Linkage of adRP to the ASCC3L1 region of chromosome 2 and identification of a mutation in hBrr2p associated with RP in this Chinese family add mutations in another splicing factor to the known causes of RP.
Abstract
Purpose.
To localize and identify the gene and mutations causing autosomal dominant retinitis pigmentosa in a Chinese Family.
Methods.
Families were ascertained and patients underwent complete ophthalmic examinations. Blood samples were collected and DNA was extracted. A linkage scan of genomic regions containing known candidate genes was performed by using 34 polymorphic microsatellite markers on genomic DNA from affected and unaffected family members, and lod scores were calculated. Candidate genes were sequenced and mutations analyzed.
Results.
A genome-wide scan yielded a lod score of 3.5 at θ = 0 for D2S2333 and 3.46 at θ = 0 for D2S2216. This region harbors the ASCC3L1 gene. Sequencing of ASCC3L1 in an affected family member showed a heterozygous single-base-pair change; c.3269G→T, predicted to result in an Arg1090Leu amino acid change.
Conclusions.
The results provide strong evidence that mutations in ASCC3L1 have resulted in autosomal dominant retinitis pigmentosa in this Chinese family.
Retinitis pigmentosa (RP) is the most common inherited retinal dystrophy, affecting approximately 1 in 4000 individuals worldwide.1 RP primarily affects the rod photoreceptors, whereas the function of the cone receptors is compromised as the disease progresses.2 Ocular findings comprise atrophic changes in the photoreceptors and retinal pigment epithelium (RPE), followed in time by attenuation of the arterioles, the appearance of melanin-containing structures in the neuroretina (bone spicules) and waxy pallor of the optic disc. Affected individuals often have severely abnormal or nondetectable rod responses in the electroretinograms (ERG) recordings, even in the early stage of the disease.2
RP may be inherited as an autosomal dominant, autosomal recessive, or X-linked recessive trait. Autosomal dominant (ad)RP represents 15% to 20% of all cases; autosomal recessive (ar)RP comprises 20% to 25% (syndromic and nonsyndromic); X-linked recessive RP makes up 10% to 15%, and the remaining 40% to 55%, in which family history is absent, are called simplex (SRP), but many of these may represent arRP.3–6
To date, 42 loci have been implicated in nonsyndromic RP, of which 33 genes are known (RetNet; URLs of data sources are provided in the Appendix). These include genes encoding components of the phototransduction cascade, proteins involved in retinoid metabolism, cell–cell interaction proteins, photoreceptor structural proteins, transcription factors, intracellular transport proteins, and splicing factors.7 The autosomal dominant forms have been associated with 17 different loci, including PRPF3(1q21.2),8 SEMA4A(1q22),9 RHO(3q22),10 GUCA1B(6p21.1),11 RDS(6p21.2),12 PAP1(7q14),13 IMPDH1(7q32),14 RP1(8q12),15 TOPORS(9p21.1),16 ROM1(11q12.3),17 NRL (14q11.2),18 PRPF8 (17p13.3),19 CA4 (17q23.2),20 FSCN2 (17q25.3),21 CRX (19q13.32),22 PRPF31 (19q13.42),23 and RP33(2cen-q12.1).24 Mutations in 16 genes, with the exception of RP33, have been identified as the underlying cause of adRP.
Mutations in ubiquitously expressed splicing factors are a well-recognized cause of RP. Mutations in the gene encoding the pre-mRNA splicing factors PRPC8, PRPF31, PRP3, and PAP-1 cause adRP.8,13,19,23 The occurrence of retina specific disease as a result of mutations in ubiquitously expressed and highly conserved splice factors has been suggested to result from the exceptionally high rate of opsin turnover in rod photoreceptors.8 It has been suggested that splicing, and particular tri-snRNP activity, is particularly sensitive in the retina because of the high level of energy consumption necessary for visual function.25
Nuclear hnRNA contains introns that are recognized and removed by the spliceosome, a ribonucleoprotein body comprising U1, U2, U4/U6, and U5 snRNPs and multiple protein splicing factors. Splicing takes place in several steps involving the binding of the U1 and U2 snRNPs to the pre-mRNA followed by binding of the U4/U6, U5 tri-snRNP complex; activation involving release of the U1 and U4 snRNPs; and two splice steps resulting in production of the mature mRNA and release of the snRNPs and splicing factors from the removed intron.26 The tri-snRNP complex contains approximately 30 distinct proteins, including PRPF31, -3, and -8. It has been proposed that PRPF3 and -31 are involved in activation of the spliceosome by unwinding of the U4/U6 duplex. PAP1 is also implicated in pre-mRNA splicing by binding with PRPF3. PRPF8 is crucial for the formation of the catalytic center in the spliceosome and interacts via its carboxyl terminus with another U5 snRNP protein hBrr2. hBrr2 is the human orthologue of yeast Brr2p and is encoded by the ASCC3L1 gene on chromosome 2, region q11.2. This 200-kDa DExH-box RNA helicase binds stably to the U5 snRNP, interacting with the hPrp6 and hSnu66 proteins and is active in disruption of the U4/U6 snRNA helices before spliceosome activation.27
Here, we report a Chinese family with isolated adRP. Clinical findings in these families are typical of early-onset severe RP. Linkage analysis shows linkage to region q11.2 of chromosome 2, including ASCC3L1. Sequencing of ASCC3L1 shows a heterozygous single-base-pair change; c.3269G→T, resulting in a R1090L amino acid change in the protein.
Materials and Methods
Patient Ascertainment
A Han Chinese family with nonsyndromic adRP (Tianjin Eye Hospital pedigree 1, TEH1) was recruited through the Ophthalmic Genetics Clinic of the Tianjin Eye Hospital. IRB approval was obtained from the National Eye Institute (Bethesda, MD) and the Tianjin Eye Hospital. The participating subjects gave informed written consent, consistent with the tenets of the Declaration of Helsinki. The family (TEH1) described in this study is from Tianjin, China. A medical history was obtained by interviewing family members. All living family members shown in the pedigree (see Fig. 3A) underwent ophthalmic examinations, including a dilated fundus examination and photography. Visual fields were recorded with an automated perimeter (Octopus 301; Haag-Streit International, Köniz, Switzerland) using a SITA-Fast strategy. Full-field ERGs were recorded according to the standards of the International Society for Clinical Electrophysiology of Vision. Multifocal ERGs were recorded with DTL electrodes using a 103 element stimulus array and results recorded as the total response in nanovolts/degree provided by the software (VERIS; Electro-Diagnostic Imaging, Inc., Redwood City, CA). Diagnosis of RP was based on abnormal electroretinography (ERG) showing decreased amplitudes of the b-waves predominating in scotopic conditions; visual field defects including loss of peripheral vision; and typical ophthalmoscopic abnormalities, including retinal pigment deposits, retinal arteriole attenuation, and pigment epithelial atrophy. Blood samples were collected from affected and unaffected family members. DNA was extracted as described by Smith et al.28
Figure 3.
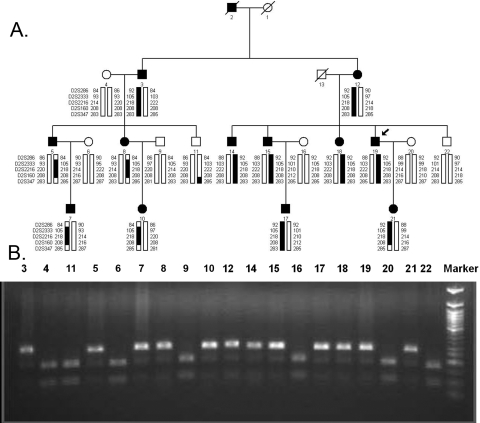
(A) Pedigree of TEH1 showing 2q11.2 haplotypes. Squares: males; circles, females; filled symbols: affected individuals; diagonal line through a symbol: a deceased family member; arrow: the proband. The haplotypes of five microsatellite markers from the ASCC3L1 region of 2q11.2 are shown with alleles forming the risk haplotype (black) and alleles not cosegregating with RP (white). (B) The c.3269G→T mutation resulted in loss of a HpaII site that cosegregated with the disease in all affected individuals.
Genotyping and Linkage Analysis
A candidate gene screen was performed with 34 highly polymorphic fluorescent markers for the 17 known adRP loci.24,29 The selected microsatellite markers flank the disease gene at each locus. These markers were D1S2726-PRPF3-D1S498, D1S484-SEMA4A-D1S2878, D2S286-RP33-D2S160, D3S1267-RHO-D3S1292, D6S422-RDS-D6S1610, D6S1650-GUCA1B-D6S257, D7S516-PAP1(RP9)-D7S484, D7S486-IMPDH1-D7S530, D8S258-RP1-D8S285, D9S171-TOPORS(RP31)-D9S161, D11S4197-ROM1-D11S987, D14S283-NRL-D14S275, D17S831-PRPF8-D17S938, D17S787-CA4-D17S944, D17S785-FSCN2-D17S784, D19S420-CRX-D19S902, and D19S572-PRPF31-D19S418.
Multiplex polymerase chain reactions (PCR) were performed as previously described.30 PCR products from each DNA sample were pooled and mixed with a loading cocktail containing size standards (HD-400; Applied Biosystems, Inc. [ABI], Foster City, CA) and loading dye. The resulting PCR products were separated on a sequencer (ABI3130; ABI) and analyzed (GeneMapper 4.0; ABI).
Two-point linkage analyses were performed with the FASTLINK version of MLINK from the LINKAGE Package.6,7,31,32 Maximum lod scores were calculated by using ILINK. Autosomal dominant RP was analyzed as a fully penetrant trait, with an affected allele frequency of 0.001. Autosomal dominant inheritance was confirmed by male-to-male transmission, and full penetrance was chosen because of the early onset of signs and symptoms (usually 7–8 years of age) relative to the age of examined family members. Also, well over half of the offspring of affected individuals are themselves affected, suggesting a high penetrance. The marker order and distances between the markers were obtained from the Généthon database and the National Center for Biotechnology Information chromosome 2 sequence maps. For the initial candidate screening, equal allele frequencies were assumed, whereas for fine mapping, allele frequencies were estimated from 96 unrelated and unaffected Han Chinese from the Tianjin region.
Mutation Screening
Individual exons of ASCC3L1 were amplified by PCR using the primer pairs shown in Table 1. Amplifications were performed as previously described33 and the PCR products were analyzed on 2% agarose gel and purified (QIAquick Gel Extraction Kit; Qiagen, Valencia, CA) according to the manufacturer's instructions followed by filtration on a manifold vacuum plate (Millipore, Billerica, MA). The PCR primers for each exon were used for bidirectional sequencing with dye termination chemistry (BigDye Terminator Ready reaction mix; ABI), according to the manufacturer's instructions. Sequencing products were resuspended in 10 μL of formamide (ABI) and denatured at 95°C for 5 minutes. Sequencing was performed on an automated sequencer (Prism 3100; ABI). Sequencing results were assembled and analyzed (Seqman software; DNAStar Inc., Madison, WI). Sequence alignment was performed using the Clustal W algorithm included in the software package.
Table 1.
Primers Used to Amplify and Sequence Exons of ASCC3L1
| Exon | Forward Primer | Reverse Primer |
|---|---|---|
| 1 | TTCTTTTCCTTCCCCAGTGC | TCAGACTGAAACTCCCTTTTTC |
| 2 | TTCCATCAGCTGGGGTAACT | TTACACCAAGCATTCCAGCA |
| 3 | TGAGGCTCAGATACCTGTTTTC | GCACTTCGAAGATTCCAAGGAT |
| 4 | AGGTGTGGCTCTTCACATCC | GGGAACTCACTGACATTAAGGTTT |
| 5 | TTTCCTGAGGAGAGGTAGTCG | GCCCAAGTCTGAAACCCATA |
| 6 | GTTTCACCCCAAAGCCATT | GACCTAGGCATGGGGAAGA |
| 7–8 | AGGTGGGCTATGGATTGTCA | GAAACAAGCACTTAGGCTGTGA |
| 9 | TTCTTTCTACCCTGAGGATGGT | AGCTTCATGCTGCAATCTTC |
| 10–11 | TTAGCTAAGCAGGGGCAGAA | CCTGGGCTCAAAATCAGAAT |
| 12 | ATCCTGGCAATCCCACAATA | GGTGGGGGTGGAAATAAAA |
| 13 | ATAAGCCCATGGAAGTTAGGG | GTCCAGGCCTGTCCACAA |
| 14 | CCAATTGCCCAAACAGAAAG | GCCTACAGAAAAGGCAGCAA |
| 15 | TTGGGATCCCTTGTCTTTTT | CCCAGTAGCACTCCTTGGAA |
| 16 | TTGGGGCAGTGAAAAAGACT | TTCTGTCAATCTTCCCCAAT |
| 17 | TGGTGCAACTTCTGCTTCCT | TGTCTGGGGCCTGAGATG |
| 18 | AGGGTCTCATGGGAGGAGAT | GAGCAAACATGGCACAAACT |
| 19–20 | AGAAGCTCTGAGGCCAACTG | ACAATAGGGACCGACCCACT |
| 21 | TGCATCCTTGTAGTTGGGAAT | GGCAGTACTCACATCAAGAGAGC |
| 22–23 | GGACAATAATGACATTGGGTCA | ACTGAGCAGGAAACCACACA |
| 24 | AGACCTGTAGCAAATCTCACCTG | AGGATGCACACAGCACACAG |
| 25 | ACCGTGTGTAGAGTGGCTCA | TTCCCATCAGACCCTTGG |
| 26 | TATTGGAAGCCATGAAGTGG | CACAGGCCTAGGAACAGGAA |
| 27–28 | GGGCTCCACTGACCATAAAG | ATCACTCTGTCCCCAGCAAG |
| 29 | CTTGCTGGGGACAGAGTGAT | AGACCTCCCAAGTGCCTGA |
| 30 | CATGTCCCAGTCCGTCTCTT | CTCCCAAAGTGCTGGGATT |
| 31 | GGTCTGTTTGGCATCTCAGG | CAGACCCAAACATTAGGTCCA |
| 31–33 | CTGTTCACAAAGGGGACCTC | TTCCTGGCAGACACTTTGC |
| 34 | TGCACAGCTGGCAAAGTG | CACCCCTCAAGTTTAACACCA |
| 35 | TTCTGAAACATAGCACAGGGTA | TCCTAGCTTACTTTCTGTGTATCTGC |
| 36 | GTCCGTGTCTCCCCGTATTA | AGCAAAAGGCAGCAAAATTC |
| 37–38 | GACCATGCCCTGTTGATTTG | CAGGCAGAGAAGGAGCAGAA |
| 39 | ATGACACTGCAGGGGACAG | CAGGGATGCCATGTGCTCT |
| 40–41 | CATCCCATTACCACCCACAT | CACCTTCGGAGGAACCATAA |
| 41–42 | AGCTCTTTGCTGCCTGTGTC | AAAAACCCCATAGCCTCAAGA |
| 44–45 | TTTCGGGAAGAAACTTTGGA | CCAGAGTGAGCAAGGGAAAG |
Annealing temperature, 56°C.
Results
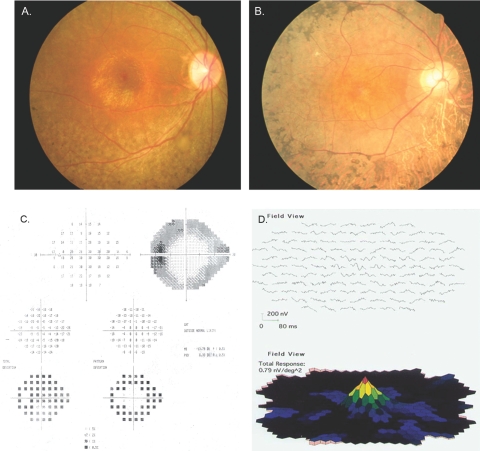
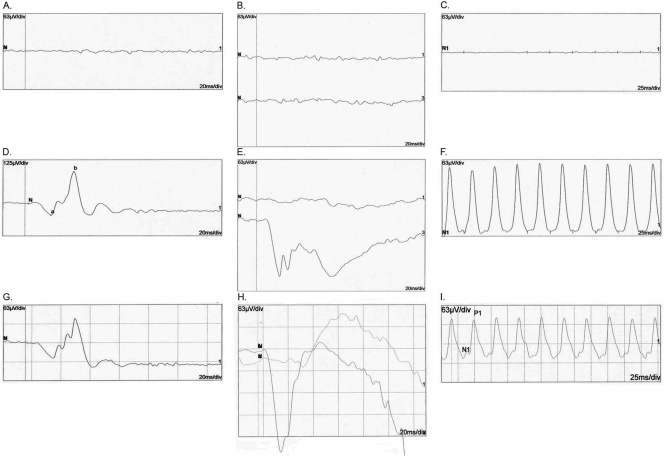
The clinical features of all affected individuals with diagnosed RP are listed in Table 2. The diagnosis of RP was made by electroretinography (ERG) and visual field testing for progressive loss in photoreceptor function. The average age at onset was 7 to 8 years, although some individuals were affected slightly later, at 10 to 11 years of age. Affected individuals have narrowing of the visual fields and night blindness accompanied by loss of visual acuity. Fundus photographs of affected individuals show changes typical of RP, including a waxy, pale optic disc, attenuation of retinal arteries, and bone spicule pigment deposits in the mid periphery of the retina, as shown in individual 21, a 14-year-old affected girl (Fig. 1A), and individual 19, a 45-year-old affected man (Fig. 1B). The visual fields of the affected individuals progressively narrowed with age, eventually leading to loss of peripheral vision with relative preservation of central vision, even in the 14-year-old girl (Figs. 1C, 1D). Affected individuals had typical RP changes on ERG including loss of both the rod and, in more advanced cases, cone responses (Fig. 2). Results from individual 19 (45 years old and severely affected) showed little detectable activity under either photopic or scotopic conditions. However, the photopic ERG and 30-MHz flicker response from individual 21 (14 years old and more mildly affected) showed preservation of cone cell function, whereas the rod response decreased under scotopic conditions, consistent with a predominantly rod pathology. A multifocal ERG in individual 21 also showed functional preservation in the central macular area and decreased signal in the peripheral macular area (Fig. 1D).
Table 2.
Clinical Characteristics of Affected TEH1 Family Members with RP
| Individual | Age/Sex | Age at Onset (y) | Visual Acuity | Fundus | Mean Visual Field Defect (dB) | Total mERG Amplitude (nV/deg) |
|---|---|---|---|---|---|---|
| 3 | 73/M | 7–8 | OD HM | NA | NA | NA |
| OS HM | ||||||
| 5 | 42/M | 10–11 | OD 0.3 | OD BP, AN, OA | OD-31.12 | OD 0.20 |
| OS 0.1 | OS BP, AN, OA | OS-32.45 | OS 0.29 | |||
| 7 | 15/M | 7–8 | OD 0.9 | OD BP, AN | OD-13.73 | OD 0.95 |
| OS 0.8 | OS BP, AN | OS-15.43 | OS 0.74 | |||
| 8 | 39/F | 10–11 | OD 0.5 | OD BP, AN, OA | OD-28.94 | OD 0.51 |
| OS 0.3 | OS BP, AN, OA | OS-27.73 | OS 0.35 | |||
| 10 | 14/F | 7–8 | OD 0.3 | OD BP, AN | OD-12.52 | OD 1.16 |
| OS 0.5 | OS BP, AN | OS-10.12 | OS 0.91 | |||
| 12 | 70/F | 8–9 | OD HM | OD NS | NA | NA |
| OS HM | OS NS | |||||
| 14 | 53/M | 7–8 | OD 0.1 | OD BP, AN, OA OS | OD-30.23 | OD 0.13 |
| OS 0.5 | BP, AN, OA | OS-30.29 | OS 0.51 | |||
| 15 | 51/M | 7–8 | OD HM | OD BP, AN, OA | OD-31.54 | NA |
| OS HM | OS BP, AN, OA | OS-32.43 | ||||
| 17 | 22/M | 7–8 | OD 0.1 | OD BP, AN | OD-13.81 | OD 0.30 |
| OS 0.2 | OS BP, AN | OS-12.84 | OS 0.39 | |||
| 18 | 48/F | 7–8 | OD 0.1 | OD BP, AN, OA | OD-33.00 | |
| OS .02 | OS BP, AN, OA | OS-33.00 | NA | |||
| 19 | 45/M | 7–8 | OD .06 | OD BP, AN, OA | OD-26.87 | OD 0.51 |
| OS 0.2 | OS BP, AN, OA | OS-24.93 | OS 0.13 | |||
| 21 | 14/F | 7–8 | OD 0.8 | OD BP, AN | OD-13.05 | OD 0.78 |
| OS 0.9 | OS BP, AN | OS-13.79 | OS 0.79 |
MD, mean defect of visual field; HM, hand motion; NS, no sight; BP, bone spicule–like pigment deposit; OA, optic atrophy; AN, narrowing of the retinal vessel; NA, not available.
Figure 1.
Fundus photographs of (A) individual 21 of family TEH1 (affected 14-year-old girl) and (B) individual 19 (affected 45-year-old man) showed changes typical of RP, including attenuation of retinal arteries and bone spicule pigment deposits in the midperiphery of the retina. (C) Retinal perimetry of individual 21 from family TEH1 showing loss of peripheral vision with relative preservation of central vision. (D) Multifocal ERG of individual 21 from family TEH1 showing decreased vision peripherally with preservation of central vision.
Figure 2.
ERG recordings of the right eye of individual 19 of family TEH1 (affected 45-year-old): (A) photopic ERG, (B) rod (top) and max (bottom) responses under scotopic conditions, and (C) cone-derived 30-Hz flicker. ERG recordings of the right eye of individual 21 of family TEH1 (affected 14-year-old): (D) photopic ERG, (E) rod (top) and max (bottom) responses under scotopic conditions, and (F) cone-derived 30-Hz flicker. ERG recordings of the right eye of a normal control: (G) photopic ERG (H) rod (top) and max (bottom) responses under scotopic conditions, and (I) cone-derived 30-Hz flicker.
A candidate gene screen was performed using a set of 34 polymorphic markers adjacent to or flanking known candidate genes for RP. In the candidate screen, all markers showed negative lod scores except that RP cosegregated with alleles of microsatellite marker D2S160, yielding a maximum lod score of 1.4 at θ = 0 (Table 3). For fine mapping additional markers, D2S233 and D2S2216 were analyzed as shown in Table 3 and Figure 3A. Two-point linkage analyses gave further evidence for linkage to markers on chromosome 2, region q11.2, with maximum lod scores of 3.5 with D2S2333 at θ = 0 and 3.47 with D2S2216 at θ = 0.
Table 3.
Two Point Lod Scores in the ASCC3L1 Gene Region
| Marker | Position | Lod Sore |
Zmax | θmax | ||||
|---|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 0.2 | 0.3 | 0.4 | ||||
| D2S286 | 75,079,610 | −∞ | −2.49 | −1.05 | −0.40 | −0.09 | 0 | 0.5 |
| D2S2333 | 85,384,880 | 3.50 | 2.88 | 2.20 | 1.45 | 0.64 | 3.5 | 0 |
| D2S2216 | 87,331,917 | 3.46 | 2.84 | 2.17 | 1.43 | 0.63 | 3.46 | 0 |
| D2S160 | 105,510,766 | 1.40 | 1.11 | 0.79 | 0.46 | 0.15 | 1.40 | 0 |
| D2S347 | 116,569,280 | −∞ | −1.38 | −0.53 | −0.18 | −0.04 | 0 | 0.5 |
Visual inspection of the haplotypes in this region support the linkage analysis (Fig. 3A), localizing the disease to a region of 2q11.2 flanked by D2S286 and D2S347. Recombination events at D2S286, either in affected individuals 5 and 8 (indicated on haplotype bars) or possibly in affected individual 3 (not indicated in haplotype bars) set the centromeric boundary. Similarly, recombination events at D2S347 in affected individual 21 and individuals 5 and 8 set the telomeric boundary of the linked interval. Once more, this recombination could have taken place either in affected individuals 5 and 8 (indicated on haplotype bars) or possibly in affected individual 3 and unaffected individual 11. In either event, the linked interval extends from D2S286 proximally to D2S347 distally.
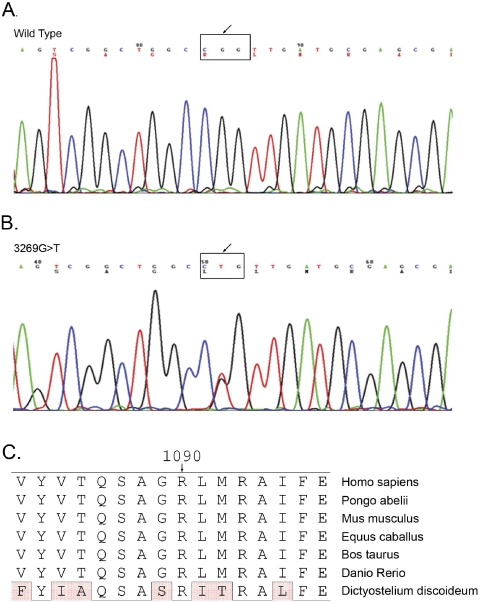
The linked region on 2q11.2 harbors ASCC3L1 (NM_014014), a known component of the tri-snRNP component of the spliceosome. The ASCC3L1 gene contains 45 exons and encodes a 2136 amino acid protein (NP_054733). Sequencing of ASCC3L1 showed a single base transversion in exon 25: c.3269G→T, predicted to result in a nonconservative change in the protein amino acid sequence: p.R1090L (Fig. 4). Sequencing of ASCC3L1 in 100 unaffected control individuals of matched (Han Chinese) ethnicity (200 chromosomes) did not reveal this sequence change. This mutation resulted in loss of a HpaII site that cosegregated with the disease in all affected individuals (Fig. 3B).
Figure 4.
Sequence chromatograms of (A) wild-type individual 20 of family TEH1 and (B) affected individual 21 of family TEH1, showing a heterozygous single-base change in exon 25: c.3269G→T. The sequence in the black box shows the codon change from CGG (ARG) to CTG (LEU). (C) Sequence alignment of ASCC3L1 gene product from human, zebra fish, horse, cow, and the DEAD/DEAH box helicase from Dictyostelium discoideum. ASCC3L1 protein sequences available from Pongo abelii, Mus musculus, Gallus gallus, Culex quinquefasciatus, and Strongylocentotus purpuratus did not align in this region of the protein and are not shown. However, R1090 was conserved when sequences predicted from the corresponding mRNAs of Pongo abelii, Mus musculus, and Gallus gallus were aligned. Conserved amino acids are boxed and differences are shaded.
R1090 is conserved among Homo sapiens, Pongo abelii, Mus musculus, Equus caballus, Bos taurus, Danio rerio, and Dictyostelium discoideum (Fig. 4C). ASCC3L1 protein sequences for Pongo abelii, Mus musculus, Gallus gallus, Culex quinquefasciatus, and Strongylocentotus purpuratus, available from NCBI, did not align in this region of the protein and are not shown. However, R1090 was conserved when sequences predicted from the corresponding mRNAs of Pongo abelii, Mus musculus, and Gallus gallus were aligned (Fig. 4C). The average percent identity for these sequences is approximately 85%, with the sequence of Dictyostelium discoideum being most divergent at 52%. The R1090L amino acid change scores a −4 on a blosum80 matrix34 and a PSIC profile score of 3.2, calculated by the POLYPHEN program.35 Both of these values indicate that this amino acid substitution is likely to have a deleterious effect on the protein.
Discussion
We report the characterization of a Han Chinese family with multiple family members affected with adRP, the localization of the adRP gene in these families to a 40-Mb region on chromosome 2m region q11.2, flanked by D2S286 and D2S347, and association of RP with an R1090L mutation in the ASCC3L1 gene. This amino acid change is a nonconservative one from a positively charged hydrophilic amino acid to a hydrophobic aliphatic one, giving a −4 on a blosum80 matrix and a 3.2 PISC score. These predictions along with the absence of this mutation in 200 unaffected chromosomes provide a high likelihood that this is a mutation that would significantly affect the function of the protein.
The ASCC3L1 gene is located in the region on 2q11.2, where an RP locus, RP33, was mapped by Zhao et al.24 in a four-generation Chinese family with adRP. They defined a 4.8-cM (9.5-Mb) interval at 2cen-q12.1 between D2S2159 and D2S1343 by haplotype analysis and obtained a maximum multipoint lod score of 4.69 at marker D2S2222. Here, we report adRP in the THE1 family also maps to this region flanked by D2S286 and D2S347 and overlapping the RP33 region. Four genes, including SEMA4C, CNGA3, HNK1ST, and MERTK, were sequenced by Zhao et al.24 as candidate genes in this region, but no mutations were found. Thus, although ASCC3L1 lies within the RP33 region, it is unproven at this time whether it corresponds to the locus previously reported in the RP33 family.
SEMA4C is a member of the semaphorin family of soluble and transmembrane proteins.36 Semaphorins are suggested to be involved in the guidance of axonal migration during neuronal development and in immune responses. Another semaphorin family member, SEMA4A, has been identified as an adRP disease gene.9,37 CNGA3 gene encodes one of a family of α subunits that form cyclic nucleotide-gated (CNG) ion channels, which are necessary for sensory transduction in rod photoreceptors and in olfactory neurons. Mutations in this gene may cause achromatopsia.38,39
HNK1ST plays a role in the biosynthesis of HNK1 carbohydrate epitope which is expressed in the human retina in both glial and neuronal cells, including photoreceptors. The carbohydrate epitope may play an important role in neuron-to-neuron and glia-to-neuron adhesion of human retinal cells.40,41 MERTK (OMIM 604705) is a known gene for ARRP. Mutations were not found after sequencing the coding regions and splice junctions of these four genes.24 However, ASCC3L1 was suggested as a candidate gene in the above region as well,24,25 mutation analysis for this gene was the first selected in this study. Although we cannot say definitively whether ASCC3L1 is the RP33 gene, it is certainly a gene within the RP33 interval that causes RP when mutated.
The ASCC3L1 gene consists of 45 exons spanning approximately 31 kb on chromosome 11, region 2, and encodes the human analogue of the yeast-splicing factor Brr2 hBrr2, a protein of 2136 amino acids. Yeast Brr2p is a DExH/DEAD-box RNA unwindase containing two Brl domains. Although it has demonstrated activity as a splicing factor, it also interacts with additional proteins involved in signal transduction, cell division, and gene transcription, all of which are consistent with its nuclear localization.39 The R1090L mutation lies in the first Sec63 motif of hBrr2, shown in yeast to be necessary for interacting with splice factor Prp8p,42 further suggesting that it might critically affect the RNA-splicing function of hBrr2.
Mutations in ubiquitously expressed splicing factors have been shown to cause a variety of tissue-specific inherited diseases including actinic porokeratosis-1,43 spinal muscular atrophy,44 myotonic dystrophy,45 as well as RP. Mutations in the gene encoding the pre-mRNA splicing factors PRPC8, PRPF31, PAP-1, and PRP3 cause adRP.8,13,19,23 Since the R1090L mutation in hBrr2 lies in a region known to be important for interacting with Prp8p, it is particularly interesting that mutations shown to cause RP in humans may cause splice defects in yeast by interfering with the interaction of Prp8p with Brr2p and blocking functional U5 snRNP and U4/U6 tri-snRNP formation in the nucleus by decreasing affinity of Brr2p for the mutant C terminus of Prp8p.46
Although the mechanisms through which some splice factor mutations act is becoming clearer, the basis for tissue specificity of these ubiquitous splicing factors remains elusive. A number of hypotheses have been advanced.47 In some disorders, including RP and spinal muscular atrophy, the explanation may lie in the elevated tissue requirements for the splicing factor due to higher metabolic or synthetic activity. It is also possible that cell-specific pre-mRNAs may be more sensitive to a deficiency of a basal splicing factor than widely expressed pre-mRNAs that must be able to participate in efficient splicing in a wide variety of nuclear environments. For example, some phenotypes caused by the absence of the splicing factors PPRP2 and CEF1 result from deficient removal of a single intron.
Linkage of adRP to the ASCC3L1 region of chromosome 2 and identification of a mutation in hBrr2p associated with RP in this Chinese family add mutations in another splicing factor to the known causes of RP. Elucidation of the specific mechanism through which this mutation acts necessitates further study of the normal and mutant hBrr2 protein and possibly the overall splicing process in the retina.
Acknowledgments
The authors thank the family members who donated samples to make this work possible.
Appendix
URLs of Data Sources
All sources are provided in the public domain.
RetNet: http://www.sph.uth.tmc.edu/Retnet/disease.htm/ University of Texas Houston Health Science Center, Houston, TX
NCBI: http://www.ncbi.nlm.nih.gov/Database/index.html/ National Center for Biotechnology Information, Bethesda, MD
POLYPHEN: http://coot.embl.de/PolyPhen/ the Bork Group, EMBL, Heidelberg, Germany
Généthon: http://www.genethon.fr/ the French Association against Myopathies, Evry, France
ISCEV: http://www.iscev.org International Society for Clinical Electrophysiology of Vision
OMIM: http://www.ncbi.nlm.nih.gov/Omim/ Online Mendelian Inheritance in Man, NCBI, Bethesda, MD
Footnotes
Supported by the National Eye Institute Intramural Program (EY-00272), National Institutes of Health, Bethesda, Maryland.
Disclosure: N. Li, None; H. Mei, None; I.M. MacDonald, None; X.-D. Jiao, and J.F. Hejtmancik, None
References
- 1.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet 2006; 368: 1795–1809 [DOI] [PubMed] [Google Scholar]
- 2.Bird AC. Retinal photoreceptor dystrophies LI. Edward Jackson Memorial Lecture. AmJ Ophthalmol 1995; 119: 543–562 [DOI] [PubMed] [Google Scholar]
- 3.Boughman JA, Conneally PM, Nance W. Population genetic studies of retinitis pigmentosa. Am J Hum Genet 1980; 32: 223–225 [PMC free article] [PubMed] [Google Scholar]
- 4.Boughman JA, Caldwell RJ. Genetic and clinical characterization of a survey population with retinitis pigmentosa. In: Clinical, Structural, and Biochemical Advances in Hereditary Eye Disorders New York: Alan R. Liss, Inc; 1982: 147–166 [PubMed] [Google Scholar]
- 5.Inglehearn CF. Molecular genetics of human retinal dystrophies. Eye 1998; 12: 571–579 [DOI] [PubMed] [Google Scholar]
- 6.Jay M. Figures and fantasies: the frequencies of the different genetic forms of retinitis pigmentosa. Birth Defects Orig Artic Ser 1982; 18: 167–173 [PubMed] [Google Scholar]
- 7.Hims MM, Daiger SP, Inglehearn CF. Retinitis pigmentosa: genes, proteins and prospects. Dev Ophthalmol 2003; 37: 109–125 [DOI] [PubMed] [Google Scholar]
- 8.Chakarova CF, Hims MM, Bolz H, et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet 2002; 11: 87–92 [DOI] [PubMed] [Google Scholar]
- 9.Abid A, Ismail M, Mehdi SQ, Khaliq S. Identification of novel mutations in the SEMA4A gene associated with retinal degenerative diseases. J Med Genet 2006; 43: 378–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dryja TP, McGee TL, Reichel E, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature 1990; 343: 364–366 [DOI] [PubMed] [Google Scholar]
- 11.Payne AM, Downes SM, Bessant DA, et al. Genetic analysis of the guanylate cyclase activator 1B (GUCA1B) gene in patients with autosomal dominant retinal dystrophies. J Med Genet 1999; 36: 691–693 [PMC free article] [PubMed] [Google Scholar]
- 12.Farrar GJ, Kenna P, Jordan SA, et al. A three-base-pair deletion in the peripherin-RDS gene in one form of retinitis pigmentosa. Nature 1991; 354: 478–480 [DOI] [PubMed] [Google Scholar]
- 13.Maita H, Kitaura H, Keen TJ, et al. PAP-1, the mutated gene underlying the RP9 form of dominant retinitis pigmentosa, is a splicing factor. Exp Cell Res 2004; 300: 283–296 [DOI] [PubMed] [Google Scholar]
- 14.Kennan A, Aherne A, Palfi A, et al. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho(−/−) mice. Hum Mol Genet 2002; 11: 547–557 [DOI] [PubMed] [Google Scholar]
- 15.Pierce EA, Quinn T, Meehan T, et al. Mutations in a gene encoding a new oxygen-regulated photoreceptor protein cause dominant retinitis pigmentosa. Nat Genet 1999; 22: 248–254 [DOI] [PubMed] [Google Scholar]
- 16.Chakarova CF, Papaioannou MG, Khanna H, et al. Mutations in TOPORS cause autosomal dominant retinitis pigmentosa with perivascular retinal pigment epithelium atrophy. Am J Hum Genet 2007; 81: 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bascom RA, Garcia-Heras J, Hsieh CL, et al. Localization of the photoreceptor gene ROM1 to human chromosome 11 and mouse chromosome 19: sublocalization to human 11q13 between PGA and PYGM. Am J Hum Genet 1992; 51: 1028–1035 [PMC free article] [PubMed] [Google Scholar]
- 18.Bessant DA, Payne AM, Mitton KP, et al. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet 1999; 21: 355–356 [DOI] [PubMed] [Google Scholar]
- 19.McKie AB, McHale JC, Keen TJ, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 2001; 10: 1555–1562 [DOI] [PubMed] [Google Scholar]
- 20.Rebello G, Ramesar R, Vorster A, et al. Apoptosis-inducing signal sequence mutation in carbonic anhydrase IV identified in patients with the RP17 form of retinitis pigmentosa. Proc Natl Acad Sci USA 2004; 101: 6617–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wada Y, Abe T, Takeshita T, et al. Mutation of human retinal fascin gene (FSCN2) causes autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 2001; 42: 2395–2400 [PubMed] [Google Scholar]
- 22.Rivolta C, Berson EL, Dryja TP. Dominant Leber congenital amaurosis, cone-rod degeneration, and retinitis pigmentosa caused by mutant versions of the transcription factor CRX. Hum Mutat 2001; 18: 488–498 [DOI] [PubMed] [Google Scholar]
- 23.Vithana EN, bu-Safieh L, Allen MJ, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 2001; 8: 375–381 [DOI] [PubMed] [Google Scholar]
- 24.Zhao C, Lu S, Zhou X, et al. A novel locus (RP33) for autosomal dominant retinitis pigmentosa mapping to chromosomal region 2cen-q12.1. Hum Genet 2006; 119: 617–623 [DOI] [PubMed] [Google Scholar]
- 25.Schmidt-Kastner R, Yamamoto H, Hamasaki D, et al. Hypoxia-regulated components of the U4/U6.U5 tri-small nuclear riboprotein complex: possible role in autosomal dominant retinitis pigmentosa. Mol Vis 2008; 14: 125–135 [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Rauhut R, Vornlocher HP, Luhrmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA 2006; 12: 1418–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laggerbauer B, Achsel T, Luhrmann R. The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA 1998; 95: 4188–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith RJH, Holcomb JD, Daiger SP, et al. Exclusion of Usher syndrome gene from much of chromosome 4. Cytogenet Cell Genet 1989; 50: 102. [DOI] [PubMed] [Google Scholar]
- 29.Papaioannou M, Chakarova CF, Prescott DC, et al. A new locus (RP31) for autosomal dominant retinitis pigmentosa maps to chromosome 9p. Hum Genet 2005; 118: 501–503 [DOI] [PubMed] [Google Scholar]
- 30.Riazuddin SA, Yasmeen A, Zhang Q, et al. A new locus for autosomal recessive nuclear cataract mapped to chromosome 19q13 in a Pakistani family. Invest Ophthalmol Vis Sci 2005; 46: 623–626 [DOI] [PubMed] [Google Scholar]
- 31.Schaffer AA, Gupta SK, Shriram K, Cottingham RW. Avoiding recomputation in genetic linkage analysis. Hum Hered 1994; 44: 225–237 [DOI] [PubMed] [Google Scholar]
- 32.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 1984; 36: 460–465 [PMC free article] [PubMed] [Google Scholar]
- 33.Riazuddin SA, Yasmeen A, Yao W, et al. Mutations in βB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci 2005; 46: 2100–2106 [DOI] [PubMed] [Google Scholar]
- 34.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 1992; 89: 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunyaev SR, Eisenhaber F, Rodchenkov IV, et al. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng 1999; 12: 387–394 [DOI] [PubMed] [Google Scholar]
- 36.Inagaki S, Ohoka Y, Sugimoto H, et al. Sema4c, a transmembrane semaphorin, interacts with a post-synaptic density protein, PSD-95. J Biol Chem 2001; 276: 9174–9181 [DOI] [PubMed] [Google Scholar]
- 37.Rice DS, Huang W, Jones HA, et al. Severe retinal degeneration associated with disruption of semaphorin 4A. Invest Ophthalmol Vis Sci 2004; 45: 2767–2777 [DOI] [PubMed] [Google Scholar]
- 38.Michalakis S, Geiger H, Haverkamp S, et al. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Invest Ophthalmol Vis Sci 2005; 46: 1516–1524 [DOI] [PubMed] [Google Scholar]
- 39.Reuter P, Koeppen K, Ladewig T, et al. Mutations in CNGA3 impair trafficking or function of cone cyclic nucleotide-gated channels, resulting in achromatopsia. Hum Mutat 2008; 29: 1228–1236 [DOI] [PubMed] [Google Scholar]
- 40.Mitsumoto Y, Oka S, Sakuma H, Inazawa J, Kawasaki T. Cloning and chromosomal mapping of human glucuronyltransferase involved in biosynthesis of the HNK-1 carbohydrate epitope. Genomics 2000; 65: 166–173 [DOI] [PubMed] [Google Scholar]
- 41.Uusitalo M, Schlotzer-Schrehardt U, Kivela T. Ultrastructural localization of the HNK-1 carbohydrate epitope to glial and neuronal cells of the human retina. Invest Ophthalmol Vis Sci 2003; 44: 961–964 [DOI] [PubMed] [Google Scholar]
- 42.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 2001; 157: 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang ZH, Niu ZM, Yuan WT, et al. A mutation in SART3 gene in a Chinese pedigree with disseminated superficial actinic porokeratosis. Br J Dermatol 2005; 152: 658–663 [DOI] [PubMed] [Google Scholar]
- 44.Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–165 [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Pegoraro E, Menegazzo E, et al. Myotonic dystrophy: evidence for a possible dominant-negative RNA mutation. Hum Mol Genet 1995; 4: 599–606 [DOI] [PubMed] [Google Scholar]
- 46.Boon KL, Grainger RJ, Ehsani P, et al. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 2007; 14: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev 2003; 17: 419–437 [DOI] [PubMed] [Google Scholar]