The internet is changing the nature of scientific publishing, both in terms of how articles are published and the very nature and presentation of scientific research itself. Andrea Rinaldi investigates current developments that may shape the article of the future.
The internet and other developments are reshaping the way science is communicated, transforming the traditional scientific article to become more interactive and more useful
Libraries, once the hub of academic life and knowledge, have started to become quiet, solitary places. Today, most scientists browse, download and read the scientific literature from the comfort of their desktop computers, laptops or a plethora of mobile devices. Yet the online revolution has not only changed where and how scientists read papers, it is also changing the format of scientific articles. Publishers and academic institutions are experimenting with new methods of presenting data that challenge the traditional, standardized way of publishing and analysing scientific results (Fig 1).
Figure 1.
The cover of the first volume of Philosophical Transactions of the Royal Society, a publication founded by the Royal Society (London, UK) in 1665. It is the oldest journal in the English-speaking world and has remained in publication to the present day. Image: The Royal Society (London, UK).
“The Internet empowers anyone to publish, disseminate and archive scientific articles, and as a result we have a tremendous opportunity to rethink how we do these things,” commented Peter Binfield, Managing Editor of the online-only journal PLoS ONE. “Because of this, we are now witnessing some interesting experimentation that exploits the capabilities of the web.” The traditional makeup of a scientific paper—verbose prose, tables, two-dimensional figures and references—is proving increasingly limiting, unable to accommodate the use of video, databases, algorithms and other types of non-static, interactive content. Modern technologies and web-based dissemination also challenge the classical structure of the ‘standard' paper—introduction, methods, results and discussion—as web formats allow data and text to be separated and recombined by the reader, enriched through the use of electronic tagging and mark-up, and linked to external resources, with a general increase in interactivity. Nevertheless, despite the many possibilities—or perhaps because of them—the future of the scientific article is far from clear: there is no overall coordination of the various initiatives from diverse publishers and institutions, many of which strike out in different directions in their attempts to transform scholarly publishing.
“[I see that] the functions of science publishing, namely record keeping on the one hand—‘keeping the minutes of science'—and the communication of results on the other, will diverge,” observed Jan Velterop, former Director of Open Access at Springer (Heidelberg, Germany), who was most recently involved in the Concept Web Alliance (http://conceptweballiance.org). “The reason is that there are too many articles to read in many areas, and reading articles therefore is not a real option any longer to stay up-to-date with new knowledge. I expect that ‘traditional' articles will survive, to a degree, as ‘annotations' to data. The actual communication of knowledge, however, will find new avenues, such as ‘nano-publication'.”
The idea behind ‘nano-publication' is the use of the Resource Description Framework (RDF). RDF can describe electronic objects, provide meta-information for text, data, figures, video and other published elements and, proponents argue, is better suited to describe the relationship between data, enabling the more efficient exchange of knowledge than traditional-format papers (Mons & Velterop, 2009). “Science publishing now is like transporting needles in huge bales of hay. There is a tremendous amount of redundancy and padding in articles—[though] much of it is needed in order to make it, including the discursive context, human-readable, of course,” Velterop explained. “Methods to separate the needles of knowledge from the hay of surrounding text are being developed, and computer-readable nano-publications will make it possible to reason, quasi-mathematically, with a vast amount of knowledge, much of which is currently under-used.”
One of the cornerstones on which the scientific article of the future will probably be constructed is semantic publishing. This enriched format of scientific dissemination makes it possible to link online articles to their underlying data sets, which makes searching for information and data integration easier, faster and more efficient. “The knowledge we seek is often fragmentary and disconnected, spread thinly across thousands of databases and millions of articles in thousands of journals,” wrote Teresa Attwood and colleagues from the University of Manchester, UK, in a key review on the subject. They stressed the current difficulties in navigating between data stored in databases and the descriptions of these data in the literature (Attwood et al, 2009). “The crux of the problem is the lack of organizational principles. The failure of online databases to interoperate seamlessly with each other, and with the literature, is ultimately a matter of standards, or lack of them.”
Various experiments and initiatives, especially in the biomedical arena, have begun to use semantic publishing to make research articles more accessible to text-mining tools (Sidebar A) and to add semantic mark-up (Pafilis et al, 2009; Shotton et al, 2009). The ChemSpider Journal of Chemistry (www.chemspider.com), for example, uses an integrated system known as ChemMantis (www.chemmantis.com) to identify, extract and mark up chemistry-related entities in documents; for instance, linking compound names to structures hosted in the ChemSpider database.
Sidebar A | Text-mining on the rocks, please!
A main driving force behind the ongoing changes in online publishing and the development of new article formats is the increasing consumption of scientific articles by computers and the need to make those articles more accessible to their machine logic. Unlike data with a mathematical foundation, scientific prose, however highly structured and stylized, is written for the subtlety of the human intellect and presents significant hurdles for computers trying to extract meaningful information. One way to overcome this problem is to add meta-information to text and other published objects to increase their computer-readability. Another way is to use sophisticated text-mining algorithms that are able to analyse text and distil the information it contains into a format more palatable to computers.
“For historical reasons, biology and/or biomedicine have developed independent databases and publications. The somehow surprising consequence is that basic information, for example the function of a protein, or its interaction with other proteins, is reported independently in database entries and scientific publications, and that the information is reproduced by different methods and persons: authors, referees, editors and journals in the latter and databases and database curators in the former,” explained Alfonso Valencia, one of the leading experts in the field of text-mining and Director of the Structural Biology and BioComputing Programme at the Spanish National Cancer Research Centre (Madrid, Spain). “The consequence is that the same information is encapsulated in different formats with different semantics, and in practice it is very difficult to trace the information from one to the other.”
Current text-mining systems thus exploit regularities in natural language to automatically extract biologically relevant information from electronic texts. Identifying words is the first step of any text-mining approach, but language processing goes much further, decoding syntax, semantic relations and biological terms including acronyms and abbreviations. The most advanced applications are able to extract specific biological attributes of genes or proteins, such as their sequences, polymorphisms, mutations, residue modifications—such as amino-acid phosphorylation and gene methylation—or their subcellular locations and biological associations—such as protein interactions, gene regulation and functional annotations (Krallinger et al., 2008).
In the recent past, Valencia's team has developed several text-mining tools (see figure, below) and has proposed their use—in particular their automatic inclusion in web servers—to mediate between authors, journals and databases, integrating the process of writing papers, submitting them for publication and extracting the basic information to be submitted to databases (Leitner & Valencia, 2008; Leitner et al., 2008). “Text-mining and information extraction systems play an essential role to close the gap between the various sources of information,” Valencia commented.
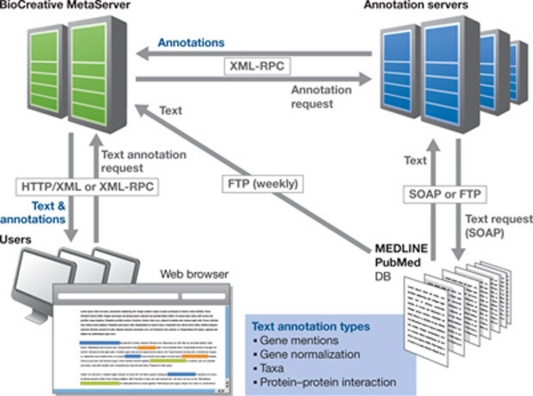 |
The BioCreative MetaServer, a suite of text-mining tools for information extraction and retrieval applications in the life sciences. This prototype platform provides automatically generated annotations for PubMed/MEDLINE abstracts. Annotation types cover gene names, gene IDs, species and protein–protein interactions. The annotations are distributed by the meta-server in both human- and machine-readable formats. Adapted from Leitner et al, 2008.
A team led by Reinhard Schneider at the European Molecular Biology Laboratory in Heidelberg, Germany, has contributed a free tool, Reflect (http://reflect.ws), which is able to recognize the names and abbreviations of proteins, genes and small molecules mentioned on a web page (Pafilis et al, 2009). Clicking on a highlighted name opens a pop-up in the browser window that provides relevant contextual information about the item without leaving the page, pulling in database entries, sequence information, synonyms and even expression data. Starting in November last year, Reflect—the 2009 winner of Elsevier's Grand Challenge competition to find innovative tools to improve the process of creating, reviewing and editing scientific content—was piloted with research articles of the journal Cell, as a step in the publication's efforts to improve the online presentation of scientific articles.
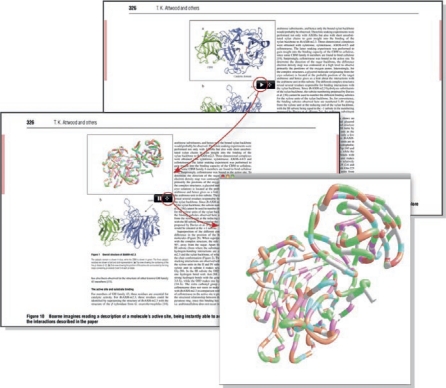
The Semantic Biochemical Journal (www.biochemj.org/bj/semantic_faq.htm) is another experiment in the integration of biological literature with databases. The project originated as a collaboration between the publisher, Portland Press, the University of Manchester and the creators of a piece of software called Utopia (http://getutopia.com) that allows the semantic enrichment of PDF files. In addition to providing access to definitions of key terms in the text, Utopia also permits items such as protein sequences or molecular structures to be linked to external websites, databases and tools for sequence alignment and three-dimensional molecular visualization (Fig 2). In addition, tables, images, videos and other elements can be linked, annotated, visualized and analysed further in a Semantic Biochemical Journal paper. “The additional data are overlaid rather than embedded in the documents, leaving their provenance and integrity intact; this means that features can be reliably associated with any version of a file,” explained Attwood and colleagues in their paper highlighting the technology (Attwood et al, 2009). “In this way, the electronic document is transformed from a digital facsimile of its printed counterpart into a gateway to related knowledge, providing the research community with focused interactive access to analysis tools, external resources and the literature.”
Figure 2.
An example of the additional information provided by the Semantic Biochemical Journal. By using the Utopia software, the reader is able to access the key structural features of the molecule on display, switching in a seamless manner from its plan description to its atomic coordinates. The three-dimensional view can be rotated freely and zoomed in and out. Taken from Attwood et al, 2009.
“I believe that semantic enrichment will have a very significant and positive impact on shaping the scientific article of the future,” Attwood said. “The true impact of ‘the Semantic Web' has yet to be felt, but I think things are starting to change as the technology begins to catch up with our dreams and visions.” Her review accordingly ends with a ‘call to arms' for learned societies and publishers, “to champion the standards for manuscript mark-up necessary to drive effective knowledge dissemination in future, and to garner community support for those standards” (Attwood et al, 2009).
In fact, the main scientific publishers have been testing new ways of communication for years. Several years ago, Nature Publishing Group (NPG; London, UK), launched Molecule Pages, the earliest example of a database with added functionality (www.signalling-gateway.org/molecule). “There's been much talk for years about the relationship between journals and databases, and how they should be two parts of a whole rather than separate endeavours. Our Signalling Gateway Molecule Pages project was our first publication bringing the two closely together,” commented Matthew Day, one of the project's developers. “You could view the Molecule Pages as a conventional review article—expert-authored and peer-reviewed—and you can view it as a highly structured database about the properties and functions of signalling proteins. It's both.” The database now provides information on more than 4,000 proteins involved in cellular signalling, highlighting information including protein–protein interactions, post-translational modifications, subcellular localization and biological function. Each ‘article' combines free text with machine-readable vocabularies and is interlinked to other databases. “Primary research articles have a lot of potential to become much more data rich too,” Day said. “When somebody is reading an article, they might just want the abstract or they might want to drill down into the hard data supporting the authors' conclusions. The article needs to serve both types of user equally well, and I predict it won't be long before far more articles do.”
In January this year, Cell launched its Article of the Future initiative, described by its Editor-in-Chief Emilie Marcus as, “a new approach to structuring the traditional sections of the article, moving away from a strictly linear organization required by print towards a more integrated and linked structure” (Marcus, 2010). Navigation through the article is enriched by the use of tabs and hyperlinks. The ‘Data' tab, for example, permits users to scan rapidly through the data in the article and links an individual figure to the related discussion of the findings; similarly, the ‘Results' tab allows the reader to zoom in or out on a figure, while its legend and associated text from the main document are visible on the same single screen. NPG plans to roll out its own system later this year for its flagship title.
The traditional makeup of a scientific paper […] is proving increasingly limiting…
The use of tabs to structure a document more dynamically on the web is certainly a nice way of presenting and organizing a scientific article, but the real challenge is to add genuine functionality and information that have previously been unavailable. “In science, we gain evidence that the results of a study represent truth when the findings are replicated by others,” said Christine Laine, Vice President of the Council of Science Editors and Editor of the Annals of Internal Medicine. “In my opinion, the greatest advance that we might see in coming years in scientific publishing will be a shift to include with publication sufficient ancillary materials in the form of protocols, statistical code and data sets that enable others working in the field to reproduce the findings.” Annals, together with several other journals, launched a ‘reproducible research' initiative, which calls for sharing protocols and additional data for every original research article published. Adding semantic mark-up to articles would then allow readers to see the original data and lab notes, which would not only increase transparency and reproducibility, but would also make it easier to spot errors and inconsistencies.
“Science publishing now is like transporting needles in huge bales of hay”
This inevitably requires more engagement by readers in post-publication annotation and evaluation. “One of the things that interests us at PLoS is the possibility of separating the act of publication from the more intangible act of evaluating the content that is published,” Binfield commented. “In order to facilitate this evaluation, we are developing a range of ‘article level metrics'—indicators such as online usage, citations, blog coverage, social bookmarks, notes and comments—which provide readers with rich contextual information about the quality of any given article. This data is not hard to compile, and so we expect the article of the future to incorporate many metrics of this type.”
PLoS is also experimenting with new avenues in web publishing, such as PLoS Currents Influenza (www.ploscurrents.org/influenza). “This site, which is based on the Google Knol platform, provides a very rapid channel for communicating new research and ideas on influenza research. Authors use Google Knol to construct their article, and as soon as it is ready it can be submitted to PLoS Currents Influenza, which is still within the Google Knol environment,” Mark Patterson, PLoS Director of Publishing, explained. “Publication in PLoS Currents Influenza does not preclude publication later on in formal journals, but shows how new technology can be used to accelerate scientific communication.”
One of the cornerstones on which the scientific article of the future will probably be constructed is semantic publishing
Visual communication of scientific research is another area under intense development, although only a few significant examples of progress exist so far. One of these is the Journal of Visualized Experiments (JoVE; www.jove.com), which focuses on lab methods and protocols in video format to show a step-by-step demonstration of the experiments, including the equipment and reagents needed. “In my opinion, whatever format will be applied to scientific articles in the future, the goal should be to make them more useful,” said Moshe Pritsker, CEO, Editor-in-Chief and co-founder of JoVE. “The article should become a ‘productivity tool' rather than what it is today: a polished story of a discovery process where truth is often concealed to ensure one got a ‘story' that integrates into the existing body of knowledge and is well accepted by peers.”
In this regard, the financial reports published by public companies could serve as an example, Pritsker reasoned. These reports typically follow a standardized format to quickly provide investors with the information they need and so, “similarly, the text part of the scientific article should be standardized to immediately answer questions interesting to practitioners (scientists): What is the problem addressed in this study? What was known before and what is known now after these studies? What about the contribution to theory of science and the contribution to practical applications?” Pritsker explained. “For rapid knowledge transfer and increasing productivity, the method part should be visualized because biological experiment is a craft more than science, as every practicing scientist knows. The format of video-presentations should be structured too.”
In light of these changes and the inevitable costs of these developments, some observers are concerned about the ability of scientific publishers to adapt and survive. Physicist and science writer, Michael Nielsen, even doomed classic scientific publishing to an almost inevitable extinction. “[S]cientific publishing is in the early days of a major disruption […] and will change radically over the next few years,” he wrote on his blog (Nielsen, 2009). The threat to scientific journals, Nielsen maintains, is “the flourishing of an ecosystem of startups that are experimenting with new ways of communicating research.” These include ChemSpider, JoVE and the growing tide of science blogs—some of which are starting to disseminate original research findings. By way of example, Nielsen quotes Tim Gowers' Mathematics related discussions (http://gowers.wordpress.com) and Richard Lipton's Gödel's Lost Letter and P=NP posts on computer science (http://rjlipton.wordpress.com).
The question is whether blurring the boundaries between journals and databases […] could bring about the end of scientific journals as we know them
“Today, scientific publishers are production companies, specializing in services such as editorial, copyediting and, in some cases, sales and marketing. My claim is that in 10–20 years, scientific publishers will be technology companies,” Nielsen wrote. “That is, their foundation will be technological innovation, and most key decision-makers will be people with deep technological expertise. Those publishers that don't become technology-driven will die off.” In other words, Nielsen argues, given the rapid changes and total interconnectedness of how science is communicated, “the people who add the most value to information are no longer the people who do production and distribution. Instead, it's the technology people, the programmers” (Nielsen, 2009). A few scientific publishers are attempting to face the revolution and become technology-driven enterprises, Nielsen acknowledged, but the rest of the guild seems largely unprepared to face the challenge.
From the point of view of Timo Hannay, Publishing Director of nature.com at NPG, the progressive convergence of publishers and technology companies is already under way. “As soon as you begin to distribute content online, there is a strong imperative to provide steadily more functionality in order to enhance its value further. The more that functionality becomes a differentiating factor, the closer the publisher comes to taking on the position of a technology player. Eventually, publishers will start—and, in some cases, have already begun—to build businesses based on functionality alone, independent of content,” he wrote (Hannay, 2009a).
The question is whether blurring the boundaries between journals and databases, which is a foreseeable side-effect of semantic publishing, could bring about the end of scientific journals as we know them. If the scientific article of the future will be interlinked with databases through a plethora of semantic tools, then one could argue that databases are the more important resource to be curated and protected by organized entities, either public or private. The dissemination of scientific information per se—which is now the realm of journals—could then be left to a loose network of academic and research institutions utilizing online publishing and in-house repositories. In essence, the question that remains to be answered is: are scientific journals really needed to filter, organize, disseminate and store information?
It is not yet clear if a new standardized format for scholarly communication will emerge […] to replace the scientific article as we know it today
Attwood and Hannay certainly do not believe that journals will disappear. “I think that this vision is unlikely to be realized soon, if ever,” Attwood commented. “For it even to come close to happening, our funding bodies will have to adopt radical new approaches to funding databases—this has been a thorny issue for many years now, and is unlikely to go away soon,” she explained. “If the financial future of databases is not secure, then a future vision of scientific information dissemination centred around them is likely to be precarious at best.”
Hannay does not see that either journals or databases will gain superiority. “Rather, as journal information becomes more structured and database information becomes more carefully curated, I think it's going to become increasingly hard to tell them apart,” he commented. “Whatever approach eventually succeeds, there will always be roles for people to make any of this information more valuable, visible and available, which is fundamentally what publishers do.” Indeed, as the amount of information grows, this role becomes even more important. “I'm quite sure that future publishers will use very different means to fulfil this goal than they do today,” Hannay added. “But whether it continues to be done by incumbent publishers, or whether others will take our place, is unclear and depends mainly on how good a job we do.”
“I think it will be a long time before the scientific journal disappears, but I don't doubt that scientific journals will evolve. They have to, in order to remain viable. What I hope will happen is that the relationship between journals and databases will evolve as they work more closely together in future,” Attwood said. “And I hope that part of that process will be a change in the way that science is reported and (peer) reviewed. In fact, I believe that there is a semantic revolution ahead that could change the face of peer review and dramatically increase the quality of reported science if we have the courage to embrace it!”
It therefore seems likely that whatever else happens, the need for genuine scientific publications to present and analyse results, put them into the context of the established literature and discuss the implications means that they will survive in some form, notwithstanding the rapid developments in disseminating information. “I think there's always going to be a place for relatively self-contained reports that describe a hypothesis, experimental results and conclusions,” Hannay said. “In that sense, the research paper still has a long future. But as we add links, interactivity, personalization and so on, the experience of ‘reading' a paper may come to feel quite different.” In this regard, Velterop agreed to a degree: “Individual articles may still be needed in accessible archives to check up on occasional facts, of course. But the primary role of traditional articles as conveyors of knowledge will greatly diminish.”
The impetus for the internet as we know it today was the need for physicists to share huge amounts of data with one another at remote locations. It is therefore apt, perhaps, that this modern communication tool has driven the evolution of scientific publishing farther and faster in the past decades than any other innovation. It is not yet clear if a new standardized format for scholarly communication will emerge from the current survival of the fittest to replace the scientific article as we know it today. It might well be that many new formats will coexist, and each discipline of science will select the mode that better fits its communication needs. Nevertheless, if one thing is certain, it is that scientific knowledge will become fully interconnected. As Hannay put it: “One link, tag, or ID at a time, the world's data are being joined together into a single seething mass that will give us not just one global computer, but also one global database” (Hannay, 2009b).
References
- Attwood TK, Kell DB, McDermott P, Marsh J, Pettifer SR, Thorne D (2009) Calling international rescue: knowledge lost in literature and data landslide! Biochem J 424: 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannay T (2009a) Walls come tumbling down. Learned Publishing 22: 153–154 [Google Scholar]
- Hannay T (2009b) From Web 2.0 to the global database. In The Fourth Paradigm: Data-Intensive Scientific Discovery Hey T, Tansley S, Tolle K (eds), pp 215–220. Redmond, WA, USA: Microsoft Research [Google Scholar]
- Krallinger M, Valencia A, Hirschman L (2008) Linking genes to literature: text mining, information extraction, and retrieval applications for biology. Genome Biol 9 (Suppl 2): S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner F, Valencia A (2008) A text-mining perspective on the requirements for electronically annotated abstracts. FEBS Lett 582: 1178–1181 [DOI] [PubMed] [Google Scholar]
- Leitner F et al. (2008) Introducing meta-services for biomedical information extraction. Genome Biol 9 (Suppl 2): S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus E (2010) Cell launches a new format for the presentation of research articles online. Cell Press Beta [online] 7 Jan [Google Scholar]
- Mons B, Velterop J (2009) Nano-publication in the e-science era. http://sunsite.informatik.rwth-aachen.de/Publications/CEUR-WS/Vol-523/Mons.pdf
- Nielsen M (2009) Is scientific publishing about to be disrupted? http://michaelnielsen.org 29 Jun
- Pafilis E, O'Donoghue SI, Jensen LJ, Horn H, Kuhn M, Brown NP, Schneider R (2009) Reflect: augmented browsing for the life scientist. Nat Biotechnol 27: 508–510 [DOI] [PubMed] [Google Scholar]
- Shotton D, Portwin K, Klyne G, Miles A (2009) Adventures in semantic publishing: exemplar semantic enhancements of a research article. PLoS Comput Biol 5: e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]