Mutations in fibulin 5 are associated with AMD and are known to cause autosomal recessive cutis laxa. This article describes the structural changes to fibulin 5 protein associated with these mutations, to help determine the pathogenicity of the AMD mutations.
Abstract
Purpose.
AMD has a complex etiology with environmental and genetic risk factors. Ten fibulin 5 sequence variants have been associated with AMD and two other fibulin 5 mutations cause autosomal-recessive cutis laxa. Fibulin 5 is a 52-kDa calcium-binding epidermal growth factor (cbEGF)–rich extracellular matrix protein that is essential for the formation of elastic tissues. Biophysical techniques were used to detect structural changes in the fibulin 5 mutants and to determine whether changes are predictive of pathogenicity.
Methods.
Native PAGE, nonreduced SDS-PAGE, size-exclusion column multiangle laser light scattering, sedimentation velocity, and circular dichroism (CD) were used to investigate the mobility, hydrodynamic radii, folding, and oligomeric states of the fibulin 5 mutants in the absence and presence of Ca2+.
Results.
CD showed that all mutants are folded, although perturbations to secondary structure contents were detected. Both cutis laxa mutants increased dimerization. Most other mutants slightly increased self-association in the absence of Ca2+ but this was also demonstrated by G202R, a polymorphism detected in a control individual. The AMD-associated mutant G412E showed lower-than-expected mobility during native-PAGE, the largest hydrodynamic radius for the monomer form and the highest levels of aggregation in both the absence and presence of Ca2+.
Conclusions.
The results identified structural differences for the disease-causing cutis laxa mutants and for one AMD variant (G412E), suggesting that this may also be pathogenic. Although the other AMD-associated mutants showed no gross structural differences, they cannot be excluded as pathogenic by differences outside the scope of this study—for example, disruption of heterointeractions.
Age related macular degeneration (AMD) has a complex etiology with both environmental risk factors1,2 and genetic associations with complement factors3–7 and the chromosome 10 locus LOC387715.8 In addition, 10 heterozygous missense fibulin 5 variants (V60L, R71Q, P87S, Q124P, V126M, I169T, G267S, R351W, A363T, and G412E; Fig. 1) have been identified in AMD patients but not in control subjects, and the variant G202R has been found in one control.9,10 Each variant occurred in only one person, and the corresponding WT fibulin 5 residue was conserved across species.9,10 A further two missense fibulin 5 mutations (C217R and S227P) are known to cause the autosomal recessive disease cutis laxa.11–13
Figure 1.
Diagram of fibulin 5 showing the location of the cbEGF domains, potential glycosylation sites, and mutations associated with cutis laxa (C217R and S227P) and age-related macular degeneration (V60L, R71Q, P87S, Q124P, V126M, I169T, G267S, R351W, A363T, and G412E). G202R is a polymorphism that was found in a control rather than in a patient.
Genetic studies of a complex disease can indicate associations between the disease and sequence variants within a candidate gene, and it can be difficult to determine whether the sequence changes confer increased disease susceptibility or the association has occurred by chance. This contrasts with the situation for monogenic disorders in which a pathogenic mutation is enough to cause the disease phenotype by altering the function of the gene or protein. We were interested in comparing the effects on fibulin 5 of the pathogenic cutis laxa mutations and the sequence variants associated with AMD.
The fibulins are seven extracellular matrix proteins that contain tandem calcium-binding epidermal growth factor domains (cbEGFs). Fibulin 5 (Fig. 1) is a 52-kDa protein containing a modified N-terminal cbEGF domain, followed by five cbEGF domains, and a fibulin-type C-terminal module.14 All its 34 cysteines are thought to be internally disulfide bonded.15 Wild-type (WT) fibulin 5 exists primarily as an L-shaped monomer in the absence of Ca2+ and as a donut-shaped dimer in the presence of extracellular physiological Ca2+ concentrations.16 Fibulins are involved in the assembly, organization, and stabilization of macromolecular complexes.17 Fibulin 5 is expressed by elastin-rich tissues,18,19 including the retinal pigmented epithelium (RPE)10 and is essential for elastinogenesis. The fibulin 5-knockout mouse exhibits severe elastinopathy, with loose skin, vascular abnormalities, and emphysematous lungs.20,21 A single mutation in fibulin 3 (also known as EFEMP1) causes the dominant disease Malattia Leventinese (Doyne's honeycomb retinal dystrophy), and the similarity between AMD and this retinal phenotype led to the initial study of fibulins in AMD and the first association of sequence variants in fibulin 5.10
Our hypothesis was that the fibulin 5 sequence variants lead to increased susceptibility to AMD by altering the structure of the fibulin 5 protein. We used a variety of biophysical techniques to detect structural changes in fibulin 5 mutants and to determine whether the results could help to predict the pathogenicity of the AMD-associated sequence changes by comparing results with those from the cutis laxa mutants. We have previously shown that the two cutis laxa mutants and four of the AMD-associated variants (G412E, G267S, I169T, and Q124P) are poorly secreted,9 and structural changes in the protein could account for this. Structural differences could also suggest a potential loss of function for those AMD-associated mutants that are secreted normally.
We investigated the two cutis laxa mutations that are known to cause disease, the G202R polymorphism that does not cause a disease, the 10 AMD variants, and the mutation R54A that should prevent binding of the fibulin 5 RGD motif (residues 54-56) to integrins, but is not associated with disease. We used EBNA 293 cells and the pCepHis vector to ensure that all mutants were secreted, followed by Ni2+-affinity chromatography for protein purification. We used a combined strategy of SEC-MALLS (size exclusion column followed by multiangle laser light scattering), sedimentation velocity, CD (circular dichroism), protein homology modeling, native-PAGE, and nonreduced SDS-PAGE, to determine the shapes, hydrodynamic radii, and secondary structure content of both monomeric and dimeric forms, and levels of self-association, for these purified fibulin 5 mutants.
Methods
Molecular Biology and Expression and Purification of Recombinant Fibulin 5 Polypeptides
Site-directed mutants were obtained (QuikChange method; Stratagene, La Jolla, CA) by using the pCepHis construct encoding wild-type fibulin 5 (residues 27-448)22,23 as a template and the primers listed (Supplementary Fig. S1; all Supplementary Figures available at http://www.iovs.org/cgi/content/full/51/5/2356/DC1). Mutations were confirmed by DNA sequencing.
WT and mutants of fibulin 5 were expressed by EBNA 293 cells stably transfected with pCepHis constructs and secreted into serum-free medium (SAFC Biosciences, Lenexa, KS). Fibulin 5 polypeptides were purified from medium to homogeneity by Ni2+-affinity chromatography.16 The presence of fibulin 5 polypeptides and the absence of impurities in Ni2+-affinity column elution fractions were demonstrated by SDS-PAGE, followed by staining with Coomassie blue and Western blot analysis (anti-3-his. antibody; Sigma-Aldrich, St. Louis, MO).16
Statistical Analysis
The appropriate statistical method for the analysis of single measurements for multiple mutants is Chebyshev's inequality24 P(x − μ ≥ δ · SD) ≤1/δ2: The probability (P) that a single measurement (x) for a mutant is greater than a number (δ) times the standard deviation (SD) for WT from the mean (μ) for WT is less than 1/δ2. For the SEC-MALLS, sedimentation velocity and CD experiments, measurements for three or five batches of WT protein and δ = 4.47 were used to construct the confidence interval μ ± 4.47 SD. This gives P ≤ 0.05 that a single measurement for a mutant lays outside this confidence interval for WT.
Size-Exclusion Column-Multiangle Laser Light Scattering
Ni2+-affinity column elution fractions containing pure fibulin 5 polypeptides were applied to a size exclusion column (Superdex 200 SEC; GE Healthcare, Piscataway, NJ) equilibrated in either buffer A (10 mM Tris [pH 7.4], 0.5 M NaCl, 2 mM EDTA, and 2 mM EGTA) or buffer B (10 mM Tris [pH 7.4], 0.5 M NaCl, and 2 mM CaCl2). A high salt concentration was used to reduce amounts of aggregated polypeptides that elute in the SEC void volume.16 The SEC eluate passed through an 18-angle MALLS detector equipped with a 688-nm laser, to determine masses, a quasi-elastic light-scattering detector (QELS) to determine hydrodynamic radii, and an r-EX refractometer to determine differential refractive index (DRI, a measure of protein concentration; all from Wyatt Technology Corp., Santa Barbara, CA). The resulting DRI, MALLS, and QELS chromatograms were baseline corrected.
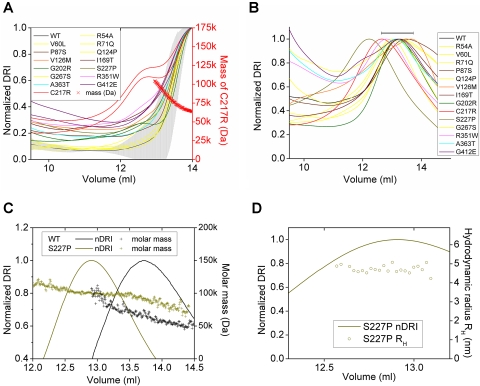
Pure fibulin 5 proteins (three batches of WT; two batches for G202R, C217R, and G412E; and one batch for the other 11 mutants) were applied to the SEC equilibrated in the absence of Ca2+ (buffer A, Fig. 2A). Figure 2A shows the mean DRI chromatogram (black line) for WT. The maxima of peaks due to monomeric protein16 were normalized to unity and aligned at 14 mL to compare peak shapes.
Figure 2.
SEC chromatograms of wild-type (WT) and mutants of fibulin 5. Chromatograms were base-line corrected and the differential refractive index (DRI, a measure of protein concentration) was normalized to 1.0 for each peak maximum. (A–D) Chromatograms for first applications to the SEC in the absence (A, buffer A) and presence (B, buffer B) of Ca2+. (A) The peak maxima were aligned at 14 mL. The mean chromatogram (line) and confidence interval (shaded area) were calculated from chromatograms for three batches of WT. Duplicates are shown for G202R, C217R, and G412E. (B) The column void peak maxima were aligned at 8.25 mL. The mean chromatogram (line) was calculated from chromatograms for five batches of WT. The error bar shows the confidence interval for the position of the WT peak maximum. (C, D) Chromatograms for the reapplication of fractions from (B) (WT 13–13.5 mL, S227P 12–12.5 mL) to the SEC in the presence of Ca2+ so that molar masses (C) and RH (D) could be determined without interference from light scattering at the column void.
Fibulin 5 proteins (five batches of WT and one batch for each of the 14 mutants) were also analyzed by SEC in the presence of 2 mM Ca2+ (buffer B, Fig. 2B). Figure 2B shows the mean DRI chromatogram (black line) for WT. In Figures 2B–D, each peak maximum was normalized to unity, and the column void peak maxima (not shown) were aligned at 8.25 mL, to enable comparison of peak positions.
SEC elution fractions containing fibulin 5 monomers in the absence of Ca2+ and dimers in the presence of Ca2+ were identified and analyzed by velocity sedimentation and CD.
Sedimentation Velocity
This technique measures the velocity at which proteins move through a buffer of known composition in response to a centrifugal force. Experiments were performed in an Optima XL-A analytical ultracentrifuge (Beckman Coulter, Hialeah, FL) equipped with an absorbance optical system. Sample and buffer were centrifuged in a two-sector centerpiece in an An-60 Ti 4-hole rotor at 45-krpm at 20°C. The sedimentation was monitored at 230 nm by 150 scans across a 1.4-cm radius with a step-size of 0.003 cm. Sedfit25 performed Lamm equation boundary modeling to calculate size (c(s)) distributions, by using floated frictional ratios (f/f0), and menisci positions, values for buffer densities and viscosities calculated with Sednterp,26 and the estimate 0.7052 mL/g for the partial specific volume of the glycosylated recombinant fibulin 5 polypeptide.16 Sednterp26 then used sedimentation coefficients for c(s) maxima to calculate f/f0s, standard sedimentation coefficients (S20,w), and hydrodynamic radii (RH).
Circular Dichroism
CD is the differential absorption of left- and right-handed circularly polarized ultraviolet light. Spectra were measured between 190 and 260 nm by a J-810 spectropolarimeter (Jasco, Easton, MD) with a 0.05-cm path length, and an average of 10 scans; spectra were then baseline and buffer corrected. Different types of protein secondary structure (helix, strand, turn, and unordered) have signature spectra. Hence, the spectra were deconvoluted by the Dichroweb27 server running the open-source CDSSTR software28 with reference set 7 to provide estimates of the percentage of each secondary structure type.
Results
Dimerization, Higher-Order Self-Association, and Mobility
Figure 2A shows the mean SEC chromatogram calculated for three batches of WT fibulin 5 and chromatograms for the mutants obtained in the absence of Ca2+. The main peak maxima were aligned at 14 mL to compare peak shape. Lower molecular weight proteins elute later from the SEC at higher volumes. Between 10 and 12 mL, chromatograms for the mutants P87S, V126M, I169T, G202R, C217R, S227P, R351W, A363T, and G412E lay above the confidence interval (shaded gray) for WT, indicating increased self-association for these mutants (P < 0.05). At 10 mL both chromatograms for G412E showed the most deviation, indicating greatly increased self-association (P < 0.001). Both chromatograms for C217R formed a distinct second peak at 12.7 mL (P < 0.005) that mass-analysis showed was due to a dimer. The chromatograms for five other mutants (P87S, V126M, I169T, S227P, and R351W) gave points of inflection between 12 and 13 mL, probably because of small amounts of dimers. In addition, chromatograms for P87S, I169T, G202R, A363T, and G412E gave points of inflection at volumes <12 mL. Chromatograms (all shown in yellow in Fig. 2A and subsequent figures) for the mutants R54A, V60L, R71Q, Q124P, and G267S lay within the confidence interval for WT, suggesting that these mutations did not significantly affect the self-association of fibulin 5 in the absence of Ca2+.
Figure 2B shows the mean SEC chromatogram calculated for five batches of WT fibulin 5 and chromatograms for the mutants obtained in the presence of Ca2+. The column-void peaks (not shown) were aligned so that main peak positions could be compared. Ca2+ causes the main peaks to shift to the left, indicating more dimers and fewer monomers, and causes increased chromatogram height at lower volumes due to increased higher-order oligomerization.16 The position of the peak maximum for S227P is outside the confidence interval (shown as the single black error bar, P < 0.05) for WT, suggesting increased self-association for S227P.
The chromatograms in Figures 2C and 2D illustrate the reapplication of SEC elution fractions (WT 13–13.5 mL, S227P 12–12.5 mL, shown in Fig. 2B) with the highest protein concentrations in the SEC in the presence of Ca2+ so that molar masses (Fig. 2C) and (Fig. 2D) could be determined easily. For WT, a dimeric molar mass (103 kDa) was approached only at the extreme left of the peak (Fig. 2C). For S227P, dimeric molar masses were measured across the whole peak (Fig. 2C), enabling us to obtain the estimate of 4.70 ± 0.04 nm for the radius for dimeric S227P (Fig. 2D).
The first chromatogram for G412E had the highest aggregation (measured by chromatogram height at ∼10 mL) of 20 chromatograms measured when Ca2+ was absent (Fig. 2A). Also, the second chromatogram for G412E showed the highest aggregation of the other 19 chromatograms measured when Ca2+ was absent (Fig. 2A). Further, G412E gave the highest aggregation of 19 chromatograms measured in the presence of Ca2+ (Fig. 2B). We suggest that these results are dependent (i.e., not a coincidence) because the following calculation gave a low probability of their being independent (i.e., occurring by chance). Given that G412E had the highest aggregation in the absence of Ca2+, the probability that independently G412E also has the second highest aggregation in the absence of Ca2+ and the highest aggregation in the presence of Ca2+ is (1/19) × (1/19) = 0.003. Further, G412E did not migrate as far toward the anode as R351W (which has a comparable electronegative substitution) during native (nondenaturing, nonreducing, pH 7.4, absence of Ca2+) PAGE (Supplementary Fig. S2A).
Hydrodynamic Parameters
The monomeric and dimeric forms of the fibulin 5 mutants in the absence and presence of Ca2+ respectively underwent velocity sedimentation, and c(s) distributions for monomers (Fig. 3A) and dimers (Fig. 3B) were calculated by the program Sedfit.25 Figure 3 shows confidence intervals (black error bars) calculated for the positions of the peak maxima for WT. The peak maxima for monomeric G202R and G412E lay outside the confidence interval for WT (P < 0.05, Fig. 3A). None of the peak maxima for the dimeric mutants were located outside the wide confidence interval calculated for WT in the presence of Ca2+ (Fig. 3B).
Figure 3.
Velocity sedimentation of wild-type (WT) and mutants of fibulin 5 purified by SEC in the absence (A, buffer A) and presence (B, buffer B) of Ca 2+. Sedfit25 provided the c(s) distributions. The horizontal error bars show the sedimentation coefficients within ±4.47 (A) and ±2 (B) SD of the three WT peak maxima.
Table 1 shows f/f0, S20,w, and RH calculated from the sedimentation velocity data (Fig. 3) for both monomeric and dimeric forms of the fibulin 5 mutants in the absence and presence of Ca2+, respectively. The f/f0 is a comparison of the shape of a protein to a sphere: The f/f0 of a sphere is unity, whereas that of a linear protein is >1. The S20,w provides information about both the shape and the mass of the protein and has units of Svedbergs (S) where 1S = 10−13 seconds. Table 1 also shows the means (μ), standard errors (SE), and confidence intervals for the aforementioned parameters calculated from measurements for three batches of WT protein. Values for the hydrodynamic parameters for the mutants (G202R and G412E) that lie outside the confidence interval for WT (P < 0.05) are in italic (smaller values) or bold (larger values).
Table 1.
Hydrodynamic Parameters (Fig. 3) for Fibulin 5 Mutants Were Determined by Analysis of the Sedimentation Velocity Data
| Monomer* |
Dimer† |
|||||
|---|---|---|---|---|---|---|
| f/f0‡ | S020,w‡ ×10−13 s | RH‡ nm | f/f0‡ | S020,w‡×10−13 s | RH‡ nm | |
| WT§ | ||||||
| Mean (μ) | 1.48 | 3.73 | 3.61 | 1.64 | 5.35 | 5.04 |
| ± SE | ± 0.01 | ± 0.02 | ± 0.02 | ± 0.07 | ± 0.25 | ± 0.23 |
| ± 4.47 SD | ± 0.07 | ± 0.17 | ± 0.17 | ± 0.57 | ± 1.90 | ± 1.74 |
| R54A | 1.54 | 3.59 | 3.74 | 1.55 | 5.66 | 4.75 |
| V60L | 1.49 | 3.70 | 3.64 | 1.65 | 5.31 | 5.06 |
| R71Q | 1.51 | 3.64 | 3.69 | 1.69 | 5.18 | 5.19 |
| P87S | 1.53 | 3.60 | 3.73 | 1.57 | 5.57 | 4.82 |
| Q124P | 1.46 | 3.78 | 3.55 | 1.85 | 4.74 | 5.67 |
| V126M | 1.50 | 3.68 | 3.65 | 1.53 | 5.71 | 4.71 |
| I169T | 1.46 | 3.79 | 3.55 | 1.57 | 5.57 | 4.82 |
| G202R | 1.56 | 3.53 | 3.81 | 1.81 | 4.85 | 5.54 |
| C217R | 1.51 | 3.65 | 3.68 | 1.50 | 5.85 | 4.59 |
| S227P | 1.54 | 3.59 | 3.74 | 1.46 | 5.98 | 4.49 |
| G267S | 1.48 | 3.73 | 3.60 | 1.65 | 5.30 | 5.07 |
| R351W | 1.43 | 3.86 | 3.48 | 1.50 | 5.85 | 4.59 |
| A363T | 1.45 | 3.80 | 3.53 | 1.57 | 5.57 | 4.82 |
| G412E | 1.58 | 3.50 | 3.84 | 1.72 | 5.09 | 5.28 |
Data for monomeric proteins were obtained in the absence of calcium (buffer A).
Data for dimeric proteins were obtained in the presence of calcium (buffer B).
Sednterp26 used the MALLS-derived masses and the S values for c(s) maxima (Fig. 3) to calculate f/f0, S20,w, and RH.
The mean (μ), SE, and 4.47 SD were calculated from results for three batches of WT protein. For the mutants, values <μ − 4.47 SD (italic) and >μ + 4.47 SD (bold) are indicated.
Analysis of Secondary Structure Content
CD is used to examine protein folding. Molar CD is expressed in units of delta epsilons. CD spectra (Fig. 4) were measured for the monomeric and dimeric forms of the fibulin 5 mutants in the absence and presence of Ca2+, respectively. Figure 4 shows the mean spectrum (black line) and confidence interval (hatched area) calculated from CD spectra for three batches of WT in the absence and presence of Ca2+. At lower wavelengths, the differences between the spectra measured in the absence of Ca2+ (Fig. 4A) were less than those measured in the presence of Ca2+ (Fig. 4B), indicating that Ca2+ interferes with measurements. The minimum in the spectrum measured for S227P in the presence of Ca2+ was shifted right to 212 nm, and lay outside the confidence interval (horizontal bar in Fig. 4B) for the WT minimum (P < 0.05). Deconvolution of the spectra provided estimates of secondary structure content (Table 2) for the monomeric and dimeric forms of the fibulin 5 mutants.
Figure 4.
CD spectra of wild-type (WT) and mutants of fibulin 5 purified by SEC in the absence (A, buffer A) and presence (B, buffer B) of Ca2+. The mean (line) and confidence interval (hatched area) for delta epsilons were calculated from spectra for three batches of WT. (B) The error bar shows the confidence interval for the wavelength of the WT delta epsilon minimum.
Table 2.
Estimates from CD Spectra (Fig. 4) of the Secondary Structure Content of the Fibulin 5 Mutants
| Monomer* |
Dimer† |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NRMSD‡ | Helix (%) | Strand (%) | Turn (%) | Unordered (%) | NRMSD‡ | Helix (%) | Strand (%) | Turn (%) | Unordered (%) | |
| WT§ | ||||||||||
| mean (μ) | 0.074 | 3.7 | 33.3 | 18.0 | 44.3 | 0.090 | 4.0 | 33.7 | 18.3 | 43.3 |
| R54A | 0.031 | 4 | 38 | 18 | 39 | 0.126 | 2 | 36 | 20 | 39 |
| V60L | 0.035 | 4 | 36 | 18 | 40 | 0.076 | 3 | 36 | 18 | 41 |
| R71Q | 0.034 | 6 | 34 | 19 | 40 | 0.065 | 4 | 37 | 18 | 40 |
| P87S | 0.066 | 4 | 34 | 18 | 43 | 0.047 | 4 | 35 | 18 | 43 |
| Q124P | 0.043 | 4 | 39 | 20 | 37 | 0.046 | 4 | 40 | 19 | 36 |
| V126M | 0.056 | 4 | 37 | 17 | 41 | 0.043 | 5 | 34 | 18 | 42 |
| I169T | 0.071 | 4 | 33 | 17 | 44 | 0.083 | 4 | 31 | 20 | 45 |
| G202R | 0.065 | 4 | 38 | 18 | 39 | 0.056 | 4 | 30 | 18 | 47 |
| C217R | 0.051 | 5 | 40 | 18 | 37 | 0.032 | 4 | 37 | 18 | 40 |
| S227P | 0.062 | 4 | 38 | 17 | 40 | 0.050 | 4 | 37 | 19 | 38 |
| G267S | 0.043 | 5 | 34 | 20 | 39 | 0.078 | 4 | 35 | 18 | 42 |
| R351W | 0.050 | 4 | 35 | 16 | 43 | 0.049 | 4 | 34 | 18 | 42 |
| A363T | 0.061 | 4 | 35 | 17 | 43 | 0.100 | 4 | 34 | 18 | 43 |
| G412E | 0.042 | 5 | 37 | 18 | 39 | 0.047 | 4 | 37 | 18 | 40 |
Data for monomeric proteins were obtained in the absence of calcium (buffer A).
Data for dimeric proteins were obtained in the presence of calcium (buffer B).
The normalized root mean square deviation (NRMSD) is a measure of the fit of the data by the CDSSTR software28 using reference set 7.
For comparison, the means (μ) were calculated from results for three batches of WT protein.
Homology Modeling
There are no high-resolution solved structures for any of fibulin 5's domains. However Figure 5A shows a putative arrangement of a fibulin 5 cbEGF based on the UniProt sequence for fibulin 515 and the consensus sequence for class I cbEGFs.29 This arrangement could represent each of fibulin 5 cbEGF-2 to -6 but not the unusual cbEGF-1. In Figure 5A, the positions of mutations associated with cutis laxa and AMD that lie within fibulin 5 cbEGF-2 to -6 are shown on this representation of a single cbEGF.
Figure 5.
(A) Schematic putative arrangement of a normal fibulin 5 cbEGF that is based on the UniProt sequence for fibulin 515 and the consensus sequence for class I cbEGFs,29 and that could represent each of fibulin 5 cbEGF-2 to -6, but not the abnormal cbEGF 1. Disulfide bonds are shown in red. Conserved residues are labeled using one-letter abbreviations. The positions of mutations associated with cutis laxa and AMD that lay within fibulin 5 cbEGFs 2 to 6 are shown on this representation of a single cbEGF. (B, C) Swiss-Model31 used the alignments produced by threading (Supplementary Fig. S3) to generate (B) an alignment of peptide backbones for fibulin 5 cbEGF-4 (green) and fibrillin 1 cbEGF 33 (blue) and therefore (C) a putative homology model for fibulin 5 WT cbEGF 4. Cysteines and disulfide bonds are shown in red with the exceptions of residue C217 (yellow) and the disulfide bond (not shown) that is broken with the C217R mutation. Blue: the position of S227.
The protein fold recognition program Threader 3.5130 assessed whether the primary sequence for fibulin 5 WT cbEGF 4 (residues 206-246) could fold like the NMR structure (1EMN) for the fibrillin 1 cbEGF 33 domain.29 The latter comprises the domain most homologous to fibulin 5 cbEGFs for which a high-resolution structure exists. Threading produced 98% alignment and 39% identity between the two sequences; all six cysteines in fibulin 5 cbEGF 4 were aligned with counterpart cysteines in fibrillin 1 cbEGF-33 (Supplementary Fig. S3). Swiss-Model31 used this alignment to generate a (good) alignment of peptide backbones (Fig. 5B) and hence a putative homology model for fibulin 5 WT cbEGF 4 (Fig. 5C). Figure 5C shows the sites of the residues C217 (yellow) and S227 (blue) that are mutated in cutis laxa. No effect on the alignments or backbone structures was predicted by Swiss-Model31 for the cutis laxa mutants C217R and S227P.
Discussion
Fibulin 5 is a calcium-binding protein of critical importance in the assembly of elastic fibers. Mutations in fibulin 5 have been shown to cause cutis laxa11–13,32 and have been associated with AMD.9,10 We used a combination of biophysical techniques to detect structural changes in these variants to help to determine the pathogenicity of the AMD-associated variants.
Results indicate that the two cutis laxa mutants (C217R and S227P) introduce differences in protein dimerization. C217R dimerizes more than the other mutants in the nonphysiological absence of Ca2+ (Fig. 2A). C217R removes a disulfide bond within cbEGF 4 (Fig. 5C), introduces a bulky charged arginine group, and is likely to reduce protein stability. S227P overdimerized in a Ca2+-mediated reaction (Figs. 2B–D), and may perturb cbEGF 4's consensus calcium-binding sequence (Fig. 5A). This may result in S227P's reduced affinity for tropoelastin and fibrillin microfibrils.33,34 Nonreduced SDS-PAGE showed that the dimerization of all mutants was not mediated by intermolecular disulfide bonds (Supplementary Fig. S2B). The AMD-associated mutant G412E gave the largest monomeric hydrodynamic radius, lower-than-expected mobility during native-PAGE, and greatly increased levels of higher-order self-association in SEC-MALLS experiments in both the presence and absence of Ca2+. Together, these results suggest that G412E aggregates more than the other mutants. The increased dimerization of C217R and S227P and aggregation of G412E may explain their reduced secretion by COS7 cells.9 It has been postulated that poor secretion leads to susceptibility to AMD.9
The three fibulin 5 mutations (R54A, V60L, and R71Q) closest to the integrin binding motif, Q124P and G267S had insignificant effects on the dimerization or aggregation of fibulin 5 in either the presence or absence of Ca2+. With the exceptions of these and G412E, the mutations appear to slightly increase self-association of fibulin 5 in the absence of Ca2+. However, that this property was exhibited by the G202R polymorphism suggests that a slight increase in self-association is not of strong predictive value when determining whether mutations cause disease. However, the G202R polymorphism is functionally different from WT because it increases binding to tropoelastin.34,35
We have presented and discussed CD spectra and estimates of secondary structure contents for WT fibulin 5 in both the absence and presence of Ca2+ previously.16 CD spectra for the mutants (Fig. 4) resemble the mean spectra for three batches of WT acquired in each buffer, providing evidence that the mutants are folded into similar conformations. The spectrum measured for the cutis laxa mutation S227P in the presence of Ca2+ (Fig. 4B) appears to be different, with a minimum that is right-shifted to 212 nm. Although this difference is not supported by calculations of secondary structure content (Table 2), more weight should be placed on the actual data (Fig 4) and less on derived quantities.
Better solvation could explain why the chemically similar fibulin 5 substitutions V126M and G267S migrated farther toward the anode than did the WT during native PAGE (Supplementary Fig. S2A). Methionine and serine are more hydrophilic than the aliphatic residues that they replaced.36 Threading predicted that the mutant G267S probably increases the solvation energy magnitude of fibulin 5.9
If we had performed these studies without the results of previous genetic studies, then we would have concluded that the two cutis laxa mutations and G412E are likely to be pathogenic. Although it remains uncertain as to whether G412E causes AMD, it is clear that our strategy detected differences for the two disease-causing cutis laxa mutations and was therefore of positive predictive value. Although the other AMD-associated mutants show no gross structural differences, the pathogenic mechanism of AMD may originate from the effects of subtle differences over a lifetime. Further, we cannot exclude the possibility that the AMD-associated mutants are pathogenic via differences outside the scope of this study—for example, intracellular effects or the disruption of intermolecular interactions.
Supplementary Material
Acknowledgments
The authors thank Adam Huffman (Department of Bioinformatics), Paul Fullwood (DNA Sequencing), Jemima Whyte and Stuart Cain (Faculty of Life Sciences), and Stephen Roberts (Faculty of Medical and Human Sciences), University of Manchester; Michael Harrison, University of Leeds, Julian Hanak (Renovo, Manchester, UK); Clive Kay (Vutrix, Weston Super Mare, UK); and Richard K. Jones.
Footnotes
Supported by Wellcome Trust Grants 123364 (DT, AJL, PNB CMK, RPOJ, CR), 072291 (CB, MCW) and VIP award (NB, RPOJ & DT); and the National Institutes of Health Research (NIHR) Manchester Biomedical Research Centre.
Disclosure: R.P.O. Jones, None; C. Ridley, None; T.A. Jowitt, None; M.-C. Wang, None; M. Howard, None; N. Bobola, None; T. Wang, None; P.N. Bishop, None; C.M. Kielty, None; C. Baldock, None; A.J. Lotery, None; D. Trump, None
References
- 1.Christen WG, Glynn RJ, Manson JE, Ajani UA, Buring JE. A prospective study of cigarette smoking and risk of age-related macular degeneration in men. JAMA 1996;276:1147–1151 [PubMed] [Google Scholar]
- 2.Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA 1996;276:1141–1146 [PubMed] [Google Scholar]
- 3.Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet 2006;38:458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards AO, Ritter R, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science 2005;308:421–424 [DOI] [PubMed] [Google Scholar]
- 5.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A 2005;102:7227–7232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005;308:419–421 [DOI] [PubMed] [Google Scholar]
- 7.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005;308:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet 2005;14:3227–3236 [DOI] [PubMed] [Google Scholar]
- 9.Lotery AJ, Baas D, Ridley C, et al. Reduced secretion of fibulin 5 in age-related macular degeneration and cutis laxa. Hum Mutat 2006;27:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone EM, Braun TA, Russell SR, et al. Missense variations in the fibulin 5 gene and age-related macular degeneration. N Engl J Med 2004;351:346–353 [DOI] [PubMed] [Google Scholar]
- 11.Elahi E, Kalhor R, Banihosseini SS, et al. Homozygous missense mutation in fibulin-5 in an Iranian autosomal recessive cutis laxa pedigree and associated haplotype. J Investig Dermatol 2006;126:1506–1509 [DOI] [PubMed] [Google Scholar]
- 12.Fischer J, Jobard F, Oudot T, et al. Identification of novel mutations in elastin and fibulin 5 in three patients affected with cutis laxa. Presented at the Third European Symposium on Elastin.Manchester, UK, June 30–July 3, 2004 [Google Scholar]
- 13.Loeys B, Van Maldergem L, Mortier G, et al. Homozygosity for a missense mutation in fibulin-5 (FBLN5) results in a severe form of cutis laxa. Hum Mol Genet 2002;11:2113–2118 [DOI] [PubMed] [Google Scholar]
- 14.Timpl R, Sasaki T, Kostka G, Chu ML. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol 2003;4:479–489 [DOI] [PubMed] [Google Scholar]
- 15.Reviewed US-P Q9UBX5 (FBLN5_HUMAN). http://wwwuniprotorg/uniprot/Q9UBX52009.
- 16.Jones RPO, Wang MC, Jowitt TA, et al. Fibulin 5 forms a compact dimer in physiological solutions. J Biol Chem 2009; 284(38):25938–25943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vega S, Iwamoto T, Yamada Y. Fibulins: Multiple roles in matrix structures and tissue functions. Cell Mol Life Sci 2009;66(11–12):1890–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowal RC, Richardson JA, Miano JM, Olson EN. EVEC, a novel epidermal growth factor-like repeat-containing protein upregulated in embryonic and diseased adult vasculature. Circ Res 1999;84:1166–1176 [DOI] [PubMed] [Google Scholar]
- 19.Nakamura T, Ruiz-Lozano P, Lindner V, et al. DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J Biol Chem 1999;274:22476–22483 [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Lozano PR, Ikeda Y, et al. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature 2002;415:171–175 [DOI] [PubMed] [Google Scholar]
- 21.Yanagisawa H, Davis EC, Starcher BC, et al. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 2002;415:168–171 [DOI] [PubMed] [Google Scholar]
- 22.Freeman LJ, Lomas A, Hodson N, et al. Fibulin-5 interacts with fibrillin-1 molecules and microfibrils. Biochem J 2005;388:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomas AC, Mellody KT, Freeman LJ, Bax DV, Shuttleworth CA, Kielty CM. Fibulin-5 binds human smooth-muscle cells through alpha5beta1 and alpha4beta1 integrins, but does not support receptor activation. Biochem J 2007;405:417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon WJ, Massey FJ. Introduction to Statistical Analysis New York: McGraw Hill; 1983;:44–152, 400,–401, 534 [Google Scholar]
- 25.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 2000;78:1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes D, Laue T, Philo J. Program Sednterp: sedimentation interpretation program. Durham, NH: University of New Hampshire; 1995. [Google Scholar]
- 27.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 2004;32:W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Compton LA, Johnson WC. Analysis of protein circular-dichroism spectra for secondary structure using a simple matrix multiplication. Anal Biochem 1986;155:155–167 [DOI] [PubMed] [Google Scholar]
- 29.Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, Handford PA. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 1996;85:597–605 [DOI] [PubMed] [Google Scholar]
- 30.Jones DT, Tress M, Bryson K, Hadley C. Successful recognition of protein folds using threading methods biased by sequence similarity and predicted secondary structure. Proteins Struct Funct Genet 1999(suppl 3);104–111 [DOI] [PubMed] [Google Scholar]
- 31.Arnold K, Bordoli L, Kopp J, Schwede T. The Swiss-Model workspace: a web-based environment for protein structure homology modelling. Bioinformatics 2006;22:195–201 [DOI] [PubMed] [Google Scholar]
- 32.Markova D, Zou Y, Ringpfeil F, et al. Genetic heterogeneity of cutis laxa: a heterozygous tandem duplication within the fibulin-5 (FBLN5) gene. Am J Hum Genet 2003;72:998–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Q, Loeys BL, Coucke PJ, et al. Fibulin-5 mutations: mechanisms of impaired elastic fiber formation in recessive cutis laxa. Hum Mol Genet 2006;15:3379–3386 [DOI] [PubMed] [Google Scholar]
- 34.Choudhury R, McGovern A, Ridley C, et al. Differential regulation of elastic fiber formation by fibulins-4 and -5. J Biol Chem 2009;284(36):24553–24567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu QR, Reymond JL, Pinel N, Zabot MT, Urban Z. Inflammatory destruction of elastic fibers in acquired cutis laxa is associated with missense alleles in the elastin and fibulin-5 genes. J Investig Dermatol 2006;126:283–290 [DOI] [PubMed] [Google Scholar]
- 36.Mathews CK, Van Holde KE. Biochemistry Reading, MA: Benjamin/Cummings. 1990:xxix, 1129 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.