Abstract
Nitric oxide synthase (NOS) enzymes synthesize nitric oxide, a signal for vasodilatation and neurotransmission at low levels, and a defensive cytotoxin at higher levels. The high active-site conservation among all three NOS isozymes hinders the design of selective NOS inhibitors to treat inflammation, arthritis, stroke, septic shock, and cancer. Our structural and mutagenesis results identified an isozyme-specific induced-fit binding mode linking a cascade of conformational changes to a novel specificity pocket. Plasticity of an isozyme-specific triad of distant second- and third-shell residues modulates conformational changes of invariant first-shell residues to determine inhibitor selectivity. To design potent and selective NOS inhibitors, we developed the anchored plasticity approach: anchor an inhibitor core in a conserved binding pocket, then extend rigid bulky substituents towards remote specificity pockets, accessible upon conformational changes of flexible residues. This approach exemplifies general principles for the design of selective enzyme inhibitors that overcome strong active-site conservation.
Keywords: nitric oxide synthase, isozyme-specific inhibitor, induced-fit, x-ray crystallography, quinazoline, aminopyridine, drug design
Nitric oxide (NO) is a small, diffusible, and transient molecule produced from amino acid L-arginine (L-Arg) by three nitric oxide synthase (NOS) enzymes1,2. The endothelial (eNOS) and neuronal (nNOS) NOS isozymes are constitutively expressed and Ca2+ regulated to provide NO for signaling, including vasodilatation, thermoregulation, neuroprotection, and endocrine function. The Ca2+-insensitive inducible NOS isozyme (iNOS) is expressed in response to cytokines or pathogens, and produces NO at a high rate to kill bacteria, viruses, and tumor cells. Insufficient NO bioavailability from eNOS and nNOS is associated with hypertension, impotence, atherosclerosis and cardiovascular disease, while excess NO from iNOS has been implicated in inflammation, rheumatoid arthritis, inflammatory bowel disease, immune-type diabetes, stroke, cancer, thrombosis, and infection susceptibilities3,4. Overproduction of NO by iNOS (and nNOS) has also been linked to neurodegenerative disorders including Parkinson’s and Alzheimer’s diseases, as well as multiple sclerosis5. Thus, the development of iNOS-specific inhibitors is highly desirable.
The three NOS isozymes share a common modular architecture and conserved active site. The N-terminal catalytic oxygenase module (NOSox) binds cofactors heme and (6R)-5,6,7,8-tetrahydro L-biopterin (H4B), substrate L-Arg, and a structural zinc ion across the dimer interface6–15. Upon calmodulin binding16, NOSox accepts electrons from the C-terminal reductase module17–19. The nearly complete amino acid conservation and structural similarity among the three NOS isozymes6–15 active sites presents a significant challenge for the design of isozyme-specific inhibitors20,21. Moreover, as NO availability is controlled at the synthesis level for signaling or cytotoxicity, NOS isozymes are a paradigmatic system to address the challenges of designing isozyme-specific inhibitors despite conserved binding pockets.
NOS inhibitors selective for iNOS are rare, and commonly exhibit only limited selectivity or significant toxicity22–25. In contrast, quinazoline26 (1–5) and aminopyridine27,28 (6–12) inhibitors possess good in vitro potency and selectivity for iNOS. In particular, the spirocyclic quinazoline (AR-C102222, 3, Fig. 1) shows excellent selectivity over eNOS (3000-fold), and exhibits significant protective, anti-inflammatory and antinociceptive activities in rodent models of adjuvant-induced arthritis, pancreatitis29, neuropathy, inflammation, and post-surgical pain30. Thus, we have chosen to focus our structural studies on quinazoline and aminopyridine inhibitors.
Figure 1. NOS inhibitors structures, inhibition and crystallographic data.
For all inhibitors, including quinazolines (left column: compounds 1–5), aminopyridines (middle column: compounds 6–12) and bicyclic thienooxazepines (right column: compounds 14–16), the chemical structure is shown in black (core with red cis-amidine nitrogens) and magenta (tail), together with IC50 values in the three human NOS isozymes. The resolution (d in Å), crystallographic R and Rfree values are indicated for each structure of murine iNOSox (unlabeled), human iNOSox (hiNOS), bovine eNOSox (beNOS) and human eNOSox (heNOS) complexes.
Here, we combined mutagenesis, biochemistry, crystallography, and drug design to elucidate the structural basis for the iNOS selectivity of some quinazoline and aminopyridine inhibitors. We demonstrate that plasticity of an isozyme-specific triad of residues distant from the active site modulates conformational changes of invariant residues nearby the active site to determine the exquisite selectivity of these inhibitors for iNOS. We design novel potent and selective iNOS inhibitors by applying an “anchored plasticity approach” (Supplementary Fig. 1 online). Selective inhibitors are designed with an inhibitor core anchored in a conserved binding pocket, and rigid bulky substituents that extend to remote specificity pockets accessible upon conformational changes of “plastic” protein residues. Fundamentally, this anchored plasticity approach is broadly applicable to the discovery of novel inhibitors against enzyme families with strong active-site conservation.
RESULTS
Inhibitor binding to iNOSox
Quinazoline (1–2), spirocyclic quinazoline (3–5), and aminopyridine (6–12) inhibitors are potent (IC50 from 10 nM to 1.2 μM) and selective (2.7- to 3000-fold) inhibitors for iNOS over eNOS and nNOS (Fig. 1 and Supplementary Table 1 online). These inhibitors share a cis-amidine derived core, but have different substituents or tails. To determine the basis for the exquisite iNOS potency of these inhibitors, we solved x-ray structures of murine iNOSox bound to compounds 1–12 and of human iNOSox bound to aminopyridine 9 (Methods).
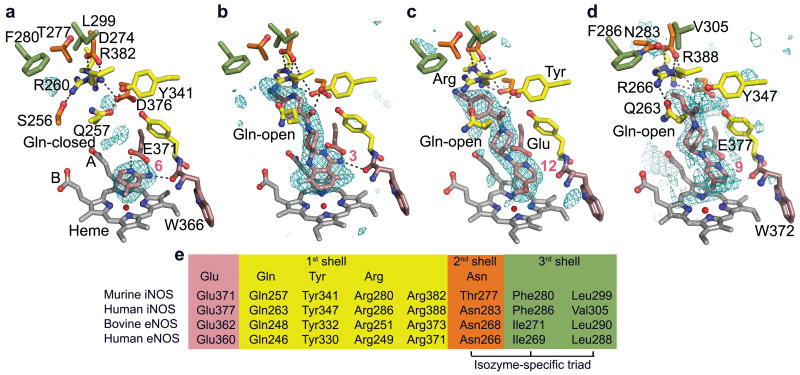
Inhibitors 1–5 and 6–12 belong to different chemotypes but all bind similarly in the iNOS active-site heme pocket (Fig. 2a–d, Supplementary Fig. 2 online). The NOSox active site is lined by the heme, invariant Glu (Glu371/377; murine/human iNOS numbering, respectively) and backwall residues (363–366/369–372). In all these inhibitor complexes, the cis-amidine moiety mimics the guanidinium group of substrate L-Arg, by making bidentate hydrogen bonds to Glu and stacking with the heme. Compounds 1–8 make an extra hydrogen bond to the main-chain carbonyl of invariant Trp366/372 and pack more parallel to the heme than compounds 9–12 (Supplementary Results). The bulky and rigid tails of compounds 2–5 and 9–12 all extend above heme propionate A and pack with invariant residues Gln (Gln257/263), Arg (Arg260/266), Pro344/350, Ala345/351 (not shown in Fig. 2), and Arg382/388. Hydrogen bonds tether the extended inhibitor tails to invariant Tyr (Tyr341/347), and either Arg382/388 (compound 2) or a water molecule (compounds 3–5 and 12). Our structural analysis thus suggests that both interactions of the inhibitor core with active-site residues and of the inhibitor tail with residues outside the active-site heme pocket mediate inhibitor binding.
Figure 2.
Quinazoline and aminopyridine binding in iNOSox and eNOSox. (a) Potent but non-selective aminopyridine compound 6 (ref. 28) bound to murine iNOSox. (b) Highly-selective quinazoline compound 3 (ref. 26) bound to murine iNOSox. (c) Selective aminopyridine compound 12 (ref. 28) bound to murine iNOSox. (d) Aminopyridine 9 (ref. 28) bound to human iNOSox. For all structures, critical hydrogen bonds (dots) and iNOS residues are shown: active-site residues (peach), first-shell residues (yellow, residues interacting directly with the inhibitor), second-shell residues (orange, residues interacting with first-shell residues) and third-shell residues (green, residues interacting with second-shell residues). The Fo-Fc electron density map contoured at 3 σ (blue mesh) is shown around each inhibitor (pink). (e) Key iNOS residues involved in inhibitor binding are colored according to a–d.
To determine the roles of residues key to inhibitor binding, we measured the binding affinity and inhibitor potency of moderately selective compound 9 for several human iNOS mutant proteins (Table 1). Mutation of active site Glu into Ala had the most dramatic effect (KD = 0.4 μM for wild type vs. ≫ 100 μM for E377A mutant), thus revealing the crucial role of this invariant charged side chain in inhibitor, as well as substrate31, binding. The close match between binding affinity (KD) in iNOSox and inhibitory potency (IC50) in full-length iNOS (Table 1) suggests that enzyme inhibition data reflects true binding affinity. Both Gln mutations (Q263A, loss of side chain and Q263N, smaller side chain with similar functionality) result in slightly decreased iNOS affinity for compound 9 (3- and 5-fold, respectively), thus corroborating the role of Gln in inhibitor tail binding. The human iNOS Y347F/Y373F double mutant displayed a ~10-fold decrease in inhibitor potency, thus revealing a key role for the Tyr hydrogen bond to the carbonyl of the inhibitor tail. Our combined structural and mutagenesis results thus suggest that heme stacking, as well as hydrogen bonds and hydrophobic interactions within and outside the active site, all significantly contribute to the binding of these inhibitors to iNOS.
Table 1.
Binding and inhibition constants for wild-type and mutant human iNOS and eNOS proteins
| Table 1. Binding and inhibition constants for iNOS and eNOS proteins with compound 9 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| KD (μM)a | IC50 (μM)b | ||||||||
|
| |||||||||
| iNOSox wt | iNOSox E377A | iNOSox F286I/V305L | iNOS wt | eNOS wt | iNOS Q263A | iNOS Q263N | eNOS Q246A | eNOS Q246N | iNOS Y347F/Y373F |
| 0.4 | ≫100 | ≫100 | 0.35 | >100 | 1.0 | 2.0 | 56 | >100 | 3.4 |
KD measured in human iNOSox wild-type (wt), E377A, and F286I/V305L mutant proteins
IC50 measured for full-length human iNOS and eNOS wild-type (wt) and mutant proteins
A novel Gln specificity pocket in iNOSox
Bulky inhibitors promote a cascade of conformational changes up to 20 Å away from the iNOSox active site, resulting in the creation of a new pocket. The comparison of our iNOS x-ray structures with small (compounds 1, 6–7) and large-tailed (2–5 and 8–12) inhibitors reveals similar overall protein structures, with all inhibitor cores anchored in the active-site heme pocket (Fig. 2a–d and Supplementary Figs. 2 and 3 online). Yet, outside this pocket, invariant first-shell and second-shell residues adopt different side-chain conformations in these complexes (Fig. 2a–e, Supplementary Fig. 3, Supplementary Results online). To prevent collision with bulky inhibitors, the first-shell Gln side chain rotates around its χ1 and χ2 torsion angles from a “Gln-closed” position with hydrogen bond to Tyr, to a “Gln-open” position with hydrogen bonds to Arg. Similarly, the Arg side chain rotates closer to second-shell residues Asp274/280 and Asn (Thr277/Asn283). Finally, side-chain rotation of Arg382/388 closer to Asp376/382 enhances hydrophobic interactions with the inhibitor tail.
Upon binding of bulky inhibitors, the coupled rotations of first-shell Gln and Arg initiate a cascade of conformational changes, which further propagate to second-shell residues. In the human iNOS Gln-open conformation, the conformational change of first-shell Arg induces rotation of second-shell Asn towards third-shell Phe286 and Val305 (Fig. 2d, Supplementary Fig. 4 and Supplementary Movie 1 online). Thus, the conformational plasticity of human iNOS second-shell Asn allows coordinated movements of first-shell Gln and Arg.
The correlated side-chain rotations of Gln, Arg, and Arg382/388 to accommodate the rigid bulky tails of compounds 2–5 and 9–12 expose a new specificity pocket for enhanced inhibitor binding in iNOS. This “Gln specificity pocket” extends from the active-site heme pocket (Fig. 3a) and is lined by residues Gln, Arg, Trp340/346, Tyr, Pro344/350, Ala345/351, Tyr367/373, Asp376/382, and Arg382/388 (Fig. 2b–d). All residues forming the iNOS Gln specificity pocket are strictly conserved among NOS isozymes, with one exception: iNOS Asp376/382, which hydrogen bonds to Arg382/388, is replaced by Asn in eNOS (Supplementary Fig. 5 online). Interestingly, all previously reported NOSox structures present the “Gln-closed” conformation7,8,11–15 or a disordered Gln conformation10. We thus conclude that the Gln-open conformation and associated cascade of conformational changes leading to the opening of the novel Gln specificity pocket are favored or induced by the binding of quinazoline and aminopyridine inhibitors bearing a rigid and extended tail.
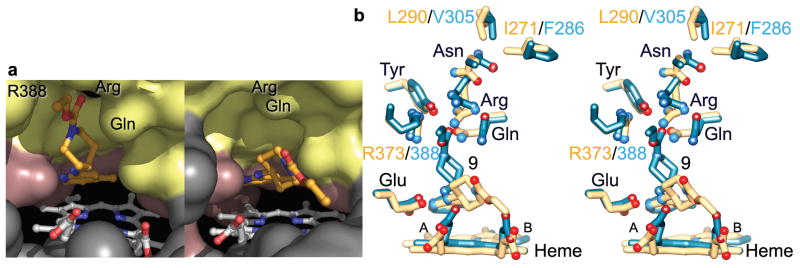
Figure 3.
Selective aminopyridine compound 9 binding to eNOS versus iNOS. (a) Solvent-accessible surfaces for the iNOS (left) and eNOS (right) active sites colored according to Fig. 2. The core of compound 9 binds closer and more parallel to the heme in eNOS. In iNOS, side-chain rotations of Gln, Arg, and Arg388 open the Gln specificity pocket for binding of the bulky inhibitor tail. (b) Stereoview of the superimposition of bovine eNOS:compound 9 (yellow) and human iNOS:compound 9 (blue) x-ray structures, highlighting the cascade of conformational changes of first-shell and second-shell residues upon inhibitor binding to iNOS.
Isozyme differences in inhibitor binding
In iNOSox and eNOSox, the binding modes for moderately selective aminopyridine 9 are dramatically different despite common overall protein structures and active sites. The inhibitor aminopyridine cores bind similarly in the active-site heme pocket of both isozymes (Fig. 3a–b and Supplementary Fig. 6 online). In eNOS, the bidentate hydrogen bonds to active-site Glu anchor the aminopyridine core almost parallel to the heme plane and place the inhibitor bulky tail between the eNOSox heme propionates. Invariant Tyr and Arg hydrogen bond to Gln, thus preventing its hydrophobic interaction with the inhibitor tail (Fig. 3b and Supplementary Fig. 6a online). As a consequence, the eNOS complex with compound 9 exhibits the Gln-closed conformation and the Gln specificity pocket is not observed (Fig. 3a). In contrast, the iNOS complex with compound 9 exhibits the Gln-open conformation allowing the inhibitor tail to bind in the Gln specificity pocket (Fig. 3b and Supplementary Fig. 6b online). Not only first-shell Gln and Arg, but also second-shell Asn, present different conformations in the two complexes.
What prevents the Gln-closed to Gln-open conversion and the opening of the Gln specificity pocket in eNOS? First, we propose that Gln gates the Gln specificity pocket and must adopt the “Gln-open” side-chain conformation to allow inhibitor access. Mutation of Gln into Ala (Q246A, loss of side chain) only marginally enhanced inhibitor potency of compound 9 for eNOS, but did not achieve the potency observed for wild-type iNOS (Table 1). The Q246N mutation had no effect. Thus, removal or rotation of the Gln gate is important, but not sufficient, for potent compound 9 binding to eNOS. Second, we followed, in eNOS, the cascade of conformational changes observed in iNOS upon binding of bulky inhibitors (Fig. 3b). An iNOS-like binding mode for compound 9 in eNOS would induce conformational changes of first-shell Gln and Arg, and second-shell Asn (Fig. 4). However, in eNOS, bulky Leu290 and β-branched rigid Ile271 in the third shell block the side-chain rotation of second-shell Asn. Consequently, this Asn conformation prevents the conformational changes of Gln and Arg necessary for the opening of the Gln specificity pocket (Figs. 3 and 4). The triad of second-shell (Asn) and third-shell (Leu and Ile) residues are the only nearby residues that are not conserved among NOS isozymes (Fig. 2e and Supplementary Fig. 5 online). To test our hypothesis for the key role of third-shell isozyme-specific residues, we made the human iNOSox F286I/V305L double mutant to mimic the corresponding eNOSox residues. Binding affinity of this mutant iNOSox enzyme for compound 9 dramatically dropped to beneath detection levels, as observed for wild-type eNOS (Table 1), while binding of non-selective inhibitor 6 was unaffected (not shown). Furthermore, the x-ray structure of the human iNOSox double mutant co-crystallized with excess compound 9 reveals a Gln-closed conformation and the absence of bound inhibitor (Supplementary Fig. 7 online). Our results thus demonstrate the crucial role of third-shell isozyme-specific residues in inhibitor binding, and provide a structural basis for the exquisite iNOS-specificity of large-tailed quinazoline and aminopyridine inhibitors.
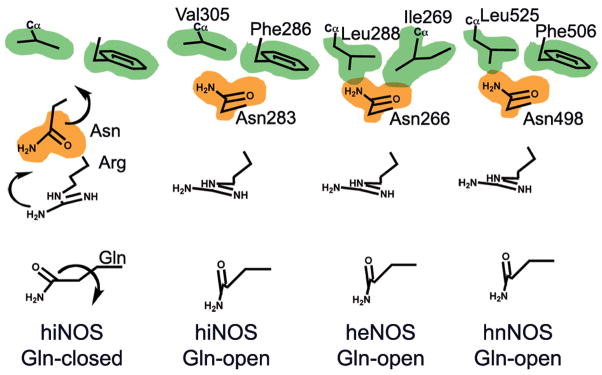
Figure 4.
Isozyme-specific induced-fit upon inhibitor binding. Schematic of the cascade of conformational changes associated with inhibitor binding in the three human NOS isozymes. The van der Waals surfaces for the isozyme-specific triads are shown in orange (second shell) and green (third shell). In human iNOS (hiNOS), inhibitor binding first induces the Gln-closed to Gln-open conformation and Arg rotation, which in turn leads to rotation of second-shell Asn towards third-shell Phe286 and Val305. In human eNOS (heNOS), bulkier third-shell residues (Ile269 and Leu288) prevent the Asn rotation (overlap of van der Waals surfaces). In human nNOS (hnNOS), partial rotation of Asn towards third-shell residues Phe506 and bulky Leu525 may be possible.
Determinants for inhibitor selectivity
Based on our combined structural and mutagenesis analyses, we propose that differences in the plasticity of second- and third-shell residues between iNOS and eNOS modulate conformational changes of invariant first-shell residues to determine inhibitor selectivity. Together, our mutational and structural results suggest that the Gln specificity pocket accounts for the excellent iNOS-selectivity of the bulky aminopyridine and quinazoline inhibitors. In turn, opening of this pocket depends not only on conformational changes of invariant first-shell Gln and Arg, but also on the plasticity of isozyme-specific second-shell Asn (Fig. 4). This hypothesis is supported by several observations. First, the potent small NOS inhibitors (1, 6–7), which do not induce the Gln-open conformation, show poor selectivity for iNOS27,28 (Figs. 1 and 2 and Supplementary Fig. 2 online). Second, the bulky, but less rigid, tail of compound 8, which neither hydrogen bonds to Tyr nor induces Arg side-chain rotation (Supplementary Fig. 2 online), binds less deeply in the Gln pocket and exhibits only modest selectivity for iNOS32. Third, eNOS third-shell residues Ile271 and Leu290 block binding of compound 9 in the Gln pocket, as evidenced by our structural and mutagenesis results on the human iNOS double mutant (Table 1 and Supplementary Fig. 7 online). Fourth, bulky quinazoline and aminopyridine inhibitors present moderate selectivity against nNOS (7- to 80-fold more selective for iNOS; Fig. 1). We predict that inhibitor binding in nNOS will induce similar side-chain rotations for first-shell Gln and Arg, and partial rotation of second-shell Asn towards third-shell Phe506 and Leu525 (Fig. 4). The substitution of small Val305 in human iNOS with bulkier Leu525 in human nNOS will likely restrict side-chain rotation of second-shell Asn. We thus conclude that the plasticity of the isozyme-specific triad tunes the inhibitor selectivity by controlling the conformational changes of invariant first-shell Gln and Arg and the formation of the new Gln specificity pocket that can be effectively used for inhibitor binding.
Our results on NOS support an anchored plasticity approach for the design of selective inhibitors. Given a protein of known structure, a set of matching protein sequences (from different species or isoforms), and a binding pocket for a common class of ligand (substrate, cofactor, inhibitor, metabolite, etc…), we propose the following procedure for selective inhibitor design: 1) Identify anchor points for binding in the conserved pocket; 2) Locate variations in sequence and structure outside this pocket; 3) Delineate pathways connecting anchor points to variations (using solvent-accessible channels, for example). 4) Design selective inhibitors that incorporate both a core for anchored binding and extended rigid substituents oriented to exploit protein plasticity along pathways leading to variations (Supplementary Fig. 1 online). The core provides binding affinity via anchoring in nonspecific binding pockets, while the extended substituents determine inhibitor selectivity. Fundamentally, this anchored plasticity approach does not necessarily require serendipitous identification of isoform-specific residue movement. Furthermore, it is readily applicable to key enzyme families, such as kinases, that exhibit overlapping specificities.
Design and synthesis of selective iNOS inhibitors
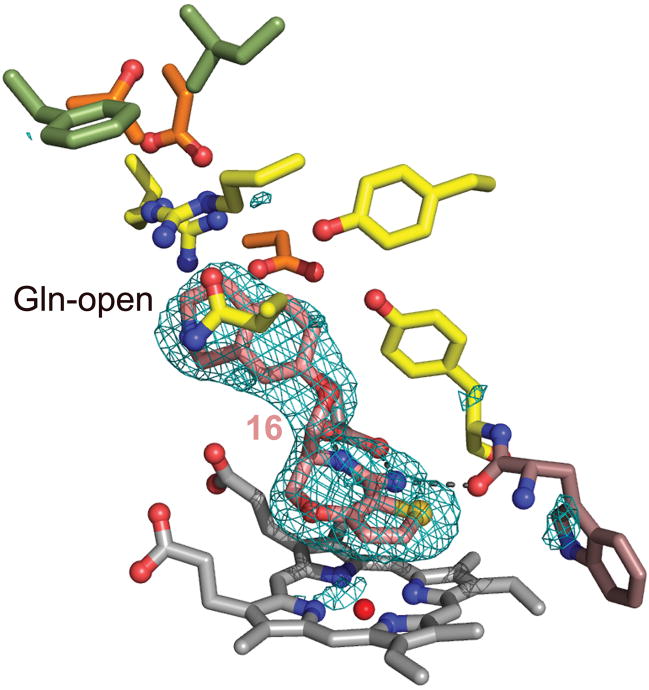
Our combined results allowed us to propose the anchored plasticity approach for the design of specific inhibitors exploiting conserved binding sites coupled to distant isozyme-specific residues via cascades of conformational changes. This approach differs from other methods to design novel NOS inhibitors that only exploit differences in first-shell residues15,33,34. To test the applicability of our results, we rationally designed potent and selective iNOS inhibitors starting from a novel and unexploited template with a 5,7-fused heterobicyclic amidine core (compound 13; Supplementary Methods online). This inhibitor is potent but not selective (IC50 = 0.2 μM for iNOS and eNOS and 0.07 μM for nNOS). Addition of small substituents (compounds 14–15) increases the potency but does not significantly affect selectivity (Fig. 1). In contrast, addition of the bulkier and rigid isoquinolinyloxy-methyl tail (compound 16) results in a significant increase in potency and selectivity for iNOS over eNOS35 (Fig. 1). We determined the x-ray structures of murine iNOSox bound to compounds 14 and 16, and of human eNOSox bound to compound 15 (Fig. 5 and Supplementary Fig. 8 online). In all structures, the inhibitor core packs above the heme and makes bidentate hydrogen bonds to invariant active-site residue Glu and to Trp366 main-chain carbonyl. As seen for bulky quinazoline (2–5) and aminopyridine (9–12) compounds, the extended tail of compound 16 packs with first-shell residues Gln, Arg, Pro344, Ala345, and Arg382, and induces the Gln-open conformation (Fig. 5). These results thus demonstrate the applicability of our anchored plasticity approach for the design of novel potent and selective NOS inhibitors.
Figure 5.
Bicyclic thioenooxazepine inhibitor binding in iNOSox. Moderately selective compound 16 binds to murine iNOSox similarly to bulky quinazoline and aminopyridine inhibitors and induces the Gln-open conformation. Residues are colored according to Fig. 2. The Fo-Fc electron density map contoured at 3 σ (blue mesh) is shown around the inhibitor (pink).
The combined inhibitor screening, structural, and mutagenesis results on iNOS and eNOS provide new insights for structure-based design of selective inhibitors. In iNOS, but not in eNOS, binding of inhibitors bearing an extended rigid tail is associated with a cascade of adaptive conformational changes, beginning with movements of invariant first-shell residues and leading to the opening of the novel Gln specificity pocket for enhanced potency and selectivity. The conformational changes reveal a specificity pocket that is separate from an otherwise conserved active-site heme pocket and not observed in previously determined NOSox x-ray structures. Indeed, others had predicted that NOS inhibitors larger than L-Arg would perturb the hydrogen bonding network with Tyr and Gln and extend into the substrate access channel20,21, in distinct contrast to our results. Here, we show that an isozyme-specific triad of second-shell (Asn) and third-shell residues in NOS tunes the plasticity of invariant first-shell residues (Gln and Arg), and thus determines the exquisite selectivity (125- to 3000-fold) of the long-tailed aminopyridine, quinazoline and bicyclic thienooxazepine inhibitors for iNOS over eNOS. The most selective spirocyclic quinazoline compound 3 (ref. 26) and aminopyridine compound 12 (ref. 28 and this study) also show good potency in whole cells and in vivo activity assays (Fig. 1; Supplementary Table 2 and Fig. 9 online). Further studies will be required to test the in vivo potency of the bicyclic thioenooxazepine compounds. Nevertheless, these highly selective NOS inhibitors are promising tools to investigate specific iNOS-mediated effects both in vivo and in vitro. More specifically, these results on iNOS and eNOS inhibitor structures can be applied to future inhibitor design for the treatment of inflammation, cancer, and other diseases, while reducing the risks of disrupting the crucial activity of eNOS in maintaining blood pressure.
DISCUSSION
The opening of an isozyme-specific pocket in iNOS results from permitted conformational changes of conserved first-shell and second-shell residues upon inhibitor binding. This selective induced-fit movement depends upon plasticity differences in conserved residues located far from the substrate-binding pocket. Exploiting such changes in flexibility to improve inhibitor potency is likely applicable to other key enzymes systems with overwhelmingly conserved active sites, including HIV reverse transcriptase36, aldose reductase37, cyclooxygenases38,39 and kinases40. In NOS, isozyme-specific second-shell and third-shell residues influence the plasticity of invariant first-shell residues, and thus determine the isozyme selectivity. In all these enzyme families, the new binding pocket is distinct from the active site and becomes accessible after adaptive conformational changes of conserved residues. Whether binding of the inhibitor induces the new conformation (induced-fit), or the inhibitor simply selects from different protein conformations in equilibrium (conformational selection) as seen in antigen recognition41, the result is an improved protein-inhibitor interaction.
Our results for prototypic NOS isozymes appear generally applicable to understanding and tuning binding affinity and specificity of enzyme inhibitors. Our structures demonstrate that differential residue plasticity can be exploited for conformational changes that create new specificity pockets suitable for the design of isozyme-specific inhibitors. Together, these results have exciting implications for drug discovery, and demonstrate that x-ray crystallography is crucial for revealing subtle, but important, differences in residue plasticity between closely related isozymes (e.g. iNOS vs. eNOS) or between homologous enzymes from different organisms. The significant roles of second- and third-shell residues in determining the plasticity of conserved first-shell residues will add to the existing challenges of accurately modeling induced-fit in proteins. Here, we show that systematic structural results combined with mutagenesis can identify selectivity-determining side-chain differences distant from the active site, thus overcoming the barriers that active-site conservation poses for isozyme-specific drug design.
METHODS
Expression and purification of NOSox proteins
Murine iNOSox Δ65 (residues 66–498) and human eNOSox (65–492, homologous to murine iNOSox Δ78) were expressed and purified as described42,12. Bovine eNOSox (53–492; homologous to murine iNOSox Δ65) was obtained via trypsinolysis of holo-eNOS, which was expressed and purified as published43. Human iNOSox wild-type and mutant constructs (82–508; homologous to murine iNOSox Δ78) were expressed and purified as described10 with slight modifications. Wild-type and mutant human iNOSox proteins were produced in a pT7 Escherichia coli expression vector based on pET-11a vector (Novagen). BL21(DE3) cells (Stratagene) were grown in the presence of ampicillin at 37 °C until reaching a cell density corresponding to A600 of 0.5–0.8. The culture was then induced with 0.5 mM IPTG, 6.125 mg l−1 ferric citrate and 450 μM ∂-amino levulinic acid, and grown for three days at 20 °C before harvesting. Pellets were re-suspended in buffer A (10 mM sodium phosphate pH 7.0, 0.1 M NaCl, 1 mM L-Arg, 2 mM DTT, 10 μM H4B), sonicated extensively and subsequently loaded on a heparin column (GE Healthcare). Protein was eluted in a single step by adding 0.3 M NaCl to buffer A. Fractions containing NOSox were concentrated, aliquoted and stored at −80 °C.
Mutagenesis
Mutations were introduced into the cDNAs of human iNOSox and full-length NOS within expression vectors by using the QuickChange™ Site-Directed Mutagenesis Kit (Stratagene). Mutagenic oligonucleotides were designed according to the manufacturer’s instructions and mutations confirmed by sequencing.
Synthesis of Compounds 1–16
Compounds 1–16 were prepared as described35,26,28. Details of the synthesis for compounds 13–16 are described in Supplementary Methods online.
Binding affinity, inhibitory potency and in vivo activity
The inhibitory potency (IC50) was determined in full-length wild-type and mutant NOS in the presence of cofactors (5 μM FAD, 5 μM FMN, 200 μM BH4, 1 mM CaCl2, 25 ng ml−1 CaM) and substrates (3 μM [3H]-arginine, 1 mM NADPH) as described26. Inhibitors were pre-incubated with NOS proteins in the presence of cofactors and NADPH for one hour before L-Arg addition.
Binding affinity (KD) of human iNOSox for compounds 6 and 9 was measured by imidazole displacement. Briefly, iNOSox was incubated for 2 hours with a matrix of concentrations of imidazole and compound 6 or 9. Samples were scanned by UV-Visible spectroscopy between 270 and 700 nm. Binding affinity was determined by plotting the absorbance difference A428–A396 as a function of the imidazole concentration. The decrease in apparent imidazole affinity as the concentration of compound increased was used to determine the binding affinity for compound 6 or 9.
Compound 12 was further tested for inhibition of NO production in an intact cell assay44 and in a rat model of inflammation via lipopolysaccharide (LPS)-induced nitric oxide production26, as described here. Compound 12 or vehicle was given orally to male conscious rats at time zero. Blood samples were taken after 2, 4 and 6 h. Total plasma nitrite and nitrate concentrations, indicative of NO production, were determined by using the Griess reaction after reduction of nitrate to nitrite by nitrate reductase. Oral administration of compound 12 to rats led to dose-dependent inhibition of elevated plasma NO levels (IC50= 1.8 μM) measured 4 h after LPS administration (Supplementary Fig. 9 online). All in vivo studies were approved by the AstraZeneca Ethical Review Committee and were conducted under licence from the UK Home Office.
Crystallization, data collection and refinement
Murine and human iNOSox, and bovine and human eNOSox were co-crystallized (with 2–5 mM inhibitor) by vapor diffusion as described7,8,10,12. Crystals grew overnight (crystallization pH was 7, 7.2, 6.5, and 6.0 for murine iNOSox, human iNOSox, bovine eNOSox, and human eNOSox, respectively). All crystals were cryo-cooled after transfer to cryoprotectant solution (murine iNOSox: 30 % glycerol, human iNOSox: 100 % MgSO4, bovine eNOSox: 15 % glycerol, human eNOSox: 15 % 2-methyl-2,4-pentanediol) in a cold nitrogen gas stream. Data were collected at 100 K at CHESS beamline F1 (compound 1), SSRL beamlines 7–1 (compounds 2–4, 8–10), 9–1 (compounds 5, 7), and 9–2 (compound 6), MAX-LAB beamline I711 (compounds 14, 16), ESRF beamlines FIP (compound 15) and ID2 (compound 12). All diffraction data sets were processed using DENZO/SCALEPACK programs45. The crystal structures of human iNOSox10 and murine iNOSox7 and human eNOSox10, with ligands, water and cofactors removed, were used as starting models for molecular replacement with AMoRe46 for human iNOSox, murine iNOSox, bovine eNOSox and human eNOSox, respectively. During crystallographic refinement of protein structures, the heme geometry, like the amino acid geometry, is restrained by a set of parameters (bond lengths, angles, dihedrals) derived from small molecules studies and high-resolution protein structures47. These parameters and their associated weights can strongly influence the resulting refined geometries for heme in protein structures48, especially those determined at lower resolution. Hence, direct comparisons of heme distortions from published x-ray structures refined in different ways can be problematic. Parameters from the HICUP database47 were used for refinement of the heme and pterin cofactors (HEC and H4B, respectively). Inhibitors were fit into Sigmaa-weighted49 Fo-Fc electron density maps, which confirmed the expected unmodified chemical structures. Overall structures were obtained by iterative cycles of refinement with CNS50 and manual fitting with O51, Xfit52, or Coot53 (Supplementary Tables 3–5 online). All superimpositions were performed for residues within a sphere of 10 Å around the inhibitor with the CCP4 program LSQKAB54. The r.m.s. deviations were calculated for all superimposed atoms.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank K. Panda and S. Ghosh for preparation of the murine iNOSox and bovine eNOSox proteins used in this study. Part of this work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), Stanford Synchrotron Radiation Laboratory (SSRL), European Synchrotron Research Facility (ESRF), and MAX-lab (Lund University). This work was supported in part by National Institute of Health Grants (E.D.G, D.J.S.), and the Skaggs Institute for Chemical Biology (E.G.).
Footnotes
Accession codes
Coordinates and structure factors were deposited with the Protein Data Bank with the following accession codes: 3E7I (1), 3E6O (2), 3E7T (3), 3E65 (4), 3E6T (5), 3E67 (6), 3E6N (7), 3E68 (8), 3E7M (murine iNOSox with 9), 3E7G (human iNOSox wild type with 9), (human iNOSox F286I/L305V mutant with 9), 3E7S (bovine eNOSox with 9), 3E6L (10), 3EAI (12), 3EBD (14), 3EAH (human eNOSox with 15), 3EBF (16).
Author contributions. E.G., crystallization, x-ray data collection and refinement, mutant design, binding assays, interpretation of structural and biochemical results, and preparation of figures and manuscript. A.S.A., crystallization, x-ray data collection and refinement. R.J.R., crystallization, x-ray data collection and refinement, and interpretation of structural results. M.D.K., mutagenesis, expression and purification of double mutant protein, binding assay, and crystallization. B.R.C., crystallization, x-ray data collection and refinement. G.A. and A.A., cloning, expression and purification of human iNOSox protein used in structural studies. G.A. and D.J.N., biochemical assays on wild type and mutant proteins, experimental design and interpretation of biochemical results. N.P.G., experimental design and interpretation of chemical data. P.J.H., A.C.T., A.M., D.R.C., and S.C., design and synthesis of novel selective inhibitors and interpretation of chemical data. P.R.M., cloning, expression and purification of wild type and mutant full-length enzymes and oxygenase modules used in biochemical assay. S.A.St-G., interpretation of chemical data. D.J.S., preparation of murine iNOSox and bovine eNOSox used for structural studies. A.V.W., experimental design, interpretation of biochemical and structural data, and mutant design. J.A.T., direction and interpretation of structural experiments. E.D.G., direction and interpretation of structural and biochemical results, and preparation of manuscript.
As specified by the author affiliations, G.A., G.A., P.J.H., P.R.M., D.J.N., S.A.St-G., A.C.T., N.P.G., A.M., D.R.C., S.C., A.Å., and A.V.W. were employed by AstraZeneca when involved in this study.
References
- 1.Geller DA, Billiar TR. Molecular Biology of nitric oxide synthases. Cancer and Metastasis Rev. 1998;17:7–23. doi: 10.1023/a:1005940202801. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C. The moving frontier in nitric oxide-dependent signaling. Science STKE. 2004;2004:pe52. doi: 10.1126/stke.2572004pe52. [DOI] [PubMed] [Google Scholar]
- 3.Bian K, Murad F. Nitric oxide (NO)--biogeneration, regulation, and relevance to human diseases. Front Biosci. 2003;8:d264–78. doi: 10.2741/997. [DOI] [PubMed] [Google Scholar]
- 4.Thippeswamy T, McKay JS, Quinn JP, Morris R. Nitric oxide, a biological double-faced Janus--is this good or bad? Histol Histopathol. 2006;21:445–458. doi: 10.14670/HH-21.445. [DOI] [PubMed] [Google Scholar]
- 5.Duncan AJ, Heales SJ. Nitric oxide and neurological disorders. Mol Aspects Med. 2005;26:67–96. doi: 10.1016/j.mam.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Crane BR, et al. The structure of nitric oxide synthase oxygenase domain and inhibitor complexes. Science. 1997;278:425–431. doi: 10.1126/science.278.5337.425. [DOI] [PubMed] [Google Scholar]
- 7.Crane BR, et al. Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- 8.Raman CS, et al. Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center. Cell. 1998;95:939–950. doi: 10.1016/s0092-8674(00)81718-3. [DOI] [PubMed] [Google Scholar]
- 9.Crane BR, et al. N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization. EMBO J. 1999;18:6271–6281. doi: 10.1093/emboj/18.22.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischmann TO, et al. Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Biol. 1999;6:233–242. doi: 10.1038/6675. [DOI] [PubMed] [Google Scholar]
- 11.Crane BR, et al. Structures of the N(omega)-hydroxy-L-arginine complex of inducible nitric oxide synthase oxygenase dimer with active and inactive pterins. Biochemistry. 2000;39:4608–4621. doi: 10.1021/bi992409a. [DOI] [PubMed] [Google Scholar]
- 12.Rosenfeld RJ, et al. Conformational changes in nitric oxide synthases induced by chlorzoxazone and nitroindazoles: crystallographic and computational analyses of inhibitor potency. Biochemistry. 2002;41:13915–13925. doi: 10.1021/bi026313j. [DOI] [PubMed] [Google Scholar]
- 13.Fedorov R, Hartmann E, Ghosh DK, Schlichting I. Structural Basis for the Specificity of the Nitric-oxide Synthase Inhibitors W1400 and N{omega}-Propyl-L-Arg for the Inducible and Neuronal Isoforms. J Biol Chem. 2003;278:45818–45825. doi: 10.1074/jbc.M306030200. [DOI] [PubMed] [Google Scholar]
- 14.Flinspach M, et al. Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase. Nat Struct Mol Biol. 2004;11:54–59. doi: 10.1038/nsmb704. [DOI] [PubMed] [Google Scholar]
- 15.Flinspach M, et al. Structures of the neuronal and endothelial nitric oxide synthase heme domain with D-nitroarginine-containing dipeptide inhibitors bound. Biochemistry. 2004;43:5181–5187. doi: 10.1021/bi0361867. [DOI] [PubMed] [Google Scholar]
- 16.Aoyagi M, Arvai AS, Tainer JA, Getzoff ED. Structural basis for endothelial nitric oxide synthase binding to calmodulin. EMBO J. 2003;22:766–775. doi: 10.1093/emboj/cdg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-Soud HM, Stuehr DJ. Nitric oxide synthases reveal a role for calmodulin in controlling electron transfer. Proc Natl Acad Sci USA. 1993;90:10769–10772. doi: 10.1073/pnas.90.22.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, et al. Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase. Comparisons with NADPH-cytochrome P450 oxidoreductase. J Biol Chem. 2001;276:37506–37513. doi: 10.1074/jbc.M105503200. [DOI] [PubMed] [Google Scholar]
- 19.Garcin ED, et al. Structural Basis for Isozyme-specific Regulation of Electron Transfer in Nitric Oxide Synthase. J Biol Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 20.Ji H, Li H, Flinspach M, Poulos TL, Silverman RB. Computer Modeling of Selective Regions in the Active Site of Nitric Oxide Synthases: Implication for the Design of Isoform-Selective Inhibitors. J Med Chem. 2003;46:5700–5711. doi: 10.1021/jm030301u. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Poulos TL. Structure-function studies on nitric oxide synthases. J Inorg Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Nikolic D, VanBreemen RB, Silverman RB. Mechanism of Inactivation of Inducible Nitric Oxide Synthase by Amidines. Irreversible Enzyme Inactivation without Inactivator Modification. J Am Chem Soc. 2005;127:858–868. doi: 10.1021/ja0445645. [DOI] [PubMed] [Google Scholar]
- 23.Strub A, et al. The novel imidazopyridine 2-[2-(4-methoxy-pyridin-2-yl)-ethyl]-3H-imidazo[4,5-b]pyridine (BYK191023) is a highly selective inhibitor of the inducible nitric-oxide synthase. Mol Pharmacol. 2006;69:328–337. doi: 10.1124/mol.105.017087. [DOI] [PubMed] [Google Scholar]
- 24.Tafi A, et al. Computational studies of competitive inhibitors of nitric oxide synthase (NOS) enzymes: towards the development of powerful and isoform-selective inhibitors. Curr Med Chem. 2006;13:1929–1946. doi: 10.2174/092986706777585031. [DOI] [PubMed] [Google Scholar]
- 25.Tinker A, Wallace AV. Selective inhibitors of inducible nitric oxide synthase: potential agents for the treatment of inflammatory diseases? Curr Top Med Chem. 2006;6:77–92. doi: 10.2174/156802606775270297. [DOI] [PubMed] [Google Scholar]
- 26.Tinker AC, et al. 1,2-dihydro-4-quinazolinamine: potent, highly selective inhibitors of inducible nitric oxide synthase which show antiinflammatory activity in vivo. J Med Chem. 2003;46:913–916. doi: 10.1021/jm0255926. [DOI] [PubMed] [Google Scholar]
- 27.Hagmann WK, et al. Substituted 2-aminopyridines as inhibitors of nitric oxide synthases. Bioorg Med Chem Lett. 2000;10:1975–1978. doi: 10.1016/s0960-894x(00)00389-9. [DOI] [PubMed] [Google Scholar]
- 28.Connolly S, et al. 2-aminopyridines as highly-selective inducible nitric oxide synthase inhibitors. Differential binding modes dependent on nitrogen substitution. J Med Chem. 2004;47:3320–3323. doi: 10.1021/jm031035n. [DOI] [PubMed] [Google Scholar]
- 29.Sandstrom P, et al. Highly selective inhibition of inducible nitric oxide synthase ameliorates experimental acute pancreatitis. Pancreas. 2005;30:e10–15. [PubMed] [Google Scholar]
- 30.LaBuda CJ, Koblish M, Tuthill P, Dolle RE, Little PJ. Antinociceptive activity of the selective iNOS inhibitor AR-C102222 in rodent models of inflammatory, neuropathic and post-operative pain. Eur J Pain. 2006;10:505–512. doi: 10.1016/j.ejpain.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Gachhui R, et al. Mutagenesis of acidic residues in the oxygenase domain of inducible nitric-oxide synthase identifies a glutamate involved in arginine binding. Biochemistry. 1997;36:5097–5103. doi: 10.1021/bi970331x. [DOI] [PubMed] [Google Scholar]
- 32.Connolly, S. & Cox, D. Compounds. C07D 213/73, A61K 31/44 edn Vol. WO 00/21934 (ed. ASTRAZENECA) 1–35 (GB, 1999).
- 33.Li H, et al. Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases. Biochemistry. 2005;44:15222–15229. doi: 10.1021/bi0513610. [DOI] [PubMed] [Google Scholar]
- 34.Ji H, et al. Minimal Pharmacophoric Elements and Fragment Hopping, an Approach Directed at Molecular Diversity and Isozyme Selectivity. Design of Selective Neuronal Nitric Oxide Synthase Inhibitors. J Am Chem Soc. 2008;130:3900–3914. doi: 10.1021/ja0772041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheshire, D. et al. 5,7-bicyclic amidine derivatives useful as nitric oxide synthase inhibitors. C07D 513/04, 498/04, 491/048, 495/05, 487/04, A61K 31/55, 31/553, 31/554, 31/5513, A61P 29/00, 19/02 edn (ed. (GB), A.A.S.A.U.L.) 1–93 (UK & Sweden, 2000).
- 36.Sluis-Cremer N, Temiz NA, Bahar I. Conformational changes in HIV-1 reverse transcriptase induced by nonnucleoside reverse transcriptase inhibitor binding. Curr HIV Res. 2004;2:323–332. doi: 10.2174/1570162043351093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urzhumtsev A, et al. A ‘specificity’ pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil. Structure. 1997;5:601–612. doi: 10.1016/s0969-2126(97)00216-5. [DOI] [PubMed] [Google Scholar]
- 38.Kurumbail RG, et al. Structural Basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature. 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 39.Garavito RM, Mulichak AM. The structure of mammalian cyclooxygenases. Annu Rev Biophys Biomol Struct. 2003;32:183–206. doi: 10.1146/annurev.biophys.32.110601.141906. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 41.James LC, Roversi P, Tawfik DS. Antibody Multispecificity Mediated by Conformational Diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 42.Ghosh DK, et al. Characterization of the inducible nitric oxide synthase oxygenase domain identifies a 49 amino acid segment required for subunit dimerization and tetrahydrobiopterin interaction. Biochemistry. 1997;36:10609–10619. doi: 10.1021/bi9702290. [DOI] [PubMed] [Google Scholar]
- 43.Martasek P, et al. Characterization of bovine endothelial nitric oxide synthase expressed in E. coli. Biochem Biophys Res Comm. 1996;219:359–365. doi: 10.1006/bbrc.1996.0238. [DOI] [PubMed] [Google Scholar]
- 44.Sherman PA, Laubach VE, Reep BR, Wood ER. Purification and cDNA sequence of an inducible nitric oxide synthase from a human tumor cell line. Biochemistry. 1993;32:11600–11605. doi: 10.1021/bi00094a017. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski A, Minor V. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Navaza J. AMoRe: An automated package for molecular replacement. Acta Cryst A. 1994;50:157–163. [Google Scholar]
- 47.Kleywegt GJ, Jones TA. Databases in protein crystallography. Acta Cryst. 1998;D54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 48.Kleywegt GJ, Henrick K, Dodson EJ, van Aalten DMF. Pound-wise but penny-foolish. How well do micromolecules fare in macromolecular refinement? Structure. 2003;11:1051–1059. doi: 10.1016/s0969-2126(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 49.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Cryst A. 1986;42:140–149. [Google Scholar]
- 50.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 51.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 52.McRee DE. XtalView/Xfit - a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Cryst D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.