Co–evolution-driven switch of J–protein specificity towards an Hsp70 partner
Molecular mechanisms by which protein–protein interactions are preserved or lost after gene duplication are not well understood. Marszalek and colleagues investigate whether changes in partner proteins accompanied specialization of gene duplicates. They report that the existence of the Hsp70 Ssq1, which arose by duplication of the gene encoding the multifunctional mtHsp70 and specializes in Fe–S cluster biogenesis, correlates with functional and structural changes within the J–domain of its J–protein partner Jac1.
Keywords: chaperone interaction network, Fe–S biogenesis, Hsp70, J proteins, protein co-evolution
Abstract
Molecular mechanisms by which protein–protein interactions are preserved or lost after gene duplication are not understood. Taking advantage of the well–studied yeast mtHsp70:J–protein molecular chaperone system, we considered whether changes in partner proteins accompanied specialization of gene duplicates. Here, we report that existence of the Hsp70 Ssq1, which arose by duplication of the gene encoding multifunction mtHsp70 and specializes in iron–sulphur cluster biogenesis, correlates with functional and structural changes in the J domain of its J–protein partner Jac1. All species encoding this shorter alternative version of the J domain share a common ancestry, suggesting that all short JAC1 proteins arose from a single deletion event. Construction of a variant that extended the length of the J domain of a ‘short' Jac1 enhanced its ability to partner with multifunctional Hsp70. Our data provide a causal link between changes in the J protein partner and specialization of duplicate Hsp70.
Introduction
Protein–protein interaction networks can expand by gene duplication and the subsequent divergence of the descendant genes to specialize in one or a subset of the functions performed by the ancestral gene (Lynch & Conery, 2000; Prince & Pickett, 2002; Zhang, 2003). We have taken advantage of the well–studied yeast mitochondrial Hsp70s (mtHsp70s) to investigate the changes that occurred during such specialization, with the goal of identifying changes in partner proteins that accompanied the sub-functionalization of a gene duplicate.
Most eukaryotes, including humans, have a single, multifunctional mtHsp70. However, a subset of fungi, including Saccharomyces cerevisiae, also contain a highly specialized mtHsp70 involved in the essential process of iron–sulphur (Fe–S) cluster biogenesis, Ssq1. Ssq1 is encoded by a gene that arose through the duplication of an mtHSP70 gene in a common ancestor of Candida albicans and S. cerevisiae (Schilke et al, 2006) about 300 million years ago (Taylor & Berbee, 2006). SSC1, the paralogue of SSQ1, encodes an abundant mtHsp70, which performs the remaining tasks of the ancestral protein, including transport of polypeptides across the mitochondrial inner membrane and protein folding (Craig & Marszalek, 2002). Similarly to other members of the Hsp70 family, both Ssq1 and Ssc1 contain an amino-terminal ATPase and a carboxy-terminal peptide-binding domain, with hydrolysis of ATP inducing conformational changes that stabilize interaction with client proteins. Ssc1, as expected for a multifunctional Hsp70, interacts with various sequences. By contrast, specialized Ssq1 seems to bind to a single protein substrate, Isu1 (Schilke et al, 2006), a scaffold on which Fe–S clusters are assembled, facilitating cluster transfer to recipient proteins (Vickery & Cupp-Vickery, 2007).
All Hsp70s, including Ssc1 and Ssq1, require a J-protein co-chaperone, which stimulates the ATPase activity of their partner Hsp70s through the conserved 70-amino-acid (approximately) J domain (Hennessy et al, 2005). Two antiparallel helices (Helices II and III), connected by a highly flexible loop, form the core of J domains. Ssc1 of S. cerevisiae is known to function with two J proteins: Pam18 and Mdj1 (Craig et al, 2006). Neither Pam18 nor Mdj1 stimulates the ATPase activity of Ssq1 (D'Silva et al, 2003; Dutkiewicz et al, 2003), reflecting the fact that Ssq1 has lost the ability to perform the protein translocation and folding activities of Ssc1. Ssq1 functions with a single J protein, Jac1. The ATPase activity of Ssc1 is stimulated by Jac1, but much less effectively than that of Ssq1, suggesting that Jac1 of S. cerevisiae has retained some residual ability to function with Ssc1 (Schilke et al, 2006).
Taking advantage of the extensive information available on fungal genomes, we determined that the existence of Ssq1 in fungal species correlates with structural and functional changes in the J domain of Jac1. We propose that this correlation uncovers a causal link between changes in the J domain and a shift of J protein specificity towards a new, specialized Hsp70 partner.
Results And Discussion
Jac1 altered properties upon co-evolution with Ssq1
The ineffective stimulation of the ATPase activity of S. cerevisiae Ssc1 by Jac1, along with the severe growth defects of strains lacking Ssq1, suggested that Jac1 might have lost the ability to function efficiently with the multifunctional Hsp70 in organisms that also express an Hsp70 specialized for Fe–S cluster biogenesis. Thus, we assessed the ability of Jac1 orthologues from several fungal species to substitute for S. cerevisiae Jac1 (Jac1Sc), focusing on three species: C. albicans, which like S. cerevisiae contains Ssq1, and Yarrowia lipolytica and Schizosaccharomyces pombe, two species that lack Ssq1 (Schilke et al, 2006). To determine whether these orthologues are functional in S. cerevisiae, we overexpressed each in Δjac1 S. cerevisiae cells. All grew as well as the control strain overexpressing Jac1Sc (Fig 1A).
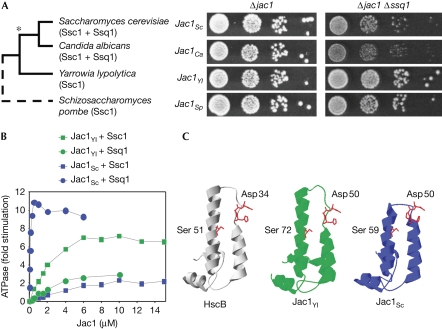
Figure 1.
Altered functional properties and structure of the J-domain distinguishes Jac1 orthologues coexisting with Ssq1. (A) The ability of JAC1 orthologues from several yeast species to support growth when Ssc1 functions in iron–sulphur biogenesis. Left panel: species phylogeny. The asterisk indicates SSC1 duplication. Right panel: Δjac1 and Δjac1 Δssq1 cells harbouring plasmid-borne copies of wild-type JAC1 from the indicated yeast species were plated as 10-fold serial dilutions (starting with approximately 5,000 cells) on rich glucose medium and incubated at 30°C for 3 days. (B) Stimulation of the ATPase activity of Ssc1 and Ssq1 from S. cerevisiae by Jac1Sc and Jac1Yl in the presence of Isu1Sc. ATPase activity in the absence of Jac1 was set to 0. (C) The structure of the J domain of HscB (PDB ID: 1fpo), a Jac1 orthologue from Escherichia coli and structural models of the J domains of Jac1Yl and Jac1Sc. Sc, Saccharomyces cerevisiae; Yl, Yarrowia lipolytica.
Similar growth rates indicated that the three JAC1 orthologues were functional in S. cerevisiae, as JAC1 is an essential gene. However, this test did not provide information as to the identity of their Hsp70 partner, as Ssq1 and Ssc1 were both present. Therefore, by using a Δjac1 Δssq1 strain we created a situation in which Jac1 could not cooperate with Ssq1. More robust growth was observed for strains expressing Jac1 from the more distant species, Y. lipolytica and S. pombe, than from the more closely related species, C. albicans, or from S. cerevisiae itself (Fig 1A). These results are consistent with the idea that Jac1 orthologues, which function with multifunctional mtHsp70 in their native environment, cooperate efficiently with Ssc1 of S. cerevisiae, whereas those that co-evolved with Ssq1 are less effective.
To determine whether the in vivo results described above could be correlated with molecular properties of Jac1, we used stimulation of the ATPase activity of Hsp70s as a biochemical measure of Jac1 interaction (Dutkiewicz et al, 2003). Two parameters can be determined in such analyses: the maximal stimulation (MS) obtainable and the concentration (C0.5) at which 50% MS is attained, a measure of apparent affinity.
Using the Hsp70s Ssc1Sc or Ssq1Sc from S. cerevisiae in conjunction with the client protein Isu1Sc, we tested two proteins: Jac1Sc, a well-characterized Jac1 that coexists with Ssq1, and Jac1 from Y. lipolytica (Jac1Yl), which works with multifunctional mtHsp70 in its native environment. As expected, Jac1Sc stimulated the ATPase activity of Ssq1 effectively and with high apparent affinity (MS=10.36-fold; C0.5=0.050 μM; Fig 1B; supplementary Table S1 online). By contrast, Jac1Sc stimulated the ATPase activity of Ssc1 threefold less efficiently (MS=3.29-fold) and had more than a 100-fold lower apparent affinity (C0.5=6.71 μM). By contrast, Jac1Yl stimulated the ATPase activity of Ssq1 poorly (MS=4.11; C0.5=6.14 μM), but was significantly better at stimulating the ATPase activity of Ssc1 (MS=8.28-fold; C0.5=2.16 μM), even though Y. lipolytica is distantly related to S. cerevisiae. This effectiveness of Jac1Yl compared with Jac1Sc in stimulating the ATPase activity of Ssc1 is consistent with its ability to outperform Jac1Sc in supporting the growth of Δjac1 Δssq1 cells (Fig 1A). Thus, together, the data support the idea that Jac1 and specialized Ssq1 have co-evolved. This co-evolution also resulted in a diminished ability of Jac1 to function with the multifunctional Ssc1.
Conversion of Jac1Sc to function better with Ssc1
Next, we considered whether the functional differences between Jac1 proteins that co-evolved with Ssq1 and those that did not could be explained by alterations in their structure. As no experimentally determined structural information for fungal Jac1 proteins exists, we turned to molecular modelling (details in supplementary information online). The size of the J domains was the most striking difference between the predicted structures (Fig 1C). The Jac1Yl J domain was significantly larger, with 21 residues in Jac1Yl between the aspartic acid of the invariant and critical HPD motif (Asp50 in both Jac1Sc and Jac1Yl) and the invariant serine in the middle of Helix-III (Ser72 and Ser59 in Jac1Yl and Jac1sc, respectively; supplementary Fig S1,S2 online). Jac1Sc has only eight residues in the homologous position (Fig 1C). Eleven of these residues in Jac1Yl are predicted to form a loop between the helices, whereas in Jac1Sc two residues form a tight loop (Fig 1C).
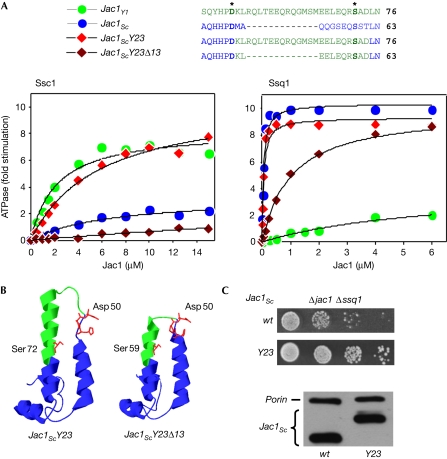
To test whether these differences near the ends of and between Helices II and III are responsible for the differences in the specificity of the J proteins, we considered whether the J domain of Jac1Sc could be altered to function more efficiently with Ssc1. We constructed a chimeric Jac1Sc–Jac1Yl gene, replacing the codons for 10 amino acids of Jac1Sc immediately carboxy-terminal of the conserved HPD motif with 23 amino acids of Jac1Yl, yielding Jac1ScY23 (Fig 2A). Jac1ScY23 stimulated the ATPase activity of Ssc1 nearly as well as Jac1Yl, increasing the MS from 3.3-fold to 10.5-fold (Fig 2A; supplementary Table S1 online). Consistent with the increase in ATPase-stimulatory ability, Δjac1 Δssq1 cells expressing Jac1ScY23 grew better than those expressing Jac1Sc (Fig 2C). Having observed that Jac1ScY23 functioned more efficiently with Ssc1 than did Jac1Sc, we next considered how these changes affected the ability of Jac1 to stimulate the ATPase activity of Ssq1. Jac1ScY23 stimulated Ssq1 almost as efficiently as wild-type Jac1Sc (Fig 2A, right panel), suggesting that Ssq1 has not lost the ability to accommodate a longer J domain over time.
Figure 2.
J-domain structure determines partnership with Ssq1 and Ssc1. (A) Stimulation of the ATPase activity of Ssc1 and Ssq1 from Saccharomyces cerevisiae by chimaeral Jac1ScY23 and Jac1ScY23Δ13 proteins in the presence of Isu1Sc. Data from Fig 1A of stimulation by Jac1Sc and Jac1Yl is included for comparison. The curves represent the best fit of the data to a Michaelis–Menten hyperbolic equation (supplementary Table S1 online). (B) Amino-acid sequence and structural model of Jac1ScY23 and Jac1ScY23Δ13. Green and blue represent sequences derived from Yarrowia lipolytica and S. cerevisiae, respectively. (C) Overexpression of Jac1ScY23 suppresses the growth defect of Δjac1 Δssq1. Top panels: 10-fold serial dilutions (starting with approximately 104 cells) of Δjac1 Δssq1 cells expressing plasmid-borne wild-type Jac1Sc (wt) or Jac1ScY23 (Y23) were plated on a rich glucose medium and incubated at 30°C for 3 days. Bottom panel: cell lysate from the indicated strains separated by electrophoresis and subjected to immunoblot analysis using antibodies specific for Jac1Sc and, as a loading control, porin. Predicted molecular weight: Jac1Sc, 21.6 kDa; Jac1ScY23, 23.3 kDa. Sc, Saccharomyces cerevisiae; Yl, Yarrowia lipolytica.
Jac1ScY23 is altered in amino-acid sequence, as well as having an extended structure, compared with Jac1Sc. To begin to understand the contribution of each, we deleted the codons for 13 amino acids from Jac1ScY23, to yield Jac1ScY23Δ13, which leaves Y. lipolytica sequences adjacent to the conserved Ser and HPD, but shortens Helix III and the loop to the size found in the S. cerevisiae protein (Fig 2A). Jac1ScY23Δ13 stimulated the ATPase activity of Ssc1 poorly and that of Ssq1 markedly less well than either Jac1Sc or Jac1ScY23. This reduced activity with both Hsp70 partners suggests that not only is the structure of the J domain important for specificity, but also that other sequences in the J domain have a role in defining Jac1–Hsp70 interactions.
Co-evolution with Ssq1 altered J-domain structure
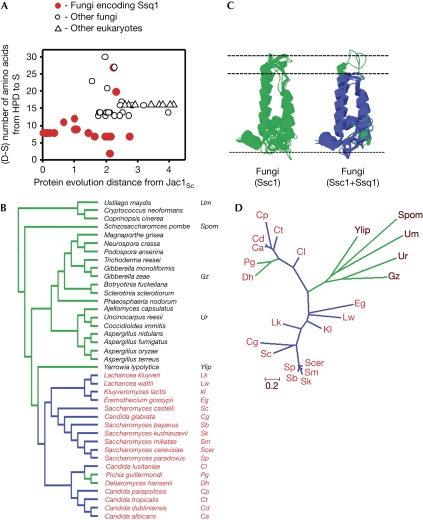
The results presented above suggest that the shortened J domain might be a common feature of the Jac1 proteins that have coexisted with Ssq1. To better assess the generality of the structural difference found between the J domains of Jac1Sc and Jac1Yl, we extended our analysis to the 39 fungal species for which genomic sequences and species phylogeny are available (Fitzpatrick et al, 2006). In particular, we were interested in whether a correlation exists between the presence of the ‘short' version of the J domain and the presence of SSQ1 across fungal phylogeny. Thus, the number of amino-acid residues from the Asp of the HPD motif to the invariant Ser in the middle of Helix III (D–S distance) were counted and plotted against the protein evolutionary distance calculated for each Jac1 orthologue relative to Jac1Sc (Fig 3A; supplementary Table S2 online). Sixteen of 18 Jac1 orthologues from species encoding SSQ1 (Fig 3B, red lettering) have ‘short' J domains, having a D–S distance between 7 and 12 amino acids. By contrast, all J domains of Jac1 orthologues from fungal species lacking SSQ1 (Fig 3B, black lettering), as well as the eight analysed non-fungal species (Fig 3; supplementary Table S2 online), are longer than 12 amino acids.
Figure 3.
Across fungal phylogeny, the presence of the ‘short' version of the J domain correlates with the presence of SSQ1. (A) For each Jac1 orthologue, the number of residues between the Asp of HPD and conserved Ser of Helix-III (D–S distance) was plotted against the evolutionary protein distance calculated for each Jac1 orthologue compared with Jac1Sc (supplementary Table S2 online). (B) Fungal phylogeny on the basis of Fitzpatrick et al (2006). Blue branches lead to species encoding a ‘short' J domain; green branches lead to species encoding J domains with a D–S distance greater than 12 amino acids (supplementary Table S2 online). Species encoding Ssq1 are marked in red. (C) The predicted structures of the J domains of Jac1 from the species listed in (B). Blue, ‘short' J domains; green, ‘regular' J domains. (D) Phylogenetic tree inferred from Jac1 structural alignment by maximum likelihood method (details in the supplementary information online). Branches are drawn to scale showing the number of amino-acid substitutions per site.
Analysis of the fungal phylogeny (Fitzpatrick et al, 2006) revealed that all species encoding a ‘short' version of the J domain belong to a monophyletic group (Fig 3B, blue branches), suggesting that this version arose in the common ancestor as the result of a single deletion event. Consistent with this idea, analysis of the topology of the phylogenetic tree on the basis of the Jac1 protein sequence indicates that all Jac1 orthologues with ‘short' J domains cluster together (Fig 3D). However, there are two outliners within this group, Pichia guillemondii and Debaromyces hansenii, two closely related species that encode SSQ1 but have ‘long' J domains, with D–S distances of 27 and 20 amino acids, respectively. Their localization on the phylogenetic tree (Fig 3B, green branches), sister to each other and nested within species encoding a ‘short' J domain (Fig 3B), implies that they are descendants of the lineage encoding a ‘short' version of the J domain, and that these ‘long' J domains are most probably derived features, acquired in the course of evolution long after the emergence of SSQ1. Such a secondary re-elongation of the J domain is consistent with our observation that the ATPase activity of Ssq1 can be stimulated efficiently by both ‘short' and ‘long' J domains (Fig 2A). In summary, the phylogenetic distribution of the ‘short' J domain implies that it arose through a single deletion event that corresponds to the timing of the origin of SSQ1.
Although this paper focuses on the co-evolution of Jac1 and Ssq1, we also considered the possibility that the third component of the system, the client protein Isu1 (Garland et al, 1999; Muhlenhoff et al, 2003), might also have co-evolved. However, analysis of an alignment of the sequences of the set of Isu1 orthologues from the species listed in Fig 3B (J.K. and J.M., unpublished results) showed no obvious signs of sequence changes that could be correlated with presence of Ssq1. Therefore, if co-evolution between Isu1 and the Jac1/Ssq1 chaperone system occurred, it is most probably a subtle phenomenon.
Hypothetical evolutionary scenario
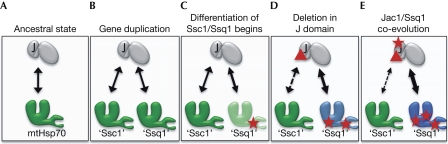
Data presented here enable us to develop a scenario regarding the evolution of the Jac1:Ssq1 specialized partnership. Although hypothetical, this scenario, as put forth in Fig 4, purports that the co-evolution of Jac1 and Ssq1 was triggered by deletion in the J domain of Jac1. About 300 million years ago, all fungal genomes encoded a single mtHsp70, Ssc1, which functioned with several J proteins, including Jac1 (Fig 4A). Duplication of the gene encoding mtHsp70 (Fig 4B; Schilke et al, 2006) presented the opportunity for division of functions between SSC1 and the paralogous gene that became SSQ1 (Fig 4C). Immediately after gene duplication, SSQ1 and JAC1 might have accumulated mutations that changed their biochemical properties but did not significantly affect the ability of Jac1 to interact with either Ssc1 or Ssq1. Only after a deletion occurred in JAC1 (Fig 4D), reducing the size of the J domain and resulting in weakened interaction with Ssc1, but not affecting interaction with Ssq1, did the Jac1–Ssq1-specific partnership begin to be established. This deletion event might have acted as an evolutionary ratchet, making reversal to the ancestral structure–function relations far more difficult, thus promoting forward co-evolution of Jac1 and Ssq1. Over time, reciprocal changes involving the sequence outside the loop region of Jac1 resulted in a highly specific and efficient interaction between Ssq1 and Jac1, forming a chaperone machinery tuned for functioning exclusively in Fe–S cluster biogenesis (Fig 4E).
Figure 4.
The hypothetical evolutionary scenario leading to the Ssq1:Jac1 specialized partnership. The lines indicate functional protein–protein interactions, with thick solid lines indicating interactions of greatest strength and dotted lines indicating the weakest interactions. The stars represent amino-acid substitutions that affected biochemical properties of interacting proteins. The triangle indicates deletion in the J domain of Jac1. (A) Before gene duplication, Jac1 functioned with multifunctional mtHsp70. (B) A duplication of the mtHsp70 gene, resulting in two Ssc1 genes that initially functioned equally well with Jac1. (C) Mutations in the Ssc1 gene copy occurred, which became SSQ1 (D). A deletion in JAC1 occurred, reducing the size of the J domain and resulting in weak interaction with Ssc1. It is noted that this deletion event could have occurred either before—as indicated in (C)—or after other alterations in the gene that became SSQ1. The hypothesis that the changes in SSQ1 that facilitated interaction with deleted Jac1 occurred before, rather than after, the JAC1 deletion is appealing, because it could possibly explain the survival of cells immediately after the occurrence of the JAC1 deletion. (E) Over time, reciprocal changes in JAC1 and SSQ1 resulted in highly specific interactions. The colour change of Hsp70 from dark green (B) to light green (C) to blue (D,E) indicates the overall changes in amino acid that occurred after the SSC1 gene duplication. The Hsp70 encoded by the gene to become SSQ1 in (B) is indicated by ‘Ssq1'.
Summary
In summary, we propose that the co-evolution-driven reciprocal changes in Ssq1 and Jac1 were triggered by a significant deletion in the J domain of Jac1, allowing selective interaction with Ssq1. Our results have implications for the multiplication and diversification of Hsp70:J-protein interaction network, beyond the specific example of Fe–S cluster assembly. In most cellular compartments, several Hsp70 proteins interact with an even higher number of J proteins (Hennessy et al, 2005), functioning in various crucial physiological processes. Thus, co-evolutionary processes in which duplicated chaperones and partner co-chaperones were optimized in a coordinated manner might have had an important role in the formation of specific partnerships. In the broader picture, our results provide experimental evidence supporting the importance of protein co-evolution as a driving force for the expansion and rewiring of the protein interaction networks (Yamada & Bork, 2009).
Methods
Yeast strains and plasmids. The S. cerevisiae strains used are haploid derivatives of PJ53 (W303), described previously (Andrew et al, 2006). JAC1 open reading frames were amplified by PCR: Jac1Yl, Y. lipolytica (CLIB99); Jac1Ca, C. albicans (SC5314); Jac1Sp, S. pombe (1457); and Jac1Sc, S. cerevisiae described by Andrew et al (2006). All were cloned into p414TEF (Mumberg et al, 1995). Chimeric JAC1Sc mutants were obtained by site-directed mutagenesis. All proteins were purified as described previously (Dutkiewicz et al, 2003; Schilke et al, 2006).
ATPase assays. ATPase activity was measured as described by Dutkiewicz et al (2003), with 0.5 μM Ssc1 or Ssq1, 10 μM Isu1, 0.5 μM Mge1 all from S. cerevisiae and variable amounts of Jac1. Robust stimulation requires inclusion of the client protein Isu1. Jac1Sc and Jac1Yl bind to Isu1Sc with similar affinity, thus validating the use of Isu1Sc for assays using both JAC1 proteins (supplementary Fig S3 online). Details are provided in the supplementary information online.
Prediction of protein structure. Homology modelling was performed using Escherichia coli HscB (PDB ID: 1fpo) as template. Alignment of the target sequence to the template structure was prepared using the GeneSilico Metaserver (Kurowski & Bujnicki, 2003). Structure models were calculated by using MODELLER (Fiser & Sali, 2003) and their potential accuracy was evaluated by using MetaMQAP (Pawlowski et al, 2008). Details are provided in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank R. Korona for critical comments. This work was supported by the Polish Ministry of Science and Higher Education Grant 2P04A 005 30 (to J.M.) and the National Institutes of Health Grants GM278709 (to E.C.) and GM081680-01 (to J.M.B.); R.D. and J.K. were supported by TEAM/2009-3/5.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andrew AJ, Dutkiewicz R, Knieszner H, Craig EA, Marszalek J (2006) Characterization of the interaction between the J-protein Jac1p and the scaffold for Fe–S cluster biogenesis, Isu1p. J Biol Chem 281: 14580–14587 [DOI] [PubMed] [Google Scholar]
- Craig EA, Marszalek J (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell Mol Life Sci 59: 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Huang P, Aron R, Andrew A (2006) The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol 156: 1–21 [DOI] [PubMed] [Google Scholar]
- D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA (2003) J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc Natl Acad Sci USA 100: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz R, Schilke B, Knieszner H, Walter W, Craig EA, Marszalek J (2003) Ssq1, a mitochondrial Hsp70 involved in iron–sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J Biol Chem 278: 29719–29727 [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374: 461–491 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland SA, Hoff K, Vickery LE, Culotta VC (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron–sulfur cluster assembly. J Mol Biol 294: 897–907 [DOI] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci 14: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski MA, Bujnicki JM (2003) GeneSilico protein structure prediction meta-server. Nucleic Acids Res 31: 3305–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Pawlowski M, Gajda MJ, Matlak R, Bujnicki JM (2008) MetaMQAP: a meta-server for the quality assessment of protein models. BMC Bioinformatics 9: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Pickett FB (2002) Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet 3: 827–837 [DOI] [PubMed] [Google Scholar]
- Schilke B, Williams B, Knieszner H, Pukszta S, D'Silva P, Craig EA, Marszalek J (2006) Evolution of mitochondrial chaperones utilized in Fe–S cluster biogenesis. Curr Biol 16: 1660–1665 [DOI] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML (2006) Dating divergences in the fungal tree of life: review and new analyses. Mycologia 98: 838–849 [DOI] [PubMed] [Google Scholar]
- Vickery LE, Cupp-Vickery JR (2007) Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron–sulfur protein maturation. Crit Rev Biochem Mol Biol 42: 95–111 [DOI] [PubMed] [Google Scholar]
- Yamada T, Bork P (2009) Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol 10: 791–803 [DOI] [PubMed] [Google Scholar]
- Zhang J (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18: 292–298 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.