IC50 values < 10 μM). Mechanistic studies revealed that the active compounds suppressed mitochondrial respiration by blocking NADH-ubiquinone oxidoreductase (complex I) at concentrations that inhibited HIF-1 activation. Under hypoxic conditions, reactive oxygen species produced by mitochondrial complex III are believed to act as a signal of cellular hypoxia that leads to HIF-1α protein induction and activation. By inhibiting electron transport (or delivery) to complex III under hypoxic conditions, lipophilic Mycale pyrroles appear to disrupt mitochondrial ROS-regulated HIF-1 signaling.
Hypoxic regions arise in solid tumors when the existing blood vessels fail to meet the increased demand for oxygen from the rapidly proliferating malignant cells. Clinical studies indicate that the extent of tumor hypoxia correlates with advanced disease stages, malignant progression, treatment resistance, and poor prognosis.1,2 Despite decades of drug discovery efforts, there is no approved drug that specifically targets tumor hypoxia. The focus of our anti-tumor hypoxia drug discovery research is to identify and characterize small molecule inhibitors of hypoxia-inducible factor-1 (HIF-1). The transcription factor HIF-1 is a heterodimer made up of two proteins – an oxygen-regulated HIF-1α subunit and a constitutively expressed HIF-1β/ARNT subunit. First discovered by Semenza and colleagues,3,4 HIF-1 has become an important molecular target for anticancer drug discovery.5–7 Under normoxic conditions, HIF-1 is inactivated due to the rapid degradation of the oxygen-regulated HIF-1α protein by the proteasome.8,9 Both the prolyl hydroxylases that tag HIF-1α protein for degradation and the asparaginyl hydroxylase that inactivates HIF-1α protein utilize oxygen as a substrate and require ferrous iron (Fe2+) as a co-factor.10–14 Hypoxic exposure, treatment with hypoxia mimetics (i.e. iron chelators, transition metals, etc.), activation of oncogenes, and inactivation of tumor suppressor genes can lead to the stabilization and activation of HIF-1α protein, and subsequent HIF-1 activation.5–7 Upon activation, HIF-1 binds to the hypoxia-response element (HRE) located in the promoter regions of target genes and regulates gene expression. The activation of HIF-1 mediated signaling pathways results in enhanced cellular adaptation and survival under hypoxic conditions.5–7 As in the case of tumor hypoxia, clinical observations have revealed that expression of the oxygen-regulated HIF-1α subunit also correlates with advanced disease stages, poor prognosis, and treatment resistance among cancer patients.5–7 In animal-based preclinical studies, the inhibition of HIF-1 by various approaches (e.g. small molecule inhibitors, RNA antagonists, etc.) leads to the suppression of tumor growth.15–17 Improved treatment outcomes have resulted when HIF-1 inhibition was combined with chemotherapeutic agents and/or radiation.18–21 Agents that inhibit HIF-1 have entered early phase clinical trials for cancer: EZN-2968, a HIF-1α RNA antagonist; topotecan, a natural product-derived topoisomerase-1/HIF-1 inhibitor; and PX-478, a small molecule that decreases HIF-1α gene expression.22 Numerous drug discovery efforts are underway to identify and develop HIF-1 inhibitors for the treatment of cancer.5–7
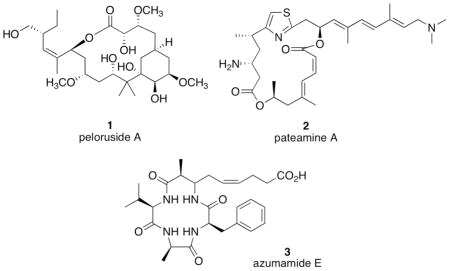
Over 20,000 extracts of plants and marine organisms have been evaluated for natural products that inhibit HIF-1 activation in a T47D human breast tumor cell-based reporter assay.23–25 The lipophilic extract of a Palau collection of the marine sponge Mycale sp. (Mycalidae) from the NCI Open Repository of marine invertebrate extracts inhibited hypoxia induced HIF-1 activation in a 96-well plate-based reporter assay. A number of cytotoxic agents have been isolated from various Mycale spp. Representative Mycale metabolites with known anti-tumor mechanisms include the microtubule stabilizer peloruside A (1),26–28 the translation inhibitor pateamine A (2) that disrupts the function of translation initiation factor eIF4A,29–31 and the histone deacetylase (HDAC) inhibitor azumamide E (3).32,33
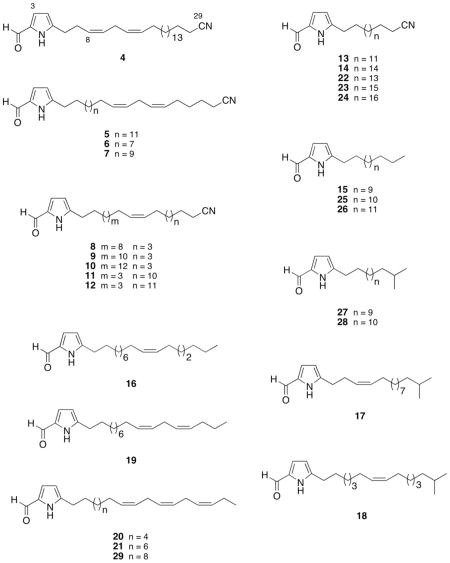
Bioassay-guided isolation of the active Mycale sp. lipid extract afforded eighteen new 5-alkylpyrrole-2-carbaldehyde metabolites 4 – 21, and eight structurally-related known compounds 22 – 29.34–36 Herein, this report describes the identification and characterization of 5-alkylpyrrole-2-carbaldehyde Mycale metabolites that inhibit HIF-1 activation. Further mechanistic investigation revealed that these compounds suppress tumor cell respiration at mitochondrial electron transport chain (ETC) complex I.
Results and Discussion
In a human breast tumor T47D cell-based reporter assay,23–25 a lipid extract of the sponge Mycale sp. inhibited hypoxia (1% O2)-induced HIF-1 activation by 53% at 5 μg mL−1. Bioassay-guided isolation and structure elucidation of the active extract (5.0 g) afforded eighteen new 2,5-disubstituted lipophilic pyrroles (4 – 21), and eight previously reported analogs (22 – 29).34–36
Compound 4 was isolated as an amorphous powder and its molecular formula, C29H46N2O, was established by HRESIMS. The presence of a disubstituted pyrrole nucleus was deduced from the 1H NMR resonances (Table 1) at δ 9.38 (br s, 1H, N-H), 6.89 (dd, 1H, J = 3.8, 2.8 Hz, H-3) and 6.11 (dd, 1H, J = 3.8, 2.8 Hz, H-4), and the 13C NMR resonances (Table 4) at δ 142.3 (C), 132.2 (C) 122.5 (CH) and 109.8 (CH). The 3.8 Hz coupling constant between the pyrrole protons, a typical value for J3,4 in pyrroles,36 indicated a 2,5-disubstitution pattern. A singlet resonance in the 1H NMR spectrum (δ 9.39, s, 1H, H-1) together with a methine 13C resonance at δ 178.4 (CH) were attributed to a formyl group conjugated to the pyrrole nucleus. The IR bands at 3260 and 1635 cm−1 and UV absorption maxima at 301 nm (ε 16,200) were characteristic of the pyrrole-2-carbaldehyde. Analysis of the HMQC spectrum indicated that four 1H NMR multiplet resonances at δ 5.47 (m, 1H), 5.42 (m, 1H), 5.38 (m, 1H) and 5.28 (m, 1H) were coupled to the 13C NMR resonances at δ 130.3 (CH), 128.1 (CH), 130.9 (CH) and 127.4 (CH), respectively. These were assigned to the olefinic protons of a 1,4-diene spin system based on observation of 1H-1H COSY correlations (i.e. H-8/H-9/H2-10/H-11/H-12). Furthermore, it was readily inferred that the double bonds were separated by a bis-allylic methylene moiety upon observation of the NMR resonances at δH 2.74 (m, 2H, H-10) and δC 25.8 (CH2), and the chemical shift of the carbon resonance was typical of a (Z,Z) 1,4-disubstituted pattern of unsaturation.37 The location of the olefinic system was determined from the 2D NMR spectra, which exhibited 1H-1H COSY correlations from H2-7 (δH 2.44) to H2-6 (δH 2.737) and H-8, as well as HMBC correlations between H2-6 and C-7 (δC 26.9) and C-8 and between H2-7 and C-5, C-6 (δC 28.1), C-8 and C-9. As in the structures of the previously reported Mycale metabolites mycalenitrile-1 (22) and mycalenitrile-2 (24),34 4 also possessed a terminal nitrile moiety at δ 120.0 (C). The molecular formula and the presence of a nitrile carbon indicated that the olefinic system had to be connected to the nitrile group through a sequence of sixteen methylene carbons. Therefore, 4 was deduced to be a new nitrile-substituted 2,5-disubstituted pyrrole and was assigned the trivial name mycalenitrile-4.
Table 1.
1H NMR Dataa δ (J, Hz) for 4, 5, 7, and 10–12
| H | 4 | 5 | 7 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|
| 1 | 9.39, s | 9.37, s | 9.37, s | 9.36, s | 9.35, s | 9.38, s |
| 3 | 6.89, dd (3.8, 2.8) | 6.90, dd (3.6, 2.8) | 6.90, dd (3.6, 2.8) | 6.90, dd (3.2, 2.8) | 6.91, dd (3.2, 2.8) | 6.89, dd (3.6, 2.4) |
| 4 | 6.11, dd (3.8, 2.8) | 6.09, dd (3.6, 2.8) | 6.08, dd (3.6, 2.8) | 6.08, dd (3.2, 2.8) | 6.08, dd (3.2, 2.8) | 6.08, dd (3.6, 2.4) |
| 6 | 2.74, m | 2.67, t (7.6) | 2.68, t (7.6) | 2.68, t (7.2) | 2.68, t (7.2) | 2.66, t (7.6) |
| 7 | 2.44, dd (7.2, 6.8) | 1.68, m | 1.68, m | 1.66, m | 1.65, m | 1.66, m |
| 8 | 5.42, m | 1.27, br s | 1.27, br s | 1.26, br s | 1.26, br s | 1.27, br s |
| 9 | 5.47, m | 1.27, br s | 1.27, br s | 1.26, br s | 1.26, br s | 1.27, br s |
| 10 | 2.74, m | 1.27, br s | 1.27, br s | 1.26, br s | 1.36, m | 1.37, m |
| 11 | 5.28, m | 1.27, br s | 1.27, br s | 1.26, br s | 2.01, m | 2.01, m |
| 12 | 5.38, m | 1.27, br s | 1.27, br s | 1.26, br s | 5.34, m | 5.36, m |
| 13 | 2.02, dd (6.8, 6.6) | 1.27, br s | 1.27, br s | 1.26, br s | 5.34, m | 5.36, m |
| 14 | 1.26, br s | 1.27, br s | 1.27, br s | 1.26, br s | 2.01, m | 2.02, m |
| 15 | 1.26, br s | 1.27, br s | 1.27, br s | 1.26, br s | 1.36, m | 1.37, m |
| 16 | 1.26, br s | 1.27, br s | 1.27, br s | 1.26, br s | 1.26, br s | 1.27, br s |
| 17 | 1.26, br s | 1.27, br s | 2.04, dt (7.2, 6.4) | 1.26, br s | 1.26, br s | 1.27, br s |
| 18 | 1.26, br s | 1.27, br s | 5.37, m | 1.26, br s | 1.26, br s | 1.27, br s |
| 19 | 1.26, br s | 2.05, dt (7.2, 6.8) | 5.37, m | 1.38, m | 1.26, br s | 1.27, br s |
| 20 | 1.26, br s | 5.37, m | 2.78, t (6.0) | 2.02, m | 1.26, br s | 1.27, br s |
| 21 | 1.26, br s | 5.37, m | 5.37, m | 5.36, m | 1.26, br s | 1.27, br s |
| 22 | 1.26, br s | 2.78, dd (6.4, 5.2) | 5.37, m | 5.36, m | 1.26, br s | 1.27, br s |
| 23 | 1.26, br s | 5.37, m | 2.10, dt (7.2, 5.6) | 2.02, m | 1.26, br s | 1.27, br s |
| 24 | 1.26, br s | 5.37, m | 1.44, m | 1.38, m | 1.43, m | 1.27, br s |
| 25 | 1.26, br s | 2.10, dt (7.2, 6.4) | 1.66, m | 1.26, br s | 1.65, m | 1.43, m |
| 26 | 1.45, m | 1.44, m | 2.33, m | 1.44, m | 2.34, t (7.2) | 1.66, m |
| 27 | 1.67, m | 1.66, m | 1.66, m | 2.34, t (7.2) | ||
| 28 | 2.34, t (7.2) | 2.34, t (7.2) | 2.35, t (6.8) | |||
| NH | 9.38, br s | 9.52, br s | 9.51, br s | 10.2, br s | 10.43, br s | 9.23, br s |
Bruker AMX 400 MHz spectrometer; Chemical shifts (ppm) referenced to CDCl3 (δH 7.27); Numerical order groups compounds of similar chain length.
Table 4.
13C NMR Dataa (δ, DEPT) for 4, 5, 7, and 10–12
| C | 4 | 5 | 7 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|
| 1 | 178.4, CH | 178.3, CH | 178.3, CH | 178.2, CH | 178.3, CH | 178.3, CH |
| 2 | 132.2, C | 132.1, C | 132.1, C | 132.0, C | 132.1, C | 132.0, C |
| 3 | 122.5, CH | 122.6, CH | 122.7, CH | 122.7, CH | 122.8, CH | 122.7, CH |
| 4 | 109.8, CH | 109.7, CH | 109.7, CH | 109.6, CH | 109.6, CH | 109.7, CH |
| 5 | 142.3, qC | 142.8, C | 143.0, C | 143.4, C | 143.4, C | 142.9, C |
| 6 | 28.1, CH2 | 28.0, CH2 | 28.0, CH2 | 28.0, CH2 | 28.1, CH2 | 28.1, CH2 |
| 7 | 26.9, CH2 | 28.7, CH2 | 28.7, CH2 | 28.9, CH2 | 29.0, CH2 | 29.0, CH2 |
| 8 | 128.1, CH | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 29.9-29.4 | 30.0-29.5b |
| 9 | 130.3, CH | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 29.9-29.4 | 30.0-29.5b |
| 10 | 25.8, CH2 | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 29.2, CH2c | 29.1, CH2 |
| 11 | 127.4, CH | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 27.4, CH2e | 27.5, CH2c |
| 12 | 130.9, CH | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 130.4, CHd | 130.5, CHd |
| 13 | 27.4, CH2 | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 129.6, CHd | 129.6, CHd |
| 14 | 29.9-29.0b | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 27.2, CH2e | 27.2, CH2c |
| 15 | 29.9-29.0b | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 29.1, CH2c | 29.1, CH2 |
| 16 | 29.9-29.0b | 29.9-29.0b | 29.9-29.0b | 29.9-29.4b | 29.9-29.4b | 30.0-29.5b |
| 17 | 29.9-29.0b | 29.9-29.0b | 27.5, CH2 | 29.9-29.4b | 29.9-29.4b | 30.0-29.5b |
| 18 | 29.9-29.0b | 29.9-29.0b | 128.0, CH | 29.9-29.4b | 29.9-29.4b | 30.0-29.5b |
| 19 | 29.9-29.0b | 27.5, CH2 | 130.6, CH | 29.2, CH2e | 29.9-29.4b | 30.0-29.5b |
| 20 | 29.9-29.0b | 127.9, CH | 25.9, CH2 | 27.4, CH2c | 29.9-29.4b | 30.0-29.5b |
| 21 | 29.9-29.0b | 130.6, CH | 128.9, CH | 130.3, CHd | 29.9-29.4b | 30.0-29.5b |
| 22 | 29.9-29.0b | 25.9, CH2 | 129.6, CH | 129.6, CHd | 29.9-29.4b | 30.0-29.5b |
| 23 | 29.9-29.0b | 128.9, CH | 27.0, CH2 | 27.2, CH2c | 29.9-29.4b | 30.0-29.5b |
| 24 | 29.9-29.0b | 129.5, CH | 28.9, CH2 | 29.1, CH2e | 28.8, CH2 | 30.0-29.5b |
| 25 | 29.9-29.0b | 27.0, CH2 | 25.6, CH2 | 29.9-29.4 | 25.6, CH2 | 28.9, CH2 |
| 26 | 28.9, CH2 | 28.9, CH2 | 17.3, CH2 | 28.8, CH2 | 17.3, CH2 | 25.6, CH2 |
| 27 | 25.6, CH2 | 25.6, CH2 | 120.1, C | 25.5, CH2 | 120.0, C | 17.4, CH2 |
| 28 | 17.3, CH2 | 17.3, CH2 | 17.3, CH2 | 120.1, C | ||
| 29 | 120.0, C | 120.1, C | 120.0, C |
Bruker AMX 100 MHz spectrometer; δ values are reported in ppm referenced to CDCl3 (δC 77.23) as internal standard; Numerical order groups compounds of similar chain length;
All carbons were CH2;
Values may be interchanged.
Compound 5 was obtained as a viscous liquid that had the same C29H46N2O molecular formula (determined by HRESIMS on the quasi-molecular ion peak [M+Na]+ at m/z 461.3506) as 4. The 1H and 13C NMR spectra (Tables 1 and 4) of 5 were very similar to those of 4, except for modest differences in the chemical shifts of several resonances. Further comparison of the NMR spectra of 5 and 4 revealed that the only difference between them was the position of the olefinic system. This gave rise to 1H NMR resonances in 5 at δ 5.37 (m, 4H) due to four olefinic protons, resonances at 2.10 (dt, 2H, J = 7.2, 6.4 Hz, H-25) and 2.05 (dt, 2H, J = 7.2, 6.8 Hz, H-19) that were attributable to two allylic methylenes, and a resonance at 2.78 (dd, 2H, J = 6.4, 5.2 Hz, H-22) assignable to a bis-allylic methylene. The location of the olefinic system in the chain was deduced from analysis of the HMBC spectrum. In particular, the H2-28 [δ 2.34 (t, 2H, J = 7.2 Hz)] resonance showed diagnostic long-range correlations with the C-26 (δ 28.9) and C-27 (δ 25.6) methylene carbons, and the C-27 resonance, in turn, was coupled to the H2-25 (δ 2.10) allylic protons. Observation of the spin system from H2-24 to H2-28 in the 1H-1H COSY spectrum confirmed the position of the olefinic system. It was therefore concluded that the diene system was at the ω-6 position,34,37 and this related compound was deduced to be another nitrile-substituted 2,5-disubstituted pyrrole that was named mycalenitrile-5 (5).
Compounds 6 and 7 were isolated as colorless oils. The HRESIMS indicated the molecular formulae C25H38N2O and C27H42N2O for each, respectively. Both compounds displayed NMR spectra that were nearly identical to those described for 5, which suggested that 6 and 7 were also pyrrole-2-carbaldehydes and that each contained a 1,4-diene side chain. Furthermore, the olefinic systems in 6 and 7 were also deduced to be at ω-6, following the same rationale above described for 5. The molecular formulae of 6 and 7 indicated that the side chains in each were shortened by either four or two methylene units, respectively, and that the site of chain truncation was between the pyrrole ring and the 1,4-diene system (relative to the structure of 5). Therefore, mycalenitrile-6 (6) and mycalenitrile-7 (7) were deduced to be the new nitrile-substituted lipophilic pyrroles.
Compound 8 was obtained as a white solid. The molecular formula C25H40N2O, was determined by HRESIMS and from analysis of the 13C NMR and DEPT spectra. Similarities in the spectroscopic data indicated that 8 was structurally similar to 6. Analysis of the 1H and 13C NMR spectra of 8 indicated that the linear side chain of 8 contained only a single double bond [δH 5.36 (m, 2H); δC 130.4 (CH) and 129.6 (CH)]. The position of the double bond was determined by examination of the GC-EIMS fragmentation pattern. In particular, compound 8 afforded a molecular ion peak at m/z 384 [M+] and two ions at m/z 234 and 288 due to fragments produced by allylic cleavage of the side chain.36,38 The ion at m/z 234 was due to the fragment containing the pyrrole nucleus (C15H24NO+), while the ion at m/z 288 arose from the fragment containing the pyrrole moiety and the olefin (C19H30NO+). These data established that the double bond was located at C-17/C-18 (ω-8). Thus, mycalenitrile-8 (8) was deduced to be a new ω-8 unsaturated lipophilic pyrrole.
Compounds 9 and 10 were isolated as a colorless oil and a white solid, respectively. Each possessed IR, UV and NMR spectra that were nearly superimposable with those of 8, indicating that the only differences between these structures was in the length of the lipophilic chain and position of the double bond. The molecular formule of 9 (C27H44N2O) and 10 (C29H48N2O), deduced from the HRESIMS data, indicated that the side chain of each compound was elongated by either two or four methylenes, respectively, with respect to the structure of 8. Compounds 9 and 10 exhibited a similar GC-EIMS cleavage pattern as observed with 8. Specifically, each produced the fragment ions (m/z 262 and 316 for 9; m/z 290 and 344 for 10), with a difference of 54 mass units. These data suggested that the side chain olefinic bond in each compound was also at the ω-8 position. Consequently, mycalenitrile-9 (9) and mycalenitrile-10 (10) were deduced to be two new lipophilic nitrile-substituted pyroles that each contained a single double bond in their respective side chains at the ω-8 position.
Compounds 11 and 12 were obtained as colorless oils of molecular formulae C27H44N2O and C28H46N2O, respectively (as deduced from analysis of the HRESIMS data). This suggested that each compound differed only by the presence of one methylene group. As in the structures of 8–10, the 1D NMR data (Tables 1 and 4) of 11 and 12 revealed that the side chains in each compound contained only a single double bond. The loss of 54 mass units between the fragment ions at m/z 164 and 218 in the GC-EIMS of each compound indicated that the double bonds in each were located at the C-12 position. Therefore, mycalenitrile-11 (11) and mycalenitrile-12 (12) were deduced to be two new lipophilic pyrroles.
Compound 13 was obtained as a yellow solid. Its molecular formula, C21H34N2O was established by HRESIMS. The presence of the pyrrole-2-carbaldehyde nucleus was confirmed upon observation of the NMR resonances at δH 9.48 (br s, 1H, N-H), 9.37 (s, 1H, H-1), 6.90 (dd, 1H, J = 3.8, 2.8 Hz, H-3) and 6.08 (dd, 1H, J = 3.8, 2.8 Hz, H-4) and δC 178.3 (CH), 143.3 (C), 132.1 (C), 122.7 (CH) and 109.7 (CH). The remaining 13C resonances were assignable to side chain methylenes, with the exception of the δ 120.0 quaternary nitrile carbon, indicating that 13 possessed a fully saturated alkyl side chain. Thus, mycalenitrile-13 (13) was deduced to be a new lipophilic nitrile-substituted pyrole with a saturated side chain.
Compound 14 was isolated as a white solid. The 1H and 13C NMR spectra of 14 were nearly identical to those of 13, indicating that 14 was also a fully unsaturated pyrrole-2-carbaldehyde. The molecular formula C24H40N2O indicated the alkyl chain was elongated by three methylenes relative to the structure of 13. Therefore, this analog of 13 with a longer side chain was assigned the trivial name mycalenitrile-14 (14).
Compound 15 was obtained as an amorphous powder with a molecular formula of C19H33NO, indicated the absence of one nitrogen atom in the structure, relative to the structures of 4 – 14. The spectroscopic data of 15 were similar to those of 13 and 14. Detailed analysis of NMR spectra established that 15 differed from 13 and 14 only by the terminal alkyl side chain substituent (i.e. a terminal methyl group rather than the nitrile moiety in 13 and 14). This methyl group gave rise to a new 1H NMR resonance at δ 0.89 (t, 3H, J = 6.4 Hz, H-19) and replacement of the downfield ω-2 and ω-3 methylene resonances (observed in 13 and 14) with only a broad second order-coupled pattern of methylene resonances at δ 1.27 (br s, 22H, H-8 to H-18). Therefore, this new compound mycalazal-14 (15) was deduced to be a new mycalazal-type 5-tetradecylpyrrole-2-carbaldehyde with a fully saturated lipophilic side chain.
Compound 16 was isolated as a colorless oil. The molecular formula C22H37NO was deduced by HRESIMS on the sodiated molecular ion at m/z 354.2748. The 1H and 13C NMR spectra indicated that, in addition to the 2,5-disubstituted pyrrole moiety and formyl group resonances, specific resonances were attributable to a linear monounsaturated alkyl side chain. In particular, the 1H NMR spectrum of 16 exhibited two olefinic proton resonances at δ 5.37 (m, 2H), one terminal methyl triplet at δ 0.89 (t, H3, J = 7.2 Hz), and one four proton multiplet at δ 2.03 (4H, H-14 and H-17) due to two allylic methylenes that were correlated in the HSQC spectrum with carbon resonances at δ 27.4 (2C). These data suggested that compound 16 contained a C17 hydrocarbon side chain with one double bond with Z-geometry. The Δ15 position of the olefinic bond was determined from the GC-EIMS, which contained fragment ions at m/z 206 and 260 that resulted from allylic cleavage of the side chain olefin. Thus, the structure of mycalazal-15 (16) was deduced to be a new lipophilic pyrrole with a single double bond in the lipophilic side chain at the ω-7 position.
Compound 17 was isolated as a colorless oil. The HRESIMS showed a quasi-molecular ion peak at m/z 340.2641 [M+Na]+ corresponding to the molecular formula C21H35NONa, which required five degrees of unsaturation. The 1H and 13C NMR spectra of 17 were similar to those of 16, except for the absence of the methyl triplet (δ 0.89 in 16), which was replaced by an isopropyl methyl doublet at δ 0.86 (d, J = 7.6 Hz) that integrated for six protons. Thus, the pyrrole-2-carbaldehyde moiety was substituted at C-5 with a monounsaturated hydrocarbon side chain with a terminal isopropyl group. The position of the double bond was assigned at C-8 by 1H-1H COSY correlations that established the H-6 to H-8 spin system. Therefore, the structure of this new Mycale metabolite mycalazal-16 (17) was deduced to be a mycalazal-type lipophic pyrrole-2-carbaldehyde with a branched side chain.
Compound 18 was isolated as a colorless oil. It was isomeric with 17, having a molecular formula of C21H35NO as determined by the HRESIMS. However, the NMR spectra of these two compounds were distinctly different. In particular, the 1H NMR spectrum of 18 exhibited both olefinic proton resonances as a single multiplet resonance centered at δ 5.37 (2H), rather than as two distinct signals at δ 5.39 and 5.46, as observed in 17. In addition, the presence of an additional higher field allylic methylene resonance at δ 2.03 (m) indicated that the difference between 18 and 17 was in the location of the olefinic bond. The position of the double bond in 18 was deduced to be at C-12 by observation of the allylic cleavage fragmention ions at m/z 164 and 218 in the GC-EIMS spectrum. Based on these data and the molecular formula, 18 was deduced to possess a Δ7-14-methylpentadecenyl side chain. Therefore, mycalazal-17 (18) was deduced to be an ω-8 unsaturated isomer of 17.
Compound 19 was obtained as a colorless oil and showed a HRESIMS quasi-molecular ion [M+Na]+ at m/z 352.2621, compatible with the molecular formula C22H35NONa. The 1H NMR spectrum contained the typical H-3, H-4 and N-H resonances of the pyrrole nucleus. Because the pyrrole-2-carbaldehyde moiety accounted for five carbons, four degrees of unsaturations and all the heteroatoms of the molecular formula, it was deduced that compound 19 contained a C17 hydrocarbon side chain with two double bonds. Furthermore, it was evident that the two double bonds were in a 1,4-diene pattern by the presence of a bis-allylic methylene 1H NMR resonances at δ 2.78 (t, 2H, J = 6.0 Hz) and δC 25.9 (CH2). The COSY 1H-1H spin systems identified the 15,18-diene functionality. Specifically, 1H-1H couplings were observed between H-19 (δ 5.37) and H2-20 (δ 2.06), between H2-21 (δ 1.43) and H2-20, and between H2-21 and H3-22 (δ 0.92). This was further confirmed by observation of long-range heteronuclear (HMBC) couplings from H3-22 to C-21 (δC 23.0) and C-22 (δC 29.4). Thus, mycalazal-18 (19) was deduced to be, a new pyrrole-2-carbaldehyde with a 1,4-diene in its lipophilic side chain.
Compound 20 was isolated as a colorless oil with a molecular formula of C22H33NO. This formula required one more degree of unsaturation, relative to the structure of 19. This indicated that 20 was pyrrole-2-carbaldehyde analog with one additional double bond in the hydrocarbon side chain. This structural assignment of 20 was further confirmed by a 1H NMR multiplet at δ 5.37 that integrated for six olefinic protons and six 13C NMR sp2 methine resonances at δ 132.0, 130.4, 128.5, 128.4, 128.0 and 127.3, which were attributable to three disubstituted double bonds. These data, together with two 1H NMR multiplets at δ 2.82 (4H) and 2.09 (4H) assignable to two bis-allylic methylenes and two allylic methylenes, respectively, were in agreement with a sequence of three methylene-interrupted double bonds (1,4,7-unsaturation pattern). Further, the combined analysis of the 1D and 2D NMR spectra indicated that the olefinic system in 20 was at the ω-3 position based on the 1H-1H COSY correlation between the terminal methyl resonance at H3-22 [δ0.98 (t, J = 7.6 Hz)] and the allylic methylene H2-21 (δ 2.09) resonance. This was further supported by long-range HMBC correlations from the H3-22 terminal methyl resonance to the C-20 (δ 132.0) olefinic carbon and the chemical shift of one of the ω-2 allylic methylene resonance at δ 20.8. Thus, the structure of this Mycale metabolite mycalazal-19 (20) was deduced to be a pyrrole-2-carbaldehyde with a 1,4,7-triene at the ω-3 position in the lipophilic side chain.
Compound 21 was isolated as a colorless oil. It exhibited IR, UV and NMR spectra nearly identical to those of 20, indicating that the two compounds differed only in side chain length. The molecular formula of C24H37NO indicated that 21 contained a side chain elongated by two methylene units between the pyrrole ring and the olefinic system, relative to the structure of 20. Therefore, mycalazal-20 (21) was deduced to be an analog of 20 with a longer side chain.
The structures of 22 – 29 were readily identified by comparison of the spectroscopic and mass spectrometric data with those reported in the literature. The structures were identified as mycalenitrile-1 (22),34 a pyrrole-2-carbaldehyde with a nitrile-substituted saturated lipophilic side chain (23),35 mycalenitrile-2 (24),34 5-pentadecylpyrrole-2-carbaldehyde (25),36 5-hexadecylpyrrole-2-carbaldehyde (26),36 a mycalazal-type lipophic pyrrole-2-carbaldehyde with a saturated side chain that contained a terminal isopropyl group (27),35 an analog of 27 with a longer side chain (28),35 and mycalazal-3 (29).34
Previous reports have indicated that certain M. cecilia mycalazals (structurally related to 15 – 17 and 29) and mycalenitriles (structurally related to 12 and 13) suppress tumor cell viability (IC50 values range from approximately 5 – 15 μM).34 However, the mechanisms responsible for these observed anti-proliferative effects are not known. Using the T47D cell-based HIF-1 activation reporter assay, 26 of the structurally related mycalazal/mycalenitrile-like metabolites (4 – 29) were titrated to determine their effects on HIF-1 activation.23–25 Based on their HIF inhibitory activities, these compounds are divided into four groups. The most active compounds (6 and 7) inhibited hypoxia-induced HIF-1 activation with IC50 values of 7.8 μM (95% confidence intervals: 6.8 to 8.8 μM) and 8.6 μM (95% confidence intervals: 7.6 to 9.9 μM), respectively (Figure 1A). The moderately active group (5, 8, 13, and 22) exhibited IC50 values between 10 and 20 μM (Figure 1B). The IC50 values for the weakly active group (15, 17 –19, 21, 27, and 29) ranged from 20 to 30 μM. The remaining thirteen compounds (4, 9 – 12, 14, 16, 20, 23 – 26, and 28) did not significantly suppress hypoxia (1% O2)-induced HIF-1 activation (< 50% inhibition at 30 μM, the highest concentration tested). In comparison to their inhibitory effects on hypoxia-induced HIF-1 activation, compounds from both the most active and the moderately active groups had less effect on chemical hypoxia-induced HIF-1 activation (1, 10-phenanthroline at 10 μM, Figure 1C). One of the well-studied HIF-1 target genes is vascular endothelial growth factor (VEGF), an angiogenic factor that is critical for tumor angiogenesis.39 Agents that inhibit VEGF are in clinical use for the treatment of cancer. The effects of 6 and 7 on hypoxia-stimulated production of secreted VEGF proteins were examined in T47D cells (Figure 1D). Compound 6 inhibited the hypoxia-induced increase in the level of secreted VEGF proteins by 50% at the concentration of 30 μM, while 7 did not significantly inhibit secreted VEGF levels at the concentrations tested (10 and 30 μM). As observed in previous studies, significantly higher concentrations of HIF-1 inhibitors are required to inhibit the induction of VEGF expression, relative to the concentrations that inhibit HIF-1 activation in the cell-based reporter assay. The most potent HIF-1 inhibitory Mycale lipophilic pyrroles appeared all possess the terminal nitrile functionality. These observations are in contrast to the SARs previously reported of LNCaP tumor cell cytotoxicity, where the nitrile-substituted mycalenitriles exhibited little or no significant activity and the ω-7 mono-unsaturated (e.g. mycalazal-8) and ω-3 tri-unsaturated mycalazals [e.g. mycalazal-3 (29)] were among the most cytotoxic.34 As an inhibitor of hypoxia-induced HIF-1 activation, 29 showed only very weak activity. One thing to note is that Zubía and coworkes only described significant SARs related to LNCaP tumor cell cytotoxicity.34 These previous studies also reported that the nitrile-substituted mycalenitriles displayed enhanced cytotoxic effects toward LoVo and HeLa cells. In our studies, few other structure-activity relationships were apparent. However, the SAR data were complicated by the highly lipophilic nature of these metabolites. It also appears that while a highly lipophilic alkyl side chain is essential for the HIF-1 inhibitory activity, extremely hydrophobic non-substituted long chain analogs were nearly inactive. This may have resulted from the limited solubility of the most lipophilic compounds in the aqueous-based media used for the cell-based bioassays.
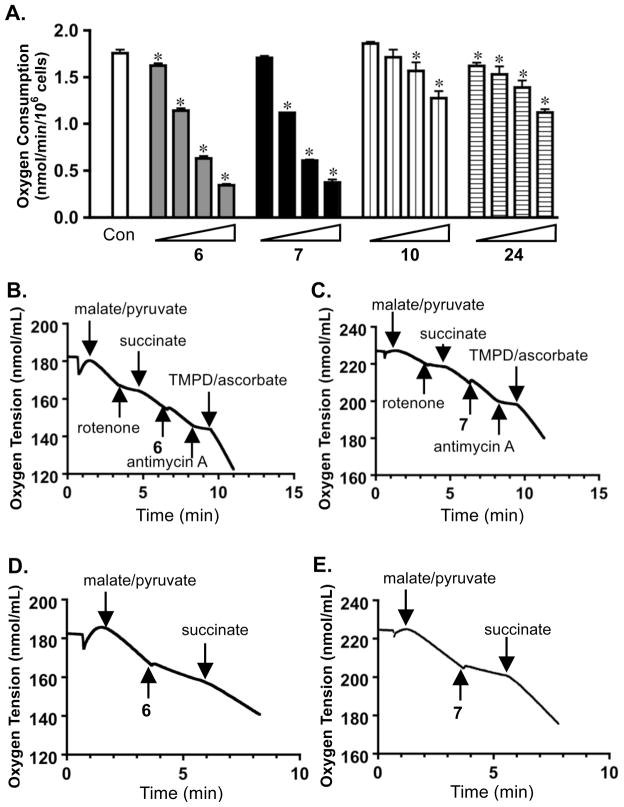
Figure 1. Inhibition of HIF-1 activation.
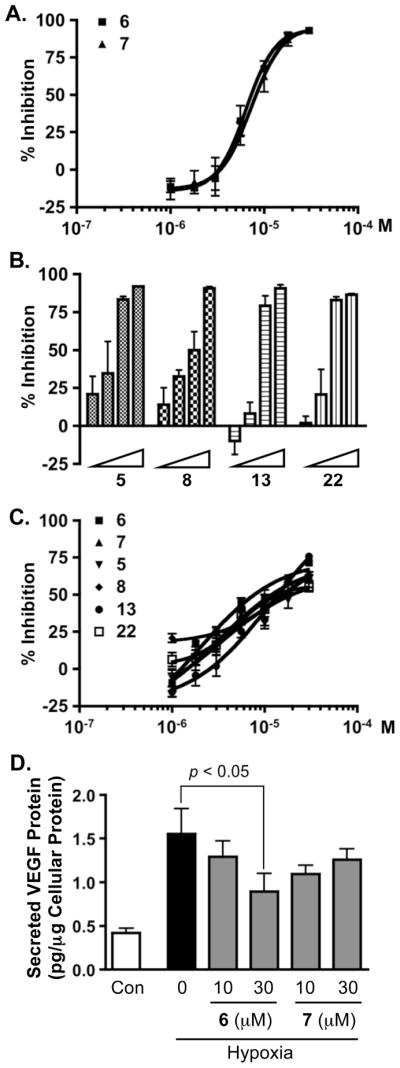
(A) Concentration-dependent inhibitory effects exerted by 6 and 7 on hypoxia (1% O2)-induced HIF-1 activation in a T47D cell-based reporter assay (mean ± SD, one experiment in triplicate). (B) Compounds 5, 8, 13 and 22 were evaluated at 5.6, 10, 17.8, and 30 μM in the reporter assay described in (A). (C) Concentration-response results of 5 – 8, 13, and 22 on 1,10-phenanthroline (10 μM)-induced HIF-1 activation in a T47D cell-based reporter assay (mean ± SD, one experiment in triplicate). (D) Levels of secreted VEGF protein in the media conditioned by T47D cells exposed to hypoxia in the presence of 6 or 7 (1% O2, 16 h), were determined by ELISA and normalized to the amounts of cellular proteins (mean ± SD, one experiment in triplicate). The p value was provided when there was a statistically significant difference between the hypoxia-induced and the compound treated samples.
Compounds that disrupt the functioning of the mitochondrial electron transport chain (ETC) represent an important group of small molecule HIF-1 inhibitors that are highly selective towards hypoxia-induced HIF-1 activation.24,25,40,41 The observation that active compounds such as 6 and 7 demonstrated greater inhibitory activity towards hypoxia induced HIF-1 activation, relative to their effects on chemical hypoxia/iron chelator induced HIF-1 activation (Figure 1), and their highly lipophilic structures, prompted us to test the hypothesis that these compounds may function as inhibitors of mitochondrial respiration. Concentration-response studies were performed to investigate the effects of 6 and 7 on T47D cell respiration (Figure 2A). Two structurally related, yet inactive, compounds 10 and 24 were included as negative controls. Compounds 6 and 7 inhibited T47D cell respiration in a concentration-dependent manner, and the concentrations that inhibited respiration correlated with those that inhibited hypoxia-induced HIF-1 activation in T47D cells (IC50 < 10 μM). As anticipated, compounds 10 and 24 did not exert significant inhibition of mitochondrial respiration (IC50 > 30 μM). Further mechanistic studies were conducted to discern the component(s) of the ETC that were affected by active compounds 6 and 7. Substrates and/or inhibitors for each ETC complex were added in a sequential manner to permeabilized T47D cells. Addition of a mixture of malate and pyruvate initiated mitochondrial respiration at complex I (NADH-ubiquinone oxidoreductase). The complex I inhibitor rotenone suppressed this respiration and the complex II substrate succinate overcame this inhibition (Figure 2B and 2C).24,25 The observation that neither 6 nor 7 affected mitochondrial respiration in the presence of succinate indicated that 6 and 7 did not inhibit complex II, III, or IV (Figure 2B and 2C). Respiration in the presence of succinate was inhibited by the complex III inhibitor antimycin A and respiration was re-initiated by addition of the complex IV substrates TMPD/ascorbate (Figure 2B and 2C). As observed in the case of mitochondrial complex I inhibitors (e.g. rotenone), the complex II substrate succinate reestablished mitochondrial respiration following inhibition exerted by 6 and 7 (Figure 2D and 2E). Thus, the data presented in Figure 2 indicate that compounds 6 and 7 appear to selectively inhibit mitochondrial respiration at complex I.
Figure 2. Compounds 6 and 7 inhibit mitochondrial respiration at complex I.
(A) Concentration-response results of compounds 6, 7, 10, and 24 on oxygen consumption by T47D cells (mean ± SD, three independent experiments). The compounds were tested at 1, 3, 10, and 30 μM. An asterisk (*) indicates statistically significant difference (p < 0.05) between the control and the compound treated sample, compared by one-way ANOVA and Bonfferoni post hoc analyses (GraphPad Prism 4.0). (B) Compound 6 (10 μM) did not affect mitochondrial electron transport chain (ETC) complex II, III, or IV. Substrates and inhibitors were added to T47D cells (5 × 106, 30 °C) in a sequential manner at the specified time point. (C) Compound 7 (10 μM) did not affect ETC complex II, III, or IV. (D) Compound 6 (10 μM) suppressed mitochondria respiration by inhibiting complex I. (E) Compound 7 (10 μM) inhibited complex I.
Under hypoxic conditions, increased production of reactive oxygen species (ROS) by the Qo site of mitochondrial complex III may act as “signal molecules” of cellular hypoxia. The ROS appear to oxidize the catalytic iron in the Fe(II)-dependent HIF-prolyl hydroxylases that are required to “tag” HIF-1α protein for degradation by the proteasome.42,43 Therefore, compounds that inhibit electron transport upstream from complex III, but do not themselves increase ROS production to the cytoplasmic site of the mitochondrial inner membrane at the site of inhibition, are able to prevent hypoxic mitochondria from releasing essential ROS signaling molecules needed to stabilize HIF-1α protein and activate HIF-mediated gene transcription.
Experimental Section
General Experimental Procedures
Melting points were measured with a Thomas Hoover capillary melting point apparatus and were uncorrected. A Bruker Tensor 27 genesis Series FTIR was used to obtain the IR spectrum, and a Varian 50 Bio spectrophotometer was used to record the UV spectra. The NMR spectra were recorded in CDCl3 on AMX-NMR spectrometers (Bruker) operating at 400 MHz for 1H and 100 MHz for 13C, respectively. Residual solvent peaks (δ 7.27 for 1H) and (δ 77.23 for 13C) were used as internal references for the NMR spectra recorded running gradients. The HRESIMS spectra were determined on a Bruker Daltonic micro TOF fitted with an Agilent 1100 series HPLC and an electrospray ionization source. The GC-EIMS study was performed on a fused-silica DB5 capillary column (i.d. = 0.80 mm; film thickness = 0.15 μm; length = 32 m) and recorded on a Hewlett-Packard HP6980 Series gas chromatograph interfaced with a HP5973 mass selective detector at 70 eV. HPLC was performed on a Waters system, equipped with 600 controller and 996 photodiode array detector. Three semi-preparative HPLC columns [(1) Phenomenex Luna RP-18, 5 μm, 250 × 10.00 mm; (2) Phenomenex Luna Si gel, 5μ, 100 Å, 250 × 10.00 mm; and (3) Phenomenex Luna Phenyl-Hexyl, 5μ, 100 Å, 250 × 10.00 mm] were employed for isolation. The TLCs were performed using Merck Si60F254 or Si60RP18F254 plates, sprayed with a 10% H2SO4 solution in H2O, heated, and visualized under UV at 254 nm.
Sponge Material
The extract was provided by the National Cancer Institute’s Open Repository Program. Mycale (Carmia) sp. was collected at −12 m depth on July 5, 1993 (collection C011823) from Turtle Bas, Palau. The sample was identified by Dr. Michele Kelly (National Institute of Water and Atmospheric Research Limited, Auckland, New Zealand). The sponge material was frozen at −20 °C and ground in a meat grinder. A voucher specimen was placed on file with the Department of Invertebrate Zoology at the National Museum of Natural History located in Washington, D.C.
Extraction and Isolation
Ground sponge material was extracted with H2O. The residual sample was lyophilized and extracted with CH2Cl2:MeOH (1:1), residual solvents were removed under vacuum, and the extract stored at −20 °C in the NCI repository at the Frederick Cancer Research and Development Center (Frederick, Maryland). The crude extract (5.0 g) inhibited hypoxia (1% O2)-induced HIF-1 activation by 53% at 5 μg mL−1 in a T47D cell-based reporter assay, and was separated into four fractions by Sephadex LH-20 column chromatography [CHCl3 in MeOH (1:1)] based on TLC analysis. The active fraction (4.4 g) inhibited hypoxia-induced HIF-1 activation by 50% at 5 μg mL−1 and was further separated into five subfractions using a Sephadex LH-20 column eluted with petroleum ether:CHCl3:MeOH at the ratio of 2:1:1. The active subfraction (3.7 g) inhibited hypoxia-induced HIF-1 activation by 48% at 5 μg mL−1 and was subjected to C18 VLC column chromatography (eluted with step gradients of 80% to 100% MeOH in H2O) that gave rise to seven fractions. The second fraction (98.1 mg, eluted with 80% MeOH in H2O) inhibited hypoxia-induced HIF-1 activation by 90% at 1 μg mL−1 and was further separated by reversed-phase HPLC (Luna 5μ, ODS-3 100 Å, 250 × 10.00 mm, isocratic 80% CH3CN in H2O, 4.0 mL min−1) to produce 6 (0.9 mg, 0.02% yield, tR 29 min), 13 (3.0 mg, 0.06% yield, tR 18 min), and 22 (4.1 mg, 0.08% yield, tR 31 min). The third fraction (1.4 g, eluted with 90% MeOH in H2O) inhibited hypoxia-induced HIF-1 activation by 56% at 5 μg mL−1 and was further separated by semi-preparative RP-HPLC eluting with isocratic 90% CH3CN in H2O that afforded eleven subfractions. Further purification was achieved through repeated semi-preparative HPLC with (1) Luna 5μ, ODS-3 100 Å, 250 × 10.00 mm, isocratic solvents of various proportions of CH3CN in H2O, 4.0 mL min−1; (2) Luna 5μ, Si gel 100 Å, 250 × 10.00 mm, isocratic solvents of various proportions of isopropanol in hexanes, 4.0 mL min−1; and (3) Luna 5μ, Phenyl-Hexyl 100 Å, 250 × 10.00 mm, isocratic solvents of various proportions of CH3CN in H2O, 4.0 mL min−1 to yield twenty-three pure compounds: 8 (7.2 mg, 0.14% yield) and 20 (1.0 mg, 0.02% yield) from subfraction #2 (13.1 mg); 7 (2.3 mg, 0.05% yield) and 14 (3.2 mg, 0.06% yield) from subfraction #3 (14.2 mg); 19 (2.8 mg, 0.06% yield) and 23 (13.5 mg, 0.27% yield) from subfraction #5 (23.5 mg); 11 (56.9 mg, 1.14% yield), 15 (2.8 mg, 0.06% yield), 18 (1.0 mg, 0.02% yield) and 21 (3.5 mg, 0.07% yield) from subfraction #6 (85.1 mg); 5 (2.8 mg, 0.07% yield), 9 (2.0 mg, 0.04% yield) and 17 (1.7 mg, 0.03% yield) from subfraction #7 (12.5 mg); 4 (2.3 mg, 0.05% yield), 12 (1.2 mg, 0.02% yield), 24 (1.0 mg, 0.02% yield) and 27 (5.5 mg, 0.11% yield) from subfraction #8 (12.6 mg); 16 (1.9 mg, 0.04% yield) and 25 (9.8 mg, 0.20% yield) from subfraction #9 (18.0 mg); 10 (49.7 mg, 1.00% yield), 26 (3.4 mg, 0.07% yield), 28 (1.3 mg, 0.03% yield) and 29 (3.8 mg, 0.08% yield) from subfraction #11 (69.0 mg).
Mycalenitrile-4 (4): amorphous powder; UV (MeOH) λmax (log ε) 201 (4.28), 250 (3.78), 301 (4.21) nm; IR (film): νmax 3260, 2912, 2848, 2246, 1635, 1496, 1352, 1185, 1040, 772, 719 cm−1; 1H and 13C NMR, see Tables 1 and 4, respectively; HRESIMS m/z 461.3514 [M+Na]+ (calcd for C29H46N2ONa, 461.3508).
Mycalenitrile-5 (5): viscous liquid; UV (MeOH) λmax (log ε) 202 (4.13), 250 (3.60), 301 (4.22) nm; IR (film): νmax 3253, 2923, 2852, 2250, 1641, 1496, 1186, 1041, 772, 720 cm−1; 1H and 13C NMR, see Tables 1 and 4, respectively; GC-EIMS (70 eV) m/z 438 [M]+ (86.5), 409 (36.3), 370 (58.0), 356 (8.3), 342 (9.8), 316 (15.1), 290 (10.3), 276 (3.0), 237 (25.5), 176 (13,9), 148 (24.4), 136 (6.8), 122 (100), 108 (54.1), 96 (15.2), 82 (3.4), 80 (63.5), 41 (10.1); HRESIMS m/z 461.3506 [M+Na]+ (calcd for C29H46N2ONa, 461.3508).
Mycalenitrile-6 (6): colorless oil; UV (MeOH) λmax (log ε) 203 (4.10), 235 (3.90), 301 (4.20) nm; IR (film): νmax 3254, 2917, 2849, 2252, 1641, 1465, 1186, 1041, 719 cm−1; 1H and 13C NMR, see Tables 2 and 5, respectively; GC-EIMS (70 eV) m/z 382 [M]+ (41.3), 353 (9.1), 300 (10.0), 260 (18.4), 234 (6.5), 202 (10.8), 162 (14.8), 148 (8.8), 136 (8.7), 122 (65.5), 108 (53.7), 96 (100), 82 (11.1), 80 (66.0), 41 (73.4); HRESIMS m/z 405.2865 [M+Na]+ (calcd for C25H38N2ONa, 405.2882).
Table 2.
1H NMR Dataa δ (J, Hz) for 6, 8, 9, and 19–21
| H | 6 | 8 | 9 | 19 | 20 | 21 |
|---|---|---|---|---|---|---|
| 1 | 9.38, s | 9.37, s | 9.38, s | 9.38, s | 9.38, s | 9.37, s |
| 3 | 6.89, dd (2.4, 1.6) | 6.90, dd (3.7, 2.6) | 6.89, dd (3.2, 2.8) | 6.90, dd (3.6, 24) | 6.89, dd (3.6, 2.4) | 6.90, dd (3.2, 2.8) |
| 4 | 6.09, br s | 6.08, dd (3.7, 2.6) | 6.08, dd (3.2, 2.8) | 6.08, dd (3.6, 2.4) | 6.08, dd (3.2, 2.4) | 6.08, dd (3.2, 2.8) |
| 6 | 2.67, t (7.6) | 2.67, t (7.6) | 2.66, t (7.6) | 2.67, t (7.6) | 2.65, t (7.6) | 2.65, t (7.6) |
| 7 | 1.70, m | 1.66, m | 1.66, m | 1.66, m | 1.65, m | 1.65, br s |
| 8 | 1.26, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s |
| 9 | 1.26, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s |
| 10 | 1.26, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s |
| 11 | 1.26, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s | 1.27, br s |
| 12 | 1.26, br s | 1.27, br s | 1.27, br s | 1.27, br s | 2.09, m | 1.27, br s |
| 13 | 1.26, br s | 1.27, br s | 1.27, br s | 1.27, br s | 5.37, m | 1.27, br s |
| 14 | 1.26, br s | 1.27, br s | 1.27, br s | 2.06, m | 5.37, m | 2.08, m |
| 15 | 2.05, dt (6.8, 6.4) | 1.27, m | 1.27, br s | 5.37, m | 2.82, m | 5.37, m |
| 16 | 5.37, m | 2.02, m | 1.27, br s | 5.37, m | 5.37, m | 5.37, m |
| 17 | 5.37, m | 5.36, m | 1.38, m | 2.78, t (6.0) | 5.37, m | 2.82, m |
| 18 | 2.78, dd (6.0, 5.6) | 5.36, m | 2.03, m | 5.37, m | 2.82, m | 5.37, m |
| 19 | 5.37, m | 2.02, m | 5.35, m | 5.37, m | 5.37, m | 5.37, m |
| 20 | 5.37, m | 1.27, m | 5.35, m | 2.06, m | 5.37, m | 2.82, m |
| 21 | 2.11, dt (6.8, 6.0) | 1.27, br s | 2.03, m | 1.43 dq (7.2, 6.0) | 2.09, m | 5.37, m |
| 22 | 1.44, m | 1.44, m | 1.38, m | 0.92, t (7.2) | 0.98, t (7.6) | 5.37, m |
| 23 | 1.65, m | 1.66, m | 1.27, br s | 2.08, m | ||
| 24 | 2.34, m | 2.34, t (6.8) | 1.45, m | 0.99, t (7.6) | ||
| 25 | 1.66, m | |||||
| 26 | 2.34, t (7.2) | |||||
| NH | 9.22, br s | 9.71, br s | 9.24, br s | 9.47, br s | 9.16, br s | 9.29, br s |
Bruker AMX 400 MHz spectrometer; Chemical shifts (ppm) referenced to CDCl3 (δH 7.27); Numerical order groups compounds of similar chain length.
Table 5.
13C NMR Dataa (δ, DEPT) for 6, 8, 9, and 19–21
| C | 6 | 8 | 9 | 19 | 20 | 21 |
|---|---|---|---|---|---|---|
| 1 | 178.3, CH | 178.3, CH | 178.3, CH | 178.3, CH | 178.3, CH | 178.4, CH |
| 2 | 132.1, C | 132.1, C | 132.1, C | 132.1, C | 132.2, C | 132.2, C |
| 3 | 122.4, CH | 122.8, CH | 122.6, CH | 122.7, CH | 122.8, CH | 122.8, CH |
| 4 | 109.7, CH | 109.6, CH | 109.6, CH | 109.6, CH | 109.6, CH | 109.6, CH |
| 5 | 142.6, C | 143.3, C | 142.9, C | 143.2, C | 142.8, C | 143.2, C |
| 6 | 28.0, CH2 | 28.1, CH2 | 28.1, CH2 | 28.1, CH2 | 28.1, CH2 | 28.1, CH2 |
| 7 | 28.7, CH2 | 29.0, CH2 | 29.0, CH2 | 29.2, CH2 | 29.1, CH2 | 29.1, CH2 |
| 8 | 29.9-29.0b | 29.9-29.5b | 30.0-29.4b | 29.9-29.5b | 29.8-29.3b | 29.8-29.4 |
| 9 | 29.9-29.0b | 29.9-29.5b | 30.0-29.4b | 29.9-29.5b | 29.8-29.3b | 29.8-29.4 |
| 10 | 29.9-29.0b | 29.9-29.5b | 30.0-29.4b | 29.9-29.5b | 29.8-29.3b | 29.8-29.4 |
| 11 | 29.9-29.0b | 29.9-29.5b | 30.0-29.4b | 29.9-29.5b | 29.8-29.3b | 29.8-29.4 |
| 12 | 29.9-29.0b | 29.9-29.5b | 30.0-29.4b | 29.9-29.5b | 27.4, CH2 | 29.8-29.4 |
| 13 | 29.9-29.0b | 29.9-29.5b | 30.0-29.4b | 29.9-29.5b | 128.0, CH | 29.8-29.4 |
| 14 | 29.9-29.0b | 29.9-29.5b | 30.0-29.4b | 27.4, CH2 | 130.4, CH | 27.4, CH2 |
| 15 | 27.5, CH2 | 29.1, CH2 | 30.0-29.4b | 128.4, CHb | 25.8, CH2c | 127.9, CH |
| 16 | 128.0, CH | 27.4, CH2c | 30.0-29.4b | 130.3, CH | 128.5, CHd | 130.6, CH |
| 17 | 130.5, CH | 130.4, CHd | 29.2, CH2e | 25.9, CH2 | 128.4, CHd | 25.8, CH2c |
| 18 | 25.9, CH2 | 129.6, CHd | 27.4, CH2c | 128.2, CHb | 25.7, CH2c | 128.5, CH |
| 19 | 128.9, CH | 27.2, CH2c | 130.5, CHd | 130.1, CH | 127.3, CH | 128.5, CH |
| 20 | 129.5, CH | 29.1, CH2 | 129.8, CHd | 29.4, CH2 | 132.0, CH | 25.7, CH2c |
| 21 | 27.0, CH2 | 29.9-29.5b | 27.2, CH2c | 23.0, CH2 | 20.8, CH2 | 127.3, CH |
| 22 | 28.9, CH2 | 28.9, CH2 | 29.1, CH2e | 14.0, CH3 | 14.5, CH3 | 132.0, CH |
| 23 | 25.6, CH2 | 25.6, CH2 | 30.0-29.4b | 20.8, CH2 | ||
| 24 | 17.4, CH2 | 17.3, CH2 | 28.9, CH2 | 14.5, CH3 | ||
| 25 | 120.1, C | 120.0, C | 25.6, CH2 | |||
| 26 | 17.3, CH2 | |||||
| 27 | 120.0, C |
Bruker AMX 100 MHz spectrometer; δ values are reported in ppm referenced to CDCl3 (δC 77.23) as internal standard; Numerical order groups compounds of similar chain length.
All carbons were CH2;
Values may be interchanged.
Mycalenitrile-7 (7): colorless oil; UV (MeOH) λmax (log ε) 201 (4.02), 242 (3.87), 301 (4.23) nm; IR (film): νmax 3256, 2924, 2853, 2248, 1640, 1496, 1186, 1041, 772, 721 cm−1; 1H and 13C NMR, see Tables 1 and 4, respectively; GC-EIMS (70 eV) m/z 410 [M]+ (64.6), 381 (25.7), 355 (23.9), 328 (19.9), 262 (4.8), 194 (19.1), 148 (32.5), 136 (2.7), 122 (100), 108 (85.0), 96 (36.1), 82 (24.1), 80 (91.2), 41 (76.9); HRESIMS m/z 433.3207 [M+Na]+ (calcd for C27H42N2ONa, 433.3195).
Mycalenitrile-8 (8): yellow solid, mp 42–44 °C; UV (MeOH) λmax (log ε) 201 (3.78), 250 (3.51), 301 (4.18) nm; IR (film): νmax 3180, 2920, 2850, 2246, 1642, 1495, 1044, 778 cm−1; 1H and 13C NMR, see Tables 2 and 5, respectively; GC-EIMS (70 eV) m/z 384 [M]+ (48.5), 355 (49.0), 302 (28.3), 288 (100), 234 (18.5), 150 (39.5), 122 (82.0), 108 (65.8), 80 (75.0), 41 (23.2); HRESIMS m/z 407.3041 [M+Na]+ (calcd for C25H40N2ONa, 407.3038).
Mycalenitrile-9 (9): colorless oil; UV (MeOH) λmax (log ε) 201 (3.78), 250 (3.53), 301 (4.17) nm; IR (film): νmax 3247, 2921, 2851, 2245, 1637, 1499, 1043, 773 cm−1; 1H and 13C NMR, see Tables 2 and 5, respectively; GC-EIMS (70 eV) m/z 412 [M]+ (85.7), 383 (27.9), 344 (2.8), 330 (12.5), 316 (9.6), 302 (2.3), 262 (18.8), 248 (5.8), 150 (18.6), 122 (100), 109 (43.0), 108 (66.6), 96 (17.2), 94 (15.9), 80 (18.6), 55 (18.5); HRESIMS m/z 435.3362 [M+Na]+ (calcd for C27H44N2ONa, 435.3371).
Mycalenitrile-10 (10): white solid, mp 42–45 °C; UV (MeOH) λmax (log ε) 202 (3.76), 250 (3.55), 301 (4.20) nm; IR (film): νmax 3217, 2920, 2851, 2246, 1642, 1495, 1043, 778 cm−1; 1H and 13C NMR, see Tables 1 and 4, respectively; GC-EIMS (70 eV) m/z 440 [M]+ (47.6), 411 (24.8), 355 (23.9), 344 (28.8), 290 (55.3), 207 (100), 192 (18.6), 150 (24.9), 122 (34.9), 109 (15.2), 108 (29.8), 96 (18.6), 94 (10.2), 80 (50.8), 55 (20.7); HRESIMS m/z 463.3672 [M+Na]+ (calcd for C29H48N2ONa, 463.3664).
Mycalenitrile-11 (11): colorless oil; UV (MeOH) λmax (log ε) 202 (3.76), 250 (3.52), 302 (4.23) nm; IR (film): νmax 3244, 2920, 2851, 2244, 1637, 1500, 1043, 774 cm−1; 1H and 13C NMR, see Tables 1 and 4, respectively; GC-EIMS (70 eV) m/z 412 [M]+ (74.4), 383 (28.8), 344 (4.9), 330 (12.8), 316 (13.0), 302 (3.3), 288 (3.0), 274 (2.9), 260 (3.7), 246 (6.5), 232 (5.5), 218 (6.5), 204 (6.5), 190 (4.9), 178 (11.6), 164 (11.3), 150 (19.5), 122 (100), 108 (58.7), 80 (48.5), 55 (16.0), 41 (11.0); HRESIMS m/z 435.3348 [M+Na]+ (calcd for C27H44N2ONa, 435.3351).
Mycalenitrile-12 (12): colorless oil; UV (MeOH) λmax (log ε) 201 (3.74), 250 (3.53), 301 (4.16) nm; IR (film): νmax 3251, 2918, 2850, 2245, 1636, 1494, 1041, 771 cm−1; 1H and 13C NMR, see Tables 1 and 4, respectively; GC-EIMS (70 eV) m/z 426 [M]+ (100), 397 (34.9), 358 (3.4), 344 (16.3), 330 (16.5), 316 (3.3), 302 (3.2), 288 (2.9), 274 (3.4), 260 (4.0), 246 (5.7), 232 (6.6), 218 (6.6), 204 (7.5), 190 (5.3), 178 (13.0), 164 (12.2), 150 (14.9), 122 (93.1), 108 (51.3), 80 (39.3), 55 (13.4), 41 (7.6); HRESIMS m/z 449.3519 [M+Na]+ (calcd for C28H46N2ONa, 449.3508).
Mycalenitrile-13 (13): white solid, mp 73–75 °C; UV (MeOH) λmax (log ε) 201 (3.78), 247 (3.43), 301 (4.20) nm; IR (film): νmax 3172, 2920, 2849, 2247, 1642, 1495, 1043, 780 cm−1; 1H and 13C NMR, see Tables 3 and 6, respectively; HRESIMS m/z 331.2750 [M]+ (calcd for C21H34N2O, 331.2749).
Table 3.
1H NMR Dataa δ (J, Hz) for 13–18
| H | 13 | 14 | 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|
| 1 | 9.37, s | 9.38, s | 9.37, s | 9.35, s | 9.38, s | 9.38, s |
| 3 | 6.90, dd (3.8, 2.8) | 6.89, br s | 6.90, dd (3.2, 2.8) | 6.90, dd (3.6, 2.8) | 6.88, dd (3.6, 2.8) | 6.89, dd (3.2, 2.8) |
| 4 | 6.08, dd (3.8, 2.8) | 6.08, br s | 6.08, dd (3.2, 2.8) | 6.09, dd (3.6, 2.8) | 6.08, dd (3.6, 2.8) | 6.08, dd (3.2, 2.8) |
| 6 | 2.66, t (7.6) | 2.66, t (7.6) | 2.66, t (7.6) | 2.66, t (7.6) | 2.71, t (7.6) | 2.66, t (7.6) |
| 7 | 1.66, m | 1.66, m | 1.66, m | 1.67, m | 2.41, t (7.2) | |
| 8 | 1.26, br s | 1.27, br s | 1.27, br s | 1.29, br s | 5.39, m | 1.27, br s |
| 9 | 1.26, br s | 1.27, br s | 1.27, br s | 1.29, br s | 5.46, m | 1.27, br s |
| 10 | 1.26, br s | 1.27, br s | 1.27, br s | 1.29, br s | 2.01, m | 1.27, br s |
| 11 | 1.26, br s | 1.27, br s | 1.27, br s | 1.29, br s | 1.26, br s | 2.03, m |
| 12 | 1.26, br s | 1.27, br s | 1.27, br s | 1.29, br s | 1.26, br s | 5.37, m |
| 13 | 1.26, br s | 1.27, br s | 1.27, br s | 1.29, br s | 1.26, br s | 5.37, m |
| 14 | 1.26, br s | 1.27, br s | 1.27, br s | 2.03, m | 1.26, br s | 2.03, m |
| 15 | 1.26, br s | 1.27, br s | 1.27, br s | 5.37, m | 1.26, br s | 1.27, br s |
| 16 | 1.26, br s | 1.27, br s | 1.27, br s | 5.37, m | 1.26, br s | 1.27, br s |
| 17 | 1.26, br s | 1.27, br s | 1.27, br s | 2.03, m | 1.26, br s | 1.27, br s |
| 18 | 1.45, m | 1.27, br s | 1.27, br s | 1.29, br s | 1.26, br s | 1.27, br s |
| 19 | 1.66, m | 1.27, br s | 0.89, t (6.4) | 1.29, br s | 1.67, m | 1.67, m |
| 20 | 2.34, t (7.2) | 1.27, br s | 1.29, br s | 0.86, d (7.6) | 0.87, d (7.6) | |
| 21 | 1.45, m | 1.29, br s | 0.86, d (7.6) | 0.87, d (7.6) | ||
| 22 | 1.66, m | 0.89, t (7.2) | ||||
| 23 | 2.34, t (7.2) | |||||
| NH | 9.48, br s | 9.49, br s | 9.44, br s | 9.25, br s | 9.17, br s | 9.16, br s |
Bruker AMX 400 MHz spectrometer; Chemical shifts (ppm) referenced to CDCl3 (δH 7.27); Numerical order groups compounds of similar chain length.
Table 6.
13C NMR Dataa (δ, DEPT) for 13–18
| C | 13 | 14 | 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|
| 1 | 178.3, CH | 178.3, CH | 178.3, CH | 178.3, CH | 178.4, CH | 178.4, CH |
| 2 | 132.1, C | 132.0, C | 132.0, C | 132.1, C | 132.2, C | 132.1, C |
| 3 | 122.7, CH | 122.8, CH | 122.8, CH | 122.6, CH | 122.8, CH | 122.8, CH |
| 4 | 109.7, CH | 109.6, CH | 109.6, CH | 109.6, CH | 109.7, CH | 109.7, CH |
| 5 | 143.3, C | 143.4, C | 143.3, C | 143.0, C | 142.8, C | 142.4, C |
| 6 | 28.1, CH2 | 28.1, CH2 | 28.1, CH2 | 28.1, CH2 | 28.1, CH2 | 28.0, CH2 |
| 7 | 29.2, CH2 | 29.2, CH2 | 29.1, CH2 | 29.1, CH2 | 26.9, CH2 | 28.7, CH2 |
| 8 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 30.0-29.2b | 127.7, CH | 29.8-29.2b |
| 9 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 30.0-29.2b | 132.4, CH | 29.8-29.2b |
| 10 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 30.0-29.2b | 27.5, CH2 | 29.8-29.2b |
| 11 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 30.0-29.2b | 30.2-29.4b | 27.5, CH2 |
| 12 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 30.0-29.2b | 30.2-29.4b | 130.8, CHc |
| 13 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 30.0-29.2b | 30.2-29.4b | 129.2, CHc |
| 14 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 27.4, CH2 | 30.2-29.4b | 27.5, CH2 |
| 15 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 130.2, CHb | 30.2-29.4b | 29.8-29.2 |
| 16 | 29.8-29.4b | 29.9-29.4b | 29.9-29.4b | 130.1, CHb | 30.2-29.4b | 29.8-29.2 |
| 17 | 29.8-29.4b | 29.9-29.4b | 32.1, CH2 | 27.4, CH2 | 30.2-29.4b | 29.8-29.2 |
| 18 | 28.9, CH2 | 29.9-29.4b | 22.9, CH2 | 30.0-29.2b | 39.2, CH2 | 39.2, CH2 |
| 19 | 25.6, CH2 | 29.9-29.4b | 14.3, CH3 | 30.0-29.2b | 27.7, CH2 | 27.7, CH2 |
| 20 | 17.3, CH2 | 29.9-29.4b | 32.0, CH2 | 22.9, CH3 | 22.9, CH3 | |
| 21 | 120.0, C | 28.9, CH2 | 22.9, CH2 | 22.9, CH3 | 22.9, CH3 | |
| 22 | 25.6, CH2 | 14.3, CH3 | ||||
| 23 | 17.3, CH2 | |||||
| 24 | 120.1, C |
Bruker AMX 100 MHz spectrometer; δ values are reported in ppm referenced to CDCl3 (δC 77.23) as internal standard; Numerical order groups compounds of similar chain length;
Carbons were CH2;
Values may be interchanged.
Mycalenitrile-14 (14): white solid, mp 74–77 °C; UV (MeOH) λmax (log ε) 201 (3.77), 249 (3.47), 301 (4.03) nm; IR (film): νmax 3178, 2921, 2848, 2245, 1645, 1494, 1043, 781, 725 cm−1; 1H and 13C NMR, see Tables 3 and 6, respectively; HRESIMS m/z 395.3012 [M+Na]+ (calcd for C24H40N2ONa, 395.3038).
Mycalazal-14 (15): amorphous powder; UV (MeOH) λmax (log ε) 201 (3.72), 250 (3.46), 301 (4.18) nm; IR (film): νmax 3174, 2921, 2849, 1645, 1494, 1043, 778, 724 cm−1; 1H and 13C NMR, see Tables 3 and 6, respectively; HRESIMS m/z 314.2450 [M+Na]+ (calcd for C19H33NONa, 314.2460).
Mycalazal-15 (16): colorless oil; UV (MeOH) λmax (log ε) 201 (3.86), 250 (3.75), 301 (4.08) nm; IR (film): νmax 3257, 2928, 2856, 1643, 1498, 1187, 1042, 772 cm−1; 1H and 13C NMR, see Tables 3 and 6, respectively; GC-EIMS (70 eV) m/z 331 [M]+ (100), 302 (12.2), 260 (7.2), 206 (3.6), 192 (4.2), 178 (8.8), 164 (9.4), 150 (23.1), 122 (59.8), 108 (78.2), 80 (70.6), 55 (21.5); HRESIMS m/z 354.2748 [M+Na]+ (calcd for C22H37NONa, 354.2773).
Mycalazal-16 (17): colorless oil; UV (MeOH) λmax (log ε) 201 (3.92), 249 (3.76), 301 (4.22) nm; IR (film): νmax 3253, 2920, 2851, 1637, 1494, 1185, 1041, 771 cm−1; 1H and 13C NMR, see Tables 3 and 6, respectively; HRESIMS m/z 340.2641 [M+Na]+ (calcd for C21H35NONa 340.2616).
Mycalazal-17 (18): colorless oil; UV (MeOH) λmax (log ε) 202 (3.87), 249 (3.72), 301 (4.18) nm; IR (film): νmax 3256, 2924, 2853, 1643, 1497, 1187, 1042, 772 cm−1; 1H and 13C NMR, see Tables 3 and 6, respectively; GC-EIMS (70 eV) m/z 317 [M]+ (40.0), 288 (18.5), 274 (5.4), 250 (2.7), 246 (2.3), 232 (3.3), 221 (51.0), 218 (7.5), 204 (12.2), 190 (11.0), 178 (2.3), 164 (9.0), 150 (22.8), 122 (100), 108 (19.8), 96 (25.1), 80 (69.2), 55 (28.2); HRESIMS m/z 340.2625 [M+Na]+ (calcd for C21H35NONa, 340.2616).
Mycalazal-18 (19): colorless oil; UV (MeOH) λmax (log ε) 201 (4.17), 241 (3.60), 301 (4.19) nm; IR (film): νmax 3255, 2923, 2853, 1641, 1496, 1186, 1042, 772, 719 cm−1; 1H and 13C NMR, see Tables 2 and 5, respectively; HRESIMS m/z 352.2621 [M+Na]+ (calcd for C22H35NONa, 352.2611).
Mycalazal-19 (20): colorless oil; UV (MeOH) λmax (log ε) 201 (4.24), 237 (3.80), 301 (4.20) nm; IR (film): νmax 3248, 2925, 2854, 1637, 1497, 1186, 1041, 772 cm−1; 1H and 13C NMR, see Tables 2 and 5, respectively; HRESIMS m/z 350.2456 [M+Na]+ (calcd for C22H33NONa, 350.2454).
Mycalazal-20 (21): colorless oil; UV (MeOH) λmax (log ε) 201 (4.29), 241 (3.82), 301 (4.17) nm; IR (film): νmax 3254, 2924, 2853, 1641, 1497, 1187, 1042, 772, 720 cm−1; 1H and 13C NMR, see Tables 2 and 5, respectively; HRESIMS m/z 378.2780 [M+Na]+ (calcd for C24H37NONa, 378.2767).
Cell-Based Reporter Assay
The maintenance of human breast tumor T47D (ATCC) cells, and the T47D cell-based reporter assay for HIF-1 activity were described previously.23–25 The extract and the fractions were dissolved in DMSO and the final solvent concentration is less than 0.5%. The pure compounds were dissolved in isopropanol at 4 mM and the stock solutions stored at −20 °C. Final solvent concentration is less than 0.75%. The following formula was used to calculate the % inhibition data:
ELISA Assay for VEGF Protein
The detailed procedure was described previously.24,25 The data were presented as the level of secreted VEGF proteins in the conditioned media normalized to the quantity of total proteins in the cell lysate determined by a micro BCA assay (Pierce).
Mitochondria Respiration Assay
As previously described,24,25 a method used to monitor the respiration of isolated mitochondria was modified to measure the level of oxygen consumption in intact or digitonin-permeabilized T47D cells and to investigate the specific target within the mitochondrial electron transport chain affected by active compounds. The known ETC inhibitors - rotenone for complex I and antimycin A for complex III were prepared as stock solutions in ethanol and added to final concentrations of 1 μM where indicated.
Cell Proliferation/Viability Assay
The maintenance of T47D and MDA-MB-231 cells (ATCC), cell plating, compound addition, exposure to specified oxygen conditions, and determination of cell proliferation/viability with the sulforhodamine B method were the same as those described.25 A BioTek Synergy plate reader was used to measure light absorbance at 490 nm with background absorbance at 690 nm. To calculate the percentage inhibition data, a formula similar to the one stated in the reporter assay was used. All compound-treated samples were compared to the media control under each specified oxygen condition.
Statistical Analysis
Data were compared using one-way ANOVA and Bonfferoni post hoc analyses (GraphPad Prism 4). Differences were considered significant when p < 0.05.
Supplementary Material
Acknowledgments
The authors thank Dr. S.L. McKnight at the University of Texas Southwestern Medical Center at Dallas for providing the pTK-HRE3-luc construct, the Natural Products Branch Repository Program at the National Cancer Institute for providing marine extract samples from the NCI Open Repository, Dr. D.J. Newman and E.C. Brown (NCI – Frederick, MD) for assistance with sample logistics and collection information. Mass spectrometry services were kindly provided by J. Gao and Drs. J. Peng and P.B. Carvalho (UM). Funding for this research was provided, in part, by the NIH grant CA98787 (DGN/YDZ) and NOAA NURP/NIUST grant NA16RU1496. This investigation was conducted in a facility constructed with Research Facilities Improvement Grant C06 RR-14503-01 from the NIH.
Footnotes
Supporting Information Available: Spectroscopic data (1H and 13C NMR) for 4 – 21. The material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Bache M, Kappler M, Said HM, Staab A, Vordermark D. Curr Med Chem. 2008;15:322–38. doi: 10.2174/092986708783497391. [DOI] [PubMed] [Google Scholar]
- 2.Tatum JL, Kelloff GJ, Gillies RJ, Arbeit JM, Brown JM, Chao KS, Chapman JD, Eckelman WC, Fyles AW, Giaccia AJ, Hill RP, Koch CJ, Krishna MC, Krohn KA, Lewis JS, Mason RP, Melillo G, Padhani AR, Powis G, Rajendran JG, Reba R, Robinson SP, Semenza GL, Swartz HM, Vaupel P, Yang D, Croft B, Hoffman J, Liu G, Stone H, Sullivan D. Int J Radiat Biol. 2006;82:699–757. doi: 10.1080/09553000601002324. [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL, Wang GL. Mol Cell Biol. 1992;12:5447–54. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GL, Semenza GL. J Biol Chem. 1995;270:1230–7. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL. IUBMB Life. 2008;60:591–7. doi: 10.1002/iub.93. [DOI] [PubMed] [Google Scholar]
- 6.Rankin EB, Giaccia AJ. Cell Death Differ. 2008;15:678–85. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza GL. Drug Discov Today. 2007;12:853–9. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Wang GL, Jiang BH, Rue EA, Semenza GL. Proc Natl Acad Sci USA. 1995;92:5510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 10.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 11.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 12.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 13.Bruick RK, McKnight SL. Science. 2001;294:1337–40. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 14.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Science. 2002;295:858–61. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 15.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Cancer Res. 2000;60:4010–5. [PubMed] [Google Scholar]
- 16.Rapisarda A, Zalek J, Hollingshead M, Braunschweig T, Uranchimeg B, Bonomi CA, Borgel SD, Carter JP, Hewitt SM, Shoemaker RH, Melillo G. Cancer Res. 2004;64:6845–8. doi: 10.1158/0008-5472.CAN-04-2116. [DOI] [PubMed] [Google Scholar]
- 17.Greenberger LM, Horak ID, Filpula D, Sapra P, Westergaard M, Frydenlund HF, Albaek C, Schroder H, Orum H. Mol Cancer Ther. 2008;7:3598–608. doi: 10.1158/1535-7163.MCT-08-0510. [DOI] [PubMed] [Google Scholar]
- 18.Unruh A, Ressel A, Mohamed HG, Johnson RS, Nadrowitz R, Richter E, Katschinski DM, Wenger RH. Oncogene. 2003;22:3213–20. doi: 10.1038/sj.onc.1206385. [DOI] [PubMed] [Google Scholar]
- 19.Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Lin X, Shoemaker AR, Albert DH, Fesik SW, Shen Y. Clin Cancer Res. 2006;12:4747–54. doi: 10.1158/1078-0432.CCR-05-2842. [DOI] [PubMed] [Google Scholar]
- 21.Cairns RA, Papandreou I, Sutphin PD, Denko NC. Proc Natl Acad Sci USA. 2007;104:9445–50. doi: 10.1073/pnas.0611662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. [accessed July 22, 2009];US NIH database – Clinical Trialsgov. http://www.clinicaltrials.gov/ct2/search.
- 23.Hodges TW, Hossain CF, Kim YP, Zhou YD, Nagle DG. J Nat Prod. 2004;67:767–71. doi: 10.1021/np030514m. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Liu R, Mao SC, Morgan JB, Jekabsons MB, Zhou YD, Nagle DG. J Nat Prod. 2008;71:1854–1860. doi: 10.1021/np800342s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Veena CK, Morgan JB, Mohammed KA, Jekabsons MB, Nagle DG, Zhou YD. J Biol Chem. 2009;284:5859–68. doi: 10.1074/jbc.M806744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West LM, Northcote PT, Battershill CN. J Org Chem. 2000;65:445–9. doi: 10.1021/jo991296y. [DOI] [PubMed] [Google Scholar]
- 27.Hood KA, West LM, Rouwe B, Northcote PT, Berridge MV, Wakefield SJ, Miller JH. Cancer Res. 2002;62:3356–60. [PubMed] [Google Scholar]
- 28.Huzil JT, Chik JK, Slysz GW, Freedman H, Tuszynski J, Taylor RE, Sackett DL, Schriemer DC. J Mol Biol. 2008;378:1016–30. doi: 10.1016/j.jmb.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Northcote PTB, JW, Munro HG. Tetrahedron Lett. 1991;32:6411–6414. [Google Scholar]
- 30.Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. Mol Cell. 2005;20:709–22. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J. Proc Natl Acad Sci USA. 2005;102:10460–5. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maulucci N, Chini MG, Micco SD, Izzo I, Cafaro E, Russo A, Gallinari P, Paolini C, Nardi MC, Casapullo A, Riccio R, Bifulco G, Riccardis FD. J Am Chem Soc. 2007;129:3007–12. doi: 10.1021/ja0686256. [DOI] [PubMed] [Google Scholar]
- 33.Nakao Y, Yoshida S, Matsunaga S, Shindoh N, Terada Y, Nagai K, Yamashita JK, Ganesan A, van Soest RW, Fusetani N. Angew Chem Int Ed Engl. 2006;45:7553–7. doi: 10.1002/anie.200602047. [DOI] [PubMed] [Google Scholar]
- 34.Ortega MJ, Zubía E, Sánchez MC, Salvá J, Carballo JL. Tetrahedron. 2004;60:2517–2524. [Google Scholar]
- 35.Compagnone RS, Oliveri MC, Piña I, Marques S, Rangel HR, Dagger F, Suárez AI, Gómez M. Nat Prod Lett. 1999;13:203–211. [Google Scholar]
- 36.Stierle DB, Faulkner DJ. J Org Chem. 1980;45:4980–4982. [Google Scholar]
- 37.Breitmaier E, Voelter W. Carbon-13 NMR Spectroscopy. 3. VCH; New York: 1989. pp. 192–194. [Google Scholar]
- 38.Venkatesham U, Rao MR, Venkateswarlu Y. J Nat Prod. 2000;63:1318–1320. doi: 10.1021/np000101a. [DOI] [PubMed] [Google Scholar]
- 39.Ferrara N, Mass RD, Campa C, Kim R. Annu Rev Med. 2007;58:491–504. doi: 10.1146/annurev.med.58.061705.145635. [DOI] [PubMed] [Google Scholar]
- 40.Nagle DG, Zhou YD. Curr Drug Targets. 2006;7:355–369. doi: 10.2174/138945006776054979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, David CA, Donnelly JB, Michaelides M, Chandel NS, Huang X, Warrior U, Weinberg F, Tormos KV, Fesik SW, Shen Y. Proc Natl Acad Sci USA. 2008;105:174–179. doi: 10.1073/pnas.0706585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Simon MC. Adv Exp Med Biol. 2006;588:165–170. doi: 10.1007/978-0-387-34817-9_15. [DOI] [PubMed] [Google Scholar]
- 44.Pan Y, Mansfield KD, Bertozzi CC, Rudenko V, Chan DA, Giaccia AJ, Simon MC. Mol Cell Biol. 2007;27:912–925. doi: 10.1128/MCB.01223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GRS, Chandel NS. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.