Abstract
Bacterial pathogens often harbour a type III secretion system (TTSS) that injects effector proteins into eukaryotic cells to manipulate host processes and cause diseases. Identification of host targets of bacterial effectors and revealing their mechanism of actions are crucial for understating bacterial virulence. We show that EspH, a type III effector conserved in enteric bacterial pathogens including enteropathogenic Escherichia coli (EPEC), enterohaemorrhagic E. coli and Citrobacter rodentium, markedly disrupts actin cytoskeleton structure and induces cell rounding up when ectopically expressed or delivered into HeLa cells by the bacterial TTSS. EspH inactivates host Rho GTPase signalling pathway at the level of RhoGEF. EspH directly binds the DH-PH domain in multiple RhoGEFs, which prevents their binding to Rho and thereby inhibits nucleotide exchange-mediated Rho activation. Consistently, infection of mouse macrophages with EPEC harbouring EspH attenuates phagocytosis of the bacteria as well as FcγR-mediated phagocytosis. EspH represents the first example of targeting RhoGEFs by bacterial effectors, and our results also reveal an unprecedented mechanism used by enteric pathogens to counteract the host defence system.
Keywords: pathogen–host interaction, Rho GTPases, signal transduction, type III effectors
Introduction
The type III secretion system (TTSS) is increasingly appreciated as a critical virulence mechanism for bacterial pathogens. Effectors secreted by the TTSS target various aspects of host signalling pathways and modulate normal host functions for the benefits of the bacteria (Galan and Wolf-Watz, 2006; Mattoo et al, 2007). Identification of host targets and revealing the biochemical mechanism of TTSS effectors are important for understanding the molecular mechanism and evolution of bacterial virulence (Shao et al, 2002, 2003; Li et al, 2007; Zhu et al, 2008; Yao et al, 2009). The TTSS is widely present and highly conserved in the large majority of Gram-negative bacterial pathogens including Yersinia, Salmonella, Shigella, Pseudomonas and pathogenic Escherichia coli. TTSS effectors from different bacteria are generally not conserved and often endowed with unique biochemical activities. Certain host molecules involved in controlling actin cytoskeleton dynamics and innate immune responses are frequent targets for TTSS effectors from different bacterial pathogens.
As models for TTSS-containing enteric pathogens, enteropathogenic E. coli (EPEC) is a serious concern of causing infantile diarrhoea in developing countries (Chen and Frankel, 2005), and the closely related enterohaemorrhagic E. coli (EHEC) is a frequent cause of food-born haemorrhagic colitis and diarrhoea in the developed world (Mead et al, 1999). On infection, both bacteria induces formation of the A/E lesions on intestinal epithelial cells (Kaper et al, 2004). The A/E lesions are characterized by localized effacement of the brush border microvilli, intimate attachment of the bacteria to the epithelium and development of pedestal-like protrusion structures comprised of actin polymer fibres beneath the bacterial attachment site. A TTSS located on a pathogenicity island known as the locus of enterocyte effacement is responsible for the A/E lesion and actin pedestal formation (Deng et al, 2004; Garmendia et al, 2005). Several of EPEC/EHEC TTSS effectors are known to directly modulate the host actin cytoskeleton signalling and dynamics (Caron et al, 2006). Tir is inserted into the host plasma membrane and serves as the receptor for bacterial outer membrane protein intimin (Kenny et al, 1997). The Tir/intimin binding mediates bacterial attachment and colonization. The carboxyl terminus of Tir inside the host cell triggers formation of the pedestal structure through multiple mechanisms (reviewed in Frankel and Phillips, 2008). In EHEC, this process is greatly aided by EspFU (also known as TccP) that directly activates the host actin nucleation-promoting factor N-WASP (Campellone et al, 2004; Garmendia et al, 2004; Cheng et al, 2008; Sallee et al, 2008). Map induces a transient filopodia formation right after bacterial attachment and before pedestal formation (Kenny et al, 2002; Alto et al, 2006; Berger et al, 2009), which results from its host-mimicking guanine nucleotide exchange factor (GEF) activity for small GTPase Cdc42 (Ohlson et al, 2008; Huang et al, 2009). EspH represses filopodia formation beneath adherent bacteria and enhances pedestal formation, suggesting its potential function of modulating the actin cytoskeleton signalling (Tu et al, 2003). In EHEC rabbit infection model, the espH− mutant has a reduced colonization throughout the intestinal tract and shows a significantly attenuated pathogenicity (Ritchie and Waldor, 2005). However, the biochemical function of EspH and its function in pathogenesis remains largely unexplored.
Actin cytoskeleton in eukaryotic cells is controlled by Rho GTPases, including RhoA, Rac and Cdc42 (Hall, 1998). The homeostatic level of cellular Rho GTPases is regulated by the ubiquitin-proteosome system in a nucleotide-dependent manner (Chen et al, 2009). GEF catalyses the exchange of GDP for GTP for activation, and GTPase-activating protein (GAP) negatively regulates the switch by enhancing the intrinsic GTPase activity. Rho signalling responds to diverse external stimuli including growth factors that engage cell surface receptors (such as G protein-coupled receptors, GPCRs) and regulate activation of downstream RhoGEFs and RhoGAPs. RGS-RhoGEFs including leukaemia-associated RhoGEF (LARG), p115-RhoGEF and PDZ-RhoGEF stimulate RhoA activation downstream of heterotrimeric G protein α subunits (Gα) and GPCRs (Hart et al, 1996; Fukuhara et al, 1999; Kourlas et al, 2000). Rho signalling is frequently targeted by bacterial effectors/toxins that usually mimic functions of the host RhoGEF or RhoGAP or covalently modify Rho GTPases (Aktories and Barbieri, 2005). Bacterial effectors directly targeting host RhoGEFs or RhoGAPs have not been reported so far.
In this study, we investigate the signalling mechanism of EspH in interfering with the host actin cytoskeleton dynamics. Through extensive biochemical and cell biological analyses, we discover that EspH directly binds to the DH-PH domain in RhoGEFs. The binding between EspH and the DH-PH domain competes with Rho binding to RhoGEFs and prevents Rho activation, thereby inhibiting downstream Rho signalling and actin cytoskeleton dynamics. Consistent with this biochemical observation, we further discover that EspH has a critical function in counteracting phagocytosis during EPEC infection of macrophages. Our study provides a new mode of hijacking the host Rho and actin cytoskeleton signalling by bacterial effectors and reveals new insights into the mechanism of EPEC/EHEC pathogenesis as well as regulation of DH-PH domain RhoGEF activities in eukaryotes.
Results
EspH disrupts the actin cytoskeleton and causes cell rounding up
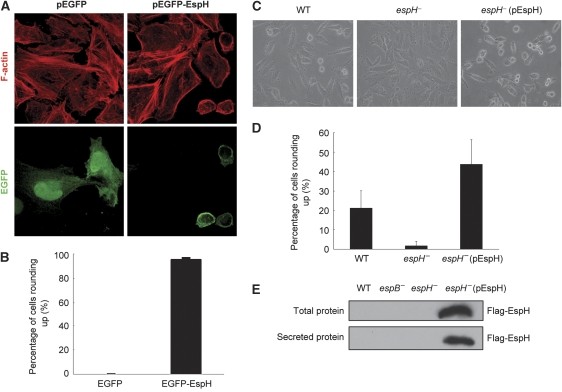
To explore the cellular function and host target of EspH, HeLa cells were transiently transfected with EGFP-EspH, in which EspH-expressing cells were identified by the coupled expression of EGFP. Consistent with the reported data (Tu et al, 2003), EGFP-EspH was found to associate with the plasma membrane and expression of EspH caused a complete disruption of the filamentous actin cytoskeleton structure (Figure 1A). The EspH-expressing cells became rounded up and tended to detach from the culture dish. The phenotype occurred in nearly 100% of EspH-transfected HeLa cells (Figure 1B) as well as other cell types such as HEK 293T cells (data not shown). Moreover, when HeLa cells were infected with wild-type EPEC strain, but not the espH− mutant strain, at a multiplicity of infection (MOI) of 100 for 1 h, about 20% of cells developed a similar rounding-up phenotype (Figure 1C and D). A more severe phenotype appeared when the deletion strain was complemented with an EspH overexpression plasmid (Figure 1E). The cell-rounding effect observed with EPEC infection seems to be cell-type specific as it did not occur on Caco-2 and T84 cells (data not shown). These results suggest that EspH, either transiently expressed or delivered by the EPEC TTSS into eukaryotic cells, harbours a potent activity of disrupting the host actin cytoskeleton structure.
Figure 1.
Cell rounding and disruption of the actin cytoskeleton structure by transfected or type III-delivered EspH. (A, B) Ectopic expression of EspH causes cell rounding up and disruption of the actin cytoskeleton structure in HeLa cells. HeLa cells were transfected with EGFP or EGFP-EspH. F-actin was stained with Rhodamine-phalloidin (red) and GFP staining marks EGFP or EGFP-EspH-expressing cells (A). Shown in (B) are statistics of transfected cells showing rounding-up phenotypes. (C, D) Rounding up of HeLa cells infected with EPEC E2348/69 WT, espH− or espH− complemented with a Flag-EspH overexpression plasmid (pEspH). Show in (C, D) are phase-contrast images of infected cells and statistics of cells showing rounding-up morphologies. At least 1000 cells were counted for each experiment and mean values±s.d. from three independent experiments are shown. (E) Secretion of EspH by EPEC. Total and secreted proteins from indicated EPEC strain were separated on SDS–PAGE followed by anti-Flag immunoblotting analyses.
Signalling pathways downstream of Rho GTPases are not affected by EspH
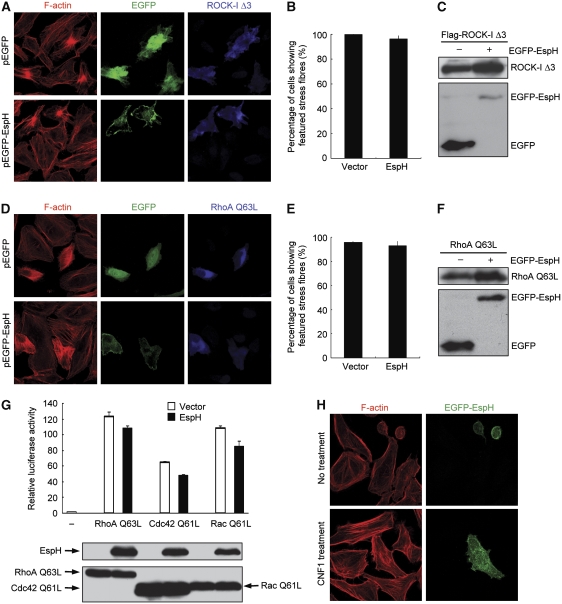
The cytoskeleton-disruption and cell-rounding phenotype caused by EspH is similar to that by Yersinia cysteine-protease type III effector YopT that cleaves the prenylated cysteine in Rho GTPases (Shao et al, 2002). We then examined whether EspH could also interfere with host Rho GTPase signal transduction. It is known that RhoA-induced stress fibres formation is through cooperative activation of two downstream effectors mDia1 and ROCK-I. In HeLa cells, a constitutively active ROCK-I mutant (ROCK-I Δ3) induced thick and sparely distributed star-like condensed actin fibres (Ishizaki et al, 1997) (Figure 2A), whereas cells expressing the constitutively active mDia1 ΔN mutant (524–1255) exhibited an unusual bipolar fusiform cell morphology and thin stress fibres that were aligned with the long axis of spindle-shaped cells (Watanabe et al, 1999) (Supplementary Figure S1A). Notably, co-expression of EGFP-EspH did not affect the development of actin stress fibres or the cell morphology changes that are induced by ROCK-I Δ3 or mDia1 ΔN (Figure 2A–C; Supplementary Figure S1). Furthermore, the dominant active variant of RhoA (RhoA Q63L) could stimulate formation of robust actin stress fibres (and slight cell-body shrinkage) despite the presence of EspH (Figure 2D–F). Similarly, distinct actin filamentous structures induced by constitutively active Rac Q61L and Cdc42 Q61L were also not affected by EspH (data not shown). These epistasis analyses indicate that signalling pathways downstream of Rho GTPases to actin polymerization remain intact in cells expressing EspH.
Figure 2.
Signalling pathways from Rho GTPases to their downstream are intact in EspH-expressing cells. (A–F) Effects of EspH on actin cytoskeleton rearrangements induced by exogenous RhoA and its effectors. HeLa cells were transfected with constitutively active ROCK-I Δ3 (A–C), or RhoA Q63L (D–F) together with a plasmid-expressing EGFP alone or EGFP-EspH. In (A, D), left panels show the Rhodamine-phalloidin staining of filamentous actin; middle panels show GFP staining that marks cells expressing EGFP or EGFP-EspH and right panels show immunofluorescence of transfected ROCK or RhoA. Shown in (B, E) are statistics of GFP-positive cells showing enhanced actin stress fibres. (C, F) show the expression of transfected plasmid using indicated antibodies. (G) Effects of EspH on Rho GTPase-stimulated transcriptional responses; 293T cells were co-transfected with luciferase reporter plasmids together with EspH and a Rho GTPase-expression construct (RhoA Q63L, Cdc42 Q61L or Rac Q61L). Luciferase activities were measured 11 h post-transfection and the ratio of firefly to Renilla-luciferase counts was calculated. The experiment was repeated for three times with each in duplicate. The relative luciferase activity from control and EspH-expressing cells has no statistically significant difference (P>0.05). Expression levels of Rho GTPases and EspH are shown at the lower panel. (H) Effects of CNF1 on EspH-induced cell rounding and disruption of the actin cytoskeleton structure. HeLa cells transfected with EGFP-EspH for 16 h were treated with 400 ng/ml of purified GST-CNF1 for 12 h. Shown are Rhodamine-phalloidin (left) and GFP staining (right) fluorescence images.
Activation of Rho GTPases turns on serum response factor (SRF)-dependent gene transcription through SRF recognition of serum response element (SRE) in the promoter (Hill et al, 1995). As expected, expression of RhoA Q63L, Rac Q61L or Cdc42 Q61L in 293T cells all resulted in potent activation of SRE luciferase, but this was not subjected to significant inhibition by co-expression of EspH (Figure 2G; Supplementary Figure S2). These data further confirm that signal transduction pathways downstream of Rho GTPases are intact and not targeted by the activity of EspH.
CNF-1 antagonizes EspH-induced cell-rounding effect and induces actin stress fibres formation in the presence of EspH
Cytotoxic necrotizing factor 1 (CNF1) is a potent cytotoxin produced by some E. coli isolates that cause extraintestinal infections in human (Caprioli et al, 1987). CNF1 catalyses irreversible deamidation of Gln63 in RhoA, thereby locking RhoA conformation in the GTP-bound state and inhibiting both intrinsic and RhoGAP-stimulated GTPase activities (Flatau et al, 1997; Schmidt et al, 1997). As shown in Figure 2H, HeLa cells treated with recombinant GST-CNF1 displayed significantly enhanced actin stress fibre formation, as expected. Expression of EspH in untreated cells caused cell rounding and disruption of actin stress fibre structures. However, GST-CNF1 treatment reversed the phenotype and EspH-expressing HeLa cells showed a flat and normal cell morphology and enhanced actin stress fibres, indistinguishable from control cells treated with GST-CNF1 (Figure 2H). These data strongly indicate that Rho GTPase itself, in addition to its downstream signalling, is fully functional and amenable to nucleotide-dependent activation even in the presence of EspH, and argue against the possibility that EspH directly targets Rho GTPases in eukaryotic host cells.
EspH inhibits signalling downstream of GPCR agonists and Gα
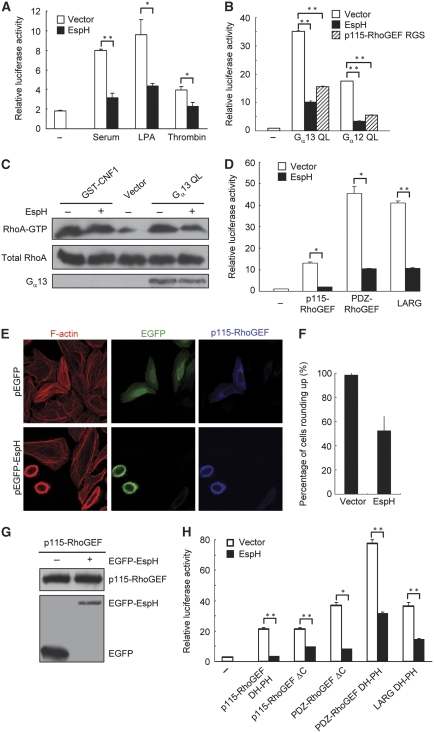
Extracellular stimuli such as serum, lysophosphatidic acid (LPA) and thrombin activate Rho signalling and induce stress fibres formation through GPCRs and their downstream Gα12 or Gα13 heterotrimeric G protein (Hart et al, 1998; Kozasa et al, 1998). We found that 20% serum, LPA and thrombin all failed to induce enhanced stress fibres in EspH-expressing NIH 3T3 cells (data not shown). Consistently, expression of EspH markedly inhibited serum- and LPA-induced SRE-luciferase activation, and to a less extent, thrombin-induced reporter activation in 293T cells (Figure 3A). The decreased reporter activation is unlikely to be a non-specific effect caused by cell rounding because the luciferase assay was performed at the time before the onset of evident cell rounding. Although EspH did not alter the expression and localization of exogenous LPA receptors (Supplementary Figure S3), we still could not completely rule out the possibility that EspH expression might lead to the loss of the GPCR receptors on the plasma membrane. Therefore, we went further to examine whether EspH could inhibit Rho signalling activated by Gα12 or Gα13. As expected, transfection of constitutively active Gα12 (Gα12 QL) and Gα13 (Gα13 QL) induced robust activation of SRE luciferase. However, this activation was largely abolished by co-expression of EspH (Figure 3B). The RGS domain of p115-RhoGEF, a dominant-negative mutant of endogenous p115-RhoGEF that can block the Gα-RhoGEF-Rho signalling (Hart et al, 1998), serves as a positive control here. In agreement with the SRE-luciferase assay, results of the RBD pulldown assay that directly measures the amount of RhoA-GTP (Ren and Schwartz, 2000) further indicated that EspH could counteract the activity of Gα13 QL, but not CNF, in elevating the cellular level of RhoA-GTP (Figure 3C). Taken together, the above analyses suggest that signalling transduction from Gα proteins to Rho GTPases downstream of the plasma membrane receptor is significantly attenuated by EspH.
Figure 3.
EspH inhibits GPCR agonist-Gα-RhoGEF-RhoA signal transduction. (A) EspH inhibits GPCR agonists-induced transcriptional responses. 293T cells were co-transfected with SRE.L-luciferase reporter plasmid together with the EspH-expression plasmid or an empty vector. Transfected cells were stimulated with serum (20%), thrombin (1 unit/ml) or LPA (5 μM) following 18-h serum starvation. Firefly and Renilla-luciferase activities were measured 6 h post-treatment and calculated ratios of firefly to Renilla-luciferase counts are shown. The experiment was repeated for three times with each in duplicate. Data were analysed using unpaired two-tailed Student's t-test, and considered statistically significant for P<0.05 (*) and P<0.005 (**). (B) Effects of EspH on Gα12 and Gα13-stimulated SRE-luciferase reporter activation. Experiments were performed similarly as those in Figure 2G and data were presented similarly as in (A). Gα12 QL and Gα13 QL are constitutively active mutants and the RGS domain of p115-RhoGEF serves as a positive control. (C) Effects of EspH on Gα13-stimulated RhoA activation. HeLa cells were transfected with Gα13 QL-expression plasmid in the presence or absence of EspH. Levels of the GTP-bound RhoA were probed by using the GST-RBD pulldown assay. RhoA activation induced by GST-CNF1 was included as a control. Expression of the EE-tagged Gα13 was analysed by western blot using the anti-EE antibody. (D) Effects of EspH on RGS-RhoGEFs-stimulated SRE-luciferase reporter activation. Expression constructs of p115-RhoGEF, PDZ-RhoGEF and LARG were transfected into 293T cells to activate the SRE-luciferase reporter. Experiments were performed and data were presented similarly as those in (B). (E–G) Effects of EspH on actin cytoskeleton rearrangements induced by p115-RhoGEF. HeLa cells were co-transfected with Flag-p115-RhoGEF-expression plasmid together with the EGFP vector or EGFP-EspH. Left panels in (E) show rhodamine-phalloidin staining of filamentous actin; middle panels in (E), GFP staining that marks transfected cells and right panels in (E), anti-Flag staining of p115-RhoGEF expression. Shown in (F, G) are statistics of transfected cells showing rounding-up phenotypes and immunoblots showing EspH and p115-RhoGEF expression, respectively. (H) Effects of EspH on SRE-luciferase reporter activation stimulated by the DH-PH domain of RGS-RhoGEFs. Experiments were performed and data were presented similarly as those in (B). p115-RhoGEF ΔC and PDZ-RhoGEF ΔC are active forms with removal of their C-terminal inhibitory sequences.
EspH inhibits RhoGEF-induced activation of Rho GTPases
Yersinia type III effector YpkA directly targets Gα and inhibits activation of downstream Rho GTPases (Navarro et al, 2007). Our initial hypothesis was that EspH might function similarly as YpkA to act on Gα. However, EspH did not bind Gα12 and Gα13 in the co-immunoprecipitation assay (data not shown). RGS-RhoGEFs such as p115-RhoGEF, PDZ-RhoGEF and LARG mediate signalling transduction between the trimeric G proteins and Rho GTPases (Mao et al, 1998). We then examined whether EspH could disrupt signalling at the level or downstream of RhoGEFs. As shown in Figure 3D, EspH markedly inhibited SRE-luciferase activation induced by p115-RhoGEF, PDZ-RhoGEF or LARG. Consistently, transfection of p115-RhoGEF into HeLa cells resulted in enhanced and thick actin filamentous bundles, but this effect was counteracted by co-expression of EspH (Figure 3E). In fact, cells expressing EspH still rounded up despite the unaffected expression level of exogenous p115-RhoGEF (Figure 3E–G). EspH also inhibited the SRE-luciferase activation stimulated by p115-RhoGEF ΔC (1–820) and PDZ-RhoGEF ΔC (1–1160) that lack the carboxyl terminal auto-inhibitory region (Figure 3H). These data suggest that EspH inhibits RhoGEF-mediated activation of Rho GTPase.
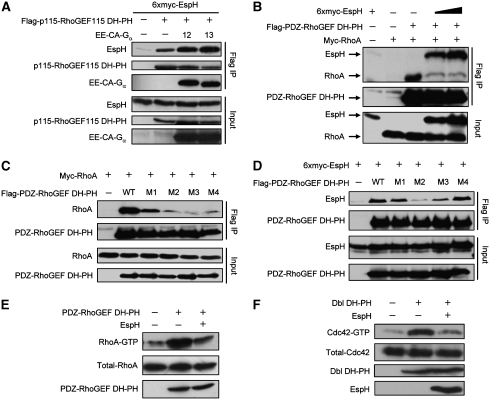
EspH directly targets RhoGEFs through binding to the DH-PH domain
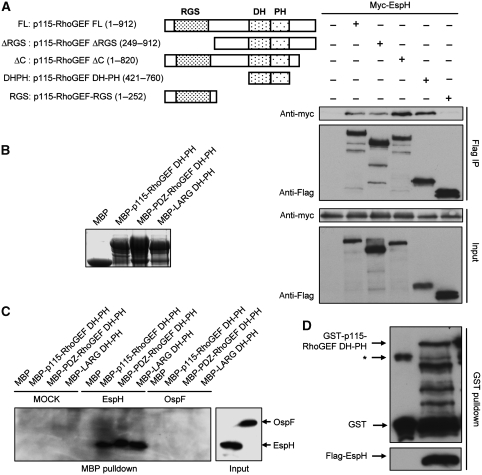
The above results promoted us to investigate whether EspH could form complexes with RhoGEF or Rho itself. When co-expressed in 293T cells, EspH efficiently co-immunoprecipitated with full-length p115-RhoGEF (Figure 4A), but not Rho GTPases (data not shown). RhoGEFs harbour a conserved DH-PH domain that catalyses the nucleotide change on Rho. In addition to that induced by full-length RGS-RhoGEFs, SRE-luciferase activation stimulated by the DH-PH domain from p115-RhoGEF, PDZ-RhoGEF or LARG was markedly inhibited by EspH (Figure 3H). Thus, we hypothesized that the DH-PH domain might be the target of EspH. To this end, we generated a series of truncations of p115-RhoGEF (Figure 4A). Notably, EspH was found to co-immunoprecipitate with all truncations that contain an intact DH-PH domain, but not the RGS domain alone (Figure 4A). Removal of the carboxyl terminal auto-inhibitory region in p115-RhoGEF resulted in a significantly enhanced interaction. A shortest fragment containing the DH-PH domain alone gave the strongest EspH binding (Figure 4A). To further confirm that EspH targets the DH-PH domain of RhoGEFs, we prepared bacterially expressed recombinant maltose-binding protein (MBP)-tagged DH-PH domain from p115-RhoGEF, PDZ-RhoGEF and LARG (Figure 4B). When these MBP proteins were incubated with lysates from intact 293T cells or 293T cells expressing EspH or OspF (Li et al, 2007), only EspH was precipitated by MBP-DH-PH and not MBP alone (Figure 4C). Furthermore, EspH also efficiently co-precipitated with p115-RhoGEF DH-PH domain when both proteins were co-expressed in EPEC (Figure 4D), indicating that the interaction does not involve any eukaryotic factors. These data suggest that EspH directly and specifically binds to the DH-PH domain of RhoGEFs both in vitro and in eukaryotic cells. The DH-PH domain is highly conserved in RhoGEFs. In the transfection assay, EspH could co-immunoprecipitate with the DH-PH domain from PDZ-RhoGEF, LARG as well as non-RGS-RhoGEFs including Dbl and p63RhoGEF. Thus, the target specificity of EspH during EPEC infection is likely mediated by other factors or a specific location effect.
Figure 4.
Binding of EspH to the DH-PH domain of RhoGEFs. (A) Co-immunoprecipitation between EspH and p115-RhoGEF in 293T cells. Myc-tagged EspH was co-transfected into 293T cells with Flag-tagged p115-RhoGEF or indicated truncation mutants illustrated on the upper left. Anti-Flag immunoprecipitates (Flag IP) and total cell lysates (Input) were analysed by immunoblotting using antibodies as indicated. (B) Coomassie blue staining of purified MBP, MBP-p115-RhoGEF DH-PH, MBP-PDZ-RhoGEF DH-PH or MBP-LARG DH-PH proteins. (C) MBP pulldown assay of binding between EspH and purified DH-PH domain from RGS-RhoGEFs. Lysates of 293T cells transfected with Flag-EspH, Flag-OspF or a control vector were incubated with MBP or MBP fusion proteins (B) immobilized on amylose beads for 2 h. Shown are anti-Flag immunoblots of the MBP pulldowns and total cell lysates (Input). (D) GST pulldown assay of binding between EspH and p115-RhoGEF DH-PH domain in bacteria. Flag-EspH was co-expressed with GST or GST-tagged p115-RhoGEF DH-PH domain in wild-type EPEC strain. Shown are immunoblots of the total lysates and GST pulldowns using Flag and GST antibodies, respectively. Asterisk (*) marks a non-specific band.
EspH competes with Rho for binding to the DH-PH domain of RhoGEFs and disrupts RhoGEF-Rho signalling
To further understand the mechanism of RhoGEF inhibition by EspH, we checked the complex formation of RhoGEF with its upstream Gα protein and downstream Rho GTPase in the presence of EspH. EspH did not interfere with the co-immunoprecipitation between Gα12/13 and p115-RhoGEF DH-PH from 293T cells (Figure 5A and data not shown). Instead, constitutively active Gα12 or Gα13 promoted the association between p115-RhoGEF DH-PH and EspH (Figure 5A). This is consistent with the observed enhanced binding between EspH and the truncated and presumably more active form of p115-RhoGEF (Figure 4A). When the interaction between the DH-PH domain and RhoA was examined, we found that EspH attenuated the co-immunoprecipitation between RhoA and the DH-PH domain of PDZ-RhoGEF or LARG in a dose-dependent manner (Figure 5B; Supplementary Figure S4). These results suggest that the interaction of EspH with the DH-PH domain interferes with the binding of RhoA to the DH-PH domain of RhoGEFs.
Figure 5.
Interaction between the DH-PH domain and EspH prevents RhoA binding and inhibits DH-PH domain-mediated RhoA activation. (A) Effects of EspH on co-immunoprecipitation between p115-RhoGEF DH-PH domain and Gα. Total 293T cell lysates (Input) and the anti-Flag immunoprecipitates (Flag IP) were analysed by immunoblotting using antibodies against myc (EspH), Flag (p115-RhoGEF DH-PH) and the EE tag (Gα) as shown (12 and 13 stand for Gα12 and Gα13, respectively). (B) Effects of EspH on co-immunoprecipitation between the DH-PH domain and RhoA. Experiments were performed and data were presented similarly as those in (A). (C, D) Effects of mutations in the PDZ-RhoGEF DH-PH domain on its interaction with RhoA (C) and EspH (D); 293T cells were co-transfected with indicated plasmid or plasmid combinations. Shown are immunoblots of total cell lysates (Input) and the anti-Flag immunoprecipitates (Flag IP) using antibodies as indicted. (E) Effects of EspH on the DH-PH domain-stimulated RhoA-GTP level; 293T cells were transfected with PDZ-RhoGEF DH-PH domain expression construct together with the EspH-expression plasmid or an empty vector. The upper panel shows anti-RhoA immunoblot of RhoA-GTP precipitated on GST-RBD beads, and the middle and lower panel show the expression of total RhoA and PDZ-RhoGEF DH-PH domain. (F) Effects of EspH on DH-PH domain-stimulated Cdc42-GTP level; 293T cells were transfected with Dbl DH-PH domain expression construct together with the EspH-expression plasmid or an empty vector. The upper panel shows anti-Cdc42 immunoblot of Cdc42-GTP precipitated on GST-PBD beads, and lower three panels show the expression of total Cdc42, Dbl DH-PH domain and EspH.
Several structures of the DH-PH domain in complex with RhoA are reported and show a conserved mode of interaction. The crystal structure of PDZ-RhoGEF DH-PH/RhoA complex reveals three major binding interfaces (Derewenda et al, 2004). E741 and S748 in the DH domain of PDZ-RhoGEF contact the switch I region in RhoA (Interface A); residues including R867, R868, L869, R872, D873 and I876 in the DH domain are involved in interactions with the switch II region centreing around RhoA W58 (Interface B); D921, E928 and N929 interact with another switch II region around R68 in RhoA (Interface C). In view of the structural information, we generated four DH-PH domain mutations including E741N/S748A (M1), R867Q/R868G/L869A (M2), R872D/D873N/I876M (M3) and D921A/E928A/N929A (M4). M1 and M4 are mutations in the Interfaces A and C, respectively, whereas M2 and M3 are expected to disrupt the Interface B. As expected, co-immunoprecipitation between either of the four mutants and RhoA were significantly compromised (M1) or almost completely abolished (M2, M3 and M4) (Figure 5C). Interestingly, the association between EspH and PDZ-RhoGEF DH-PH was also largely attenuated by the M2 mutation, and to a less extent the M3 mutation (Figure 5D), predicting an overlapping surface in the DH-PH domain for binding to EspH and RhoA. These data strongly suggest that EspH competes with RhoA for binding to the DH-PH domain of RhoGEFs.
The above analyses further predict that recombinant EspH should directly inhibit nucleotide exchange of RhoA catalysed by the DH-PH domain of RhoGEFs. Unfortunately, we were not able to obtain recombinant EspH protein despite extensive tries in several different expression systems. We then turned to measure the DH-PH domain-stimulated higher level of RhoA-GTP in cells expressing EspH or not. As shown in Figure 5E, overexpression of the DH-PH domain increased the level of RhoA-GTP, which was counteracted by co-expression of EspH. Similarly, EspH also attenuated activation of Cdc42 induced by the DH-PH domain of Dbl when the level of CDC42-GTP was measured by the PBD pulldown assay (Figure 5F). These results support the above hypothesis that EspH inhibits the DH-PH domain-catalysed Rho activation by competing with Rho for binding to the DH-PH domain in RhoGEFs.
EspH inhibits macrophage phagocytosis
The espH− mutant of EHEC has a reduced colonization throughout the intestinal tract and shows a significantly attenuated pathogenicity in EHEC rabbit infection model (Ritchie and Waldor, 2005). EspH has been proposed to have a function in bacterial adherence or resistance to host defences (Ritchie and Waldor, 2005). Macrophage phagocytosis serves as a first line of host defence again invading bacterial pathogens. Phagocytosis is driven by extensive local reorganization of the actin cytoskeleton and is known to be dependent on the activity of Rho GTPases that are spatially and temporally regulated by Rho GTPase regulators including RhoGEFs (Caron and Hall, 1998; Patel et al, 2002). EPEC also effectively inhibits its own uptake by blocking F-actin polymerization at the site of bacterial contact in its TTSS-dependent manner (Goosney et al, 1999; Celli et al, 2001; Quitard et al, 2006; Iizumi et al, 2007). These information promoted us to test whether EspH could indeed have a function in counteracting macrophage phagocytosis.
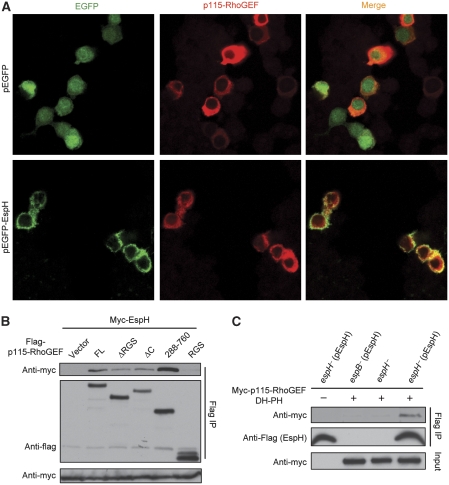
We first investigated whether EspH could target the DH-PH domain RhoGEF in macrophage cells. When transfected into macrophage-like RAW264.7 cells, EGFP-EspH was found to co-localize with p115-RhoGEF at the cell membrane region (Figure 6A). EspH also co-immunoprecipitated with p115-RhoGEF in macrophages, and also similarly to that observed in 293T cells, only DH-PH domain-containing fragments could pulldown EspH in RAW264.7 cells (Figure 6B). Furthermore, the DH-PH domain of p115-RhoGEF co-immunoprecipitated with EspH translocated into RAW264.7 cells by the EPEC TTSS during EPEC infection (Figure 6C). These data suggest that EspH could potentially target the DH-PH domain RhoGEFs in macrophage cells.
Figure 6.
EspH targets the DH-PH domain of RhoGEF in macrophages. (A) Co-localization of EspH and p115-RhoGEF in RAW264.7 cells. Flag-p115-RhoGEF and EGFP-EspH or EGFP control plasmid were co-transfected into RAW264.7 cells. Shown are GFP and anti-Flag fluorescence staining of transfected cells. (B) Co-immunoprecipitation between EspH and p115-RhoGEF in RAW264.7 cells. Myc-EspH was co-transfected with Flag-tagged p115-RhoGEF or indicated truncation mutants into RAW264.7 cells. Shown are immunoblots of anti-Flag immunoprecipitates (Flag IP) and total cell lysates (Input) using antibodies as indicated. (C) Co-immunoprecipitation between EspH and p115-RhoGEF DH-PH domain during EPEC infection. RAW264.7 cells expressing Flag-tagged p115-RhoGEF DH-PH domain were infected with indicated pre-activated EPEC strains. Cells were lysed in 0.05% Triton X-100 buffer that has been verified not to lyse the bacteria. Cell lysates were subjected to Flag immunoprecipitation in the presence of 1% Triton X-100. Shown are immunoblots of the immunoprecipitates (Flag IP) and total cell lysates (Input).
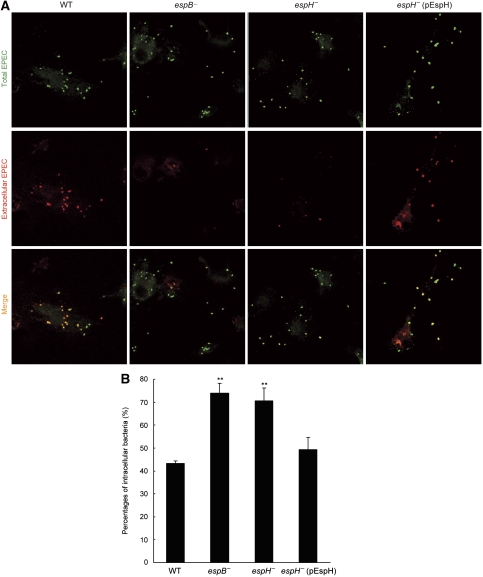
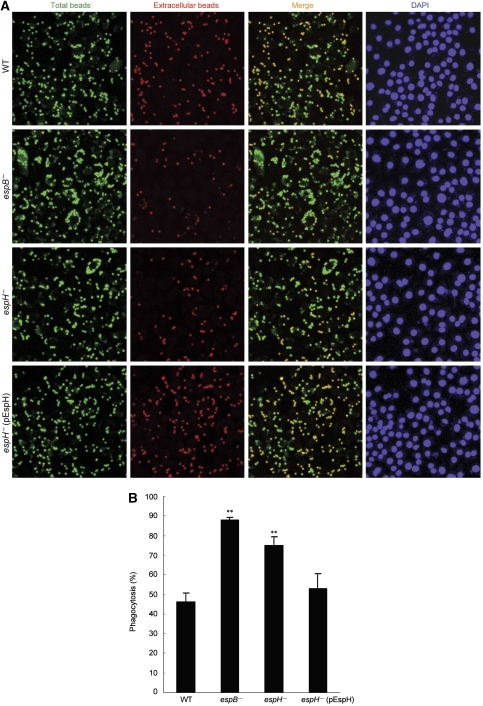
Secondly, C57BL/6 mice-derived bone marrow macrophages were infected with wild-type or mutant EPEC strains. Extracellular bacteria and total cell-associated bacteria were stained by red- and green-coloured secondary antibodies, respectively. As expected, most of the wild-type bacteria stayed extracellularly after infection (Figure 7; Supplementary Figure S5). Similarly to the type III deficient espB− mutant earlier shown to lose the antiphagocytosis activity (Luo and Donnenberg, 2006; Iizumi et al, 2007), deletion of EspH from EPEC resulted in significantly attenuated staining signal of extracellular bacteria. Statistics of percentages of internalized bacteria further confirmed the impaired antiphagocytosis capability of espH− mutant bacteria, which was rescued by complementation with the EspH-expression plasmid (Figure 7; Supplementary Figure S5). Using a general and more classical phagocytosis assay, deletion of EspH as well as EspB resulted in enhanced phagocytosis of IgG-coated latex beads by EPEC-infected J774A.1 cells (Figure 8). This phenotype could also be efficiently rescued by complementation with the EspH-expression plasmid. These data clearly show that EspH secreted by EPEC TTSS has an important function in antagonizing phagocytosis during infection of macrophages.
Figure 7.
EspH cis-inhibits macrophage phagocytosis during EPEC infection. Bone marrow macrophages from C57BL/6 mice were infected with wild type or indicated mutant EPEC E2348/69 strains (MOI of 20:1). Bacteria were stained by using anti-E. coli O127 antibody. Extracellular and total cell-associated bacteria were distinguished by red and green-coloured secondary antibody staining (A), respectively. Zoom-in pictures showing a clearer overlap of the red and green fluorescence are in Supplementary Figure S5. Shown in (B) are statistics (mean±s.d.) of percentages of internalized bacteria from three independent experiments. **Denotes a statistically significant difference compared with the wild-type strain infection.
Figure 8.
EspH inhibits FcγR-mediated phagocytosis during EPEC infection. J774A.1 macrophages infected with wild type or indicated mutant EPEC E2348/69 strains (MOI of 20:1) were challenged with mouse IgG-opsonized latex beads for 40 min. Total and extracellular beads were differentially stained as described in Materials and methods. Shown in (A) are fluorescence images of IgG-beads and DAPI staining of nuclei (cell and bacteria). Statistics (mean±s.d.) of percentages of internalized beads are in (B). **Denotes a statistically significant difference compared with the wild-type strain infection. About 500 cells were counted for each assay.
Discussion
The type III effector EspH from EPEC/EHEC is a small protein of 20 kDa and does not show sequence homologies with any known proteins. Here, we discover that EspH directly binds to the DH-PH domain in RhoGEFs and disrupts the host Gα-RhoGEF-RhoA signalling. A number of bacterial effectors or toxins capable of modulating the host Rho pathway are reported. These bacterial proteins usually target the Rho GTPase itself and modulate or modify the nucleotide-bound state, effector-binding region or membrane association of the GTPase (Aktories and Barbieri, 2005). The activity of EspH is unique and it represents the first bacterial effector that acts directly on RhoGEFs. Notably, modulation of RhoGEF activity also exists in eukaryotic cells. A host protein known as MLK3 binds to the DH-PH domain in p63RhoGEF and negatively regulates Gαq-mediated RhoA activation (Swenson-Fields et al, 2008). In this regard, EspH functions similarly to MLK3 in that EspH could also bind to p63RhoGEF in a co-immunoprecipitation assay (Supplementary Figure S6). As opposed to EspH, MLK3 seems to compromise the interaction between p63RhoGEF and Gαq. This is consistent with the fact that Gαq and RhoA bind to different sides of the DH-PH domain of p63RhoGEF (Lutz et al, 2007). Thus, EspH and MLK3 are similar in binding the DH-PH domain in RhoGEF and inhibiting Rho activation, but are distinct regarding their binding surfaces on the DH-PH domain as well as the mechanism of inhibition. Many DH-PH domain RhoGEFs are auto-inhibitory under the resting state. For instance, an N-terminal helical extension in two RhoGEFs (VAV and Tim) binds to DH domain active sites and blocks its access to GTPases (Aghazadeh et al, 2000; Yohe et al, 2007). It is certainly possible that the mode of EspH function in inhibiting RhoGEF activity might be reminiscent of RhoGEF auto-inhibition. Future efforts in this regard will further our understanding of the biochemical mechanism of EspH inhibition of RhoGEF activity.
Another interesting question that remains to be answered is the target specificity of EspH. In transfection assays, EspH could interact with DH-PH domains from multiple RhoGEFs including the three RGS-RhoGEFs and p63RhoGEF and Dbl (Supplementary Figure S6A and data not shown). This is not unexpected as the DH-PH domain is highly conserved in sequence and structure. The human genome contains >50 DH-PH domain RhoGEFs. As the amounts of EspH secreted by EPEC into host cells are low, it is reasonable to speculate that EspH might only target a specific pool of a particular RhoGEF(s) during infection given the model of EspH function as a competitive inhibitor of RhoGEF. Interestingly, recent studies show that EPEC type III effector Map mimics the host Dbl and catalyses the exchange of GDP for GTP in Cdc42 both in vitro and in host cells (Ohlson et al, 2008; Huang et al, 2009). It will be interesting to determine whether EspH is also capable of binding and inhibiting Map or other RhoGEF-like effectors from EPEC itself.
An earlier study shows that inhibition of Rho GTPase activity accelerates the kinetics of actin pedestal elongation during infection (Ben-Ami et al, 1998). Our discovery that EspH attenuates the host Rho GTPase signalling through antagonizing the DH-PH domain RhoGEF provides a possible mechanistic understanding of the function of EspH in aiding pedestal formation. Meanwhile, EPEC infection also induces an immediate and transient formation of filopodia (Kenny et al, 2002). This requires activation of Cdc42, which is mediated by the Dbl-mimicking Cdc42 GEF activity of Map (Ohlson et al, 2008; Berger et al, 2009; Huang et al, 2009) and possibly also involves a host Cdc42 GEF(s). Therefore, inhibition of filopodia formation by EspH observed by Tu et al (2003) might result from EspH inactivation of RhoGEF revealed in our study. Inhibition of actin polymerization during filopodia formation by EspH might increase the local concentration of actin monomers, which could facilitate actin pedestal formation that requires high local concentration of the building block.
In EHEC rabbit infection, the espH− mutant has a significantly reduced colonization throughout the intestinal tract and only the infected animals have mild diarrhoea. These suggest that EspH might have a function in bacterial adherence or resistance to host defence (Mundy et al, 2004; Ritchie and Waldor, 2005). Consistent with this hypothesis, we show that EspH is critical for resisting macrophage phagocytosis. Three other EPEC TTSS effectors have been proposed to have antiphagocytosis function. EspF, a small-size effector associated with a dozen of different pathogenic functions, is required for EPEC antiphagocytosis with the mechanism undefined (Celli et al, 2001; Quitard et al, 2006; Dean and Kenny, 2009). EspJ has been proposed to inhibit both IgG- and complement receptor-mediated phagocytosis, but the mechanism remains completely unknown (Marches et al, 2008). The TTSS translocator protein EspB does this by targeting the host myosin (Iizumi et al, 2007), but it is neither solely responsible nor sufficient for the antiphagocytosis function of EPEC (Dean and Kenny, 2009). Therefore, EspH might function together with EspF and EspB to block macrophage phagocytosis by targeting different host processes, thereby promoting bacterial survival in the host. Antiphagocytosis function of EspH likely results from its inhibitory activity towards RhoGEFs and actin cytoskeleton dynamics. Until now, no DH-PH domain RhoGEFs are established in controlling Rho activation and actin cytoskeleton arrangements in phagocytosis. Identification of the specific RhoGEF targeted by EspH during EPEC infection will likely help to define the function of RhoGEFs in regulating macrophage phagocytosis.
Materials and methods
Plasmids, antibodies and reagents
EspH DNA (accession number GI: 47118301) was amplified from EHEC strain O157:H7. For expression in mammalian cells, the PCR fragment was cloned into pCDNA3 vector with an N-terminal flag tag, pCS2 vector with six copies of myc epitope at the N-terminus and the pEGFP-C1 vector. For complementation of the deletion mutant, the PCR fragment was cloned into the pZLQ vector with the trc promote (Luo et al, 2003). pRK5-myc-RhoGTPases constructs were described earlier (Shao et al, 2002). pFL-mDia1 and pCAG-myc-ROCK-I provided by Dr Shuh Narumiya were used as the PCR template for subcloning into the pCS2-Flag-expression vector. All RhoGEFs and their truncations were cloned into pCS2-Flag-expression vector. EE-tagged constitutively active hetero GTPase Gα13 Q226L, Gα12 Q231L and Gαq Q209L were purchased from Missouri S&T cDNA Resource Center. All the point mutation constructs were generated by QuikChange Site-Directed Mutagenesis kit (Stratagene). All the plasmids were verified by DNA sequencing. Antibodies for RhoA (26C4) and myc were purchased from Santa Cruz Biotechnology. Anti-Flag M2 antibody and Flag M2 agarose affinity gel were from Sigma. Anti-E. coli serum (serotype O127) (Tianjin Biochip Corporation) was used for immunostaining of EPEC E2348/69. Rhodamine-phalloidin, Alexa-488- and Alexa-546-conjugated goat anti-rabbit IgG secondary antibodies were purchased from Invitrogen. Cell culture products were also from Invitrogen and all other chemicals were Sigma-Aldrich products unless noted.
Cell culture and fluorescence microscopy
All cell lines were obtained from ATCC; 293T, NIH 3T3, HeLa cells, RAW264.7 and J774A.1 were grown in Dulbecco's modified Eagle's medium (DMEM) (Hyclone) supplemented with 10% foetal bovine serum (FBS), 2 mM L-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. Caco-2 cells were grown in Minimum Essential Medium supplemented with 20% FBS and 2 mM L-glutamine. T84 cells were grown in a 1:1 mixture of Ham's F12 medium and DMEM supplemented with 5% FBS and 2 mM L-glutamine. Bone marrow macrophages were isolated from femurs (C57BL/6 mice) as described earlier (Boyden and Dietrich, 2006) and maintained in RPMI 1640 medium containing 20% FBS and 30% L929 cell-conditioned supernatant. On day 7, macrophages were re-plated into RPMI 1640 medium containing 10% FBS. Cells were cultivated in a humidified atmosphere containing 5% CO2 at 37°C.
HeLa cells cultured on coverslips were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 10 min at room temperature (RT), washed with PBS and then permeabilized for 10 min in PBS containing 0.5% Triton X-100. Fixed cells were blocked with 1% BSA for 30 min. Filamentous actin were stained for 1 h by using Rhodamine-phalloidin supplemented with 0.1% BSA. Images were acquired using a Zeiss LSM510 META laser scanning confocal microscopy.
Transfection, immunoprecipitation and luciferase assays
The 293T and HeLa cells were transfected with an efficiency of ∼85–90% using the standard calcium phosphate method. Lipofectamine 2000 (Invitrogen) and Vigorous transfection reagent (Vigorous Biotechnology Beijing Co., Ltd) were used to transfect NIH 3T3 and RAW264.7 cells, respectively. For immunoprecipitation and GST co-precipitation assays, 293T and RAW264.7 cells at a confluence of 80% were transfected with indicated plasmids (5 μg for 293T cells and 10 μg for RAW264.7 cells). Transfected cells were lysed 24 h later in a buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate and a protease inhibitor mixture (Roche Molecular Biochemicals). Cell lysates were incubated with Flag M2 beads or glutathione beads for 2 h at 4°C followed by extensive wash with the lysis buffer. Proteins bound on the beads were eluted with the SDS loading buffer and analysed by immunoblotting. For infected RAW264.7 cells, cells were washed three times with PBS after 2 h of infection and lysed in a buffer containing 25 mM Tris (pH 7.4), 150 mM NaCl, 30 mM MgCl2, 1 mM DTT, 0.05% Triton X-100 and the protease inhibitor mixture for 30 min at 4°C. A total of 1% Triton X-100 was added to the centrifuged lysates before subjection to immunoprecipitation.
For luciferase assays, 293T cells were serum starved (0.5% serum) for 1 h before transfection with luciferase reporter plasmid, and luciferase activity was determined 11 h post-transfection using the dual-luciferase assay kit (Promega). As luciferase reporter plasmids are co-transfected together with EspH, the luciferase counts only measures the reporter activity from transfected cells. For stimulation, 293T cells were serum starved for 18 h after transfection and then stimulated with LPA (50 μM), serum (20%) or thrombin (10 unit/ml) for 6 h before the assay time.
EPEC manipulation and phagocytosis assays
Nalidixic acid-resistant EPEC strain E2348/68 strains (wild type and the EspB deletion mutant UMD864 and the espH− mutant) were cultured in LB broth. The espH− strain was constructed as described earlier (Donnenberg and Kaper, 1991) and confirmed by PCR analyses. For infection, bacteria were cultured overnight at 37°C without shaking, and the LB culture was diluted 1:40 into DMEM containing 2 mM L-glutamine. Before infection, bacteria were pre-activated by incubation in a humidified atmosphere containing 5% CO2 at 37°C for 3 h. To determine EPEC secretion of EspH, bacteria cultured in DMEM were induced with 1 mM IPTG after OD600 reached 0.6. A total of 1.5 ml of induced culture was centrifuged, and the filtered supernatant (0.2 μm pore size) was precipitated with 10% trichloroacetic acid for 1 h at 4°C. The pellet was washed twice with ice-cold acetone and suspended in SDS loading buffer. Samples were analysed by anti-flag immunoblotting analyses.
To examine cell-rounding effects, HeLa cells were infected with the wild-type or espH− strain (MOI, 100:1). Rounding cells were identified and photographed using a phase contrast microscope. To examine phagocytosis of the bacteria by macrophages, bone marrow macrophages were seeded onto coverslips in six-well dishes 1 day before infection at a density of 7 × 105 cells/well. J774A.1 cells were seeded at a density of 4 × 105 cells/well. Both cells were incubated for 1 h in serum-free medium and then infected for 1 h with indicated EPEC strains (MOI of 20:1). Infected cells were washed with PBS, fixed in 4% paraformaldehyde for 30 min, and blocked with 10% normal goat serum for 1 h at RT. Phagocytosis assays were performed by differential staining of extracellular and total macrophage-associated EPEC (Goosney et al, 1999). Briefly, infected macrophages were sequentially stained with anti-E. coli O127 antibody and Alexa-546-conjugated goat anti-rabbit IgG antibody (extracellular bacteria). Afterwards, cells were washed with PBS, fixed in 4% paraformaldehyde for another 10 min and permeabilized with 0.5% Triton X-100. Total cell-associated bacteria were identified by staining of permeabilized cells with the same primary antibody followed by Alexa-488-conjugated goat anti-rabbit IgG antibody. At least 50 cells per coverslip were randomly selected and the number of extracellular and total macrophage-associated EPEC was counted. The experiment was repeated for at least three times.
For phagocytosis of the latex beads (3 μm), J774A.1 cells were seeded at a density of 9 × 105 cells/well 12 h before infection. Latex particles were opsonized with 1 mg/ml IgG (3 h at RT), washed with PBS and re-suspended in serum-free DMEM. After 1.5 h of starvation in serum-free DMEM, J774A.1 cells were pre-treated with 150 ng/ml of PMA for 20 min, and then infected with pre-activated EPEC strain (MOI, 20:1); 2 h later, cells were challenged with latex beads for 40 min at 37°C, extensively washed and then fixed in 4% paraformaldehyde for 10 min. Extracellular beads were stained with Alexa-546-conjugated goat anti-mouse IgG antibody. After permeabilization with 0.5% Triton X-100, total beads were then stained with Alexa-488-conjugated goat anti-mouse IgG antibody. The phagocytosis index was determined by counting the number of latex beads using the Image J software.
Supplementary Material
Acknowledgments
We are grateful to Dr Gary M Bokoch for providing pGEX-rhotekin-RBD construct; Dr Shuh Narumiya for pFL-mDia1-, pCAG-myc-ROCK-I- and pCAG-myc-ROCK-II-expression plasmids; Dr Klaus Aktories for GST-CNF1 bacterial-expression vector; Drs Dianqing Wu and Zhong Li for p115-RhoGEF and SRE.L-luciferase reporter plasmid; Dr J Silvio Gutkind for PDZ-RhoGEF and LARG cDNAs and Dr Anthony R Means for MLK3 and p63RhoGEF-expression constructs. We thank Dr Michael S Donnenberg and Dr Wensheng Luo for the gift of pCVD442 suicide vector and EPEC E2348/69 wild type and the EspB deletion mutant (UMD864) strains. We also thank the members of the Shao laboratory for helpful discussions and technical assistance. This work was supported by Chinese Ministry of Science and Technology ‘863' Grant (2008AA022309) and ‘973' grant (2006CB806502).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aghazadeh B, Lowry WE, Huang XY, Rosen MK (2000) Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102: 625–633 [DOI] [PubMed] [Google Scholar]
- Aktories K, Barbieri JT (2005) Bacterial cytotoxins: targeting eukaryotic switches. Nat Rev Microbiol 3: 397–410 [DOI] [PubMed] [Google Scholar]
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, Dixon JE (2006) Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124: 133–145 [DOI] [PubMed] [Google Scholar]
- Ben-Ami G, Ozeri V, Hanski E, Hofmann F, Aktories K, Hahn KM, Bokoch GM, Rosenshine I (1998) Agents that inhibit Rho, Rac, and Cdc42 do not block formation of actin pedestals in HeLa cells infected with enteropathogenic Escherichia coli. Infect Immun 66: 1755–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger CN, Crepin VF, Jepson MA, Arbeloa A, Frankel G (2009) The mechanisms used by enteropathogenic Escherichia coli to control filopodia dynamics. Cell Microbiol 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ED, Dietrich WF (2006) Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet 38: 240–244 [DOI] [PubMed] [Google Scholar]
- Campellone KG, Robbins D, Leong JM (2004) EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell 7: 217–228 [DOI] [PubMed] [Google Scholar]
- Caprioli A, Falbo V, Ruggeri FM, Baldassarri L, Bisicchia R, Ippolito G, Romoli E, Donelli G (1987) Cytotoxic necrotizing factor production by hemolytic strains of Escherichia coli causing extraintestinal infections. J Clin Microbiol 25: 146–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E, Crepin VF, Simpson N, Knutton S, Garmendia J, Frankel G (2006) Subversion of actin dynamics by EPEC and EHEC. Curr Opin Microbiol 9: 40–45 [DOI] [PubMed] [Google Scholar]
- Caron E, Hall A (1998) Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282: 1717–1721 [DOI] [PubMed] [Google Scholar]
- Celli J, Olivier M, Finlay BB (2001) Enteropathogenic Escherichia coli mediates antiphagocytosis through the inhibition of PI 3-kinase-dependent pathways. EMBO J 20: 1245–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HD, Frankel G (2005) Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev 29: 83–98 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F (2009) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 35: 841–855 [DOI] [PubMed] [Google Scholar]
- Cheng HC, Skehan BM, Campellone KG, Leong JM, Rosen MK (2008) Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U). Nature 454: 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P, Kenny B (2009) The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr Opin Microbiol 12: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB (2004) Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci USA 101: 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda U, Oleksy A, Stevenson AS, Korczynska J, Dauter Z, Somlyo AP, Otlewski J, Somlyo AV, Derewenda ZS (2004) The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle. Structure 12: 1955–1965 [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59: 4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P (1997) Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature 387: 729–733 [DOI] [PubMed] [Google Scholar]
- Frankel G, Phillips AD (2008) Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell Microbiol 10: 549–556 [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind JS (1999) A novel PDZ domain containing guanine nucleotide exchange factor links heterotrimeric G proteins to Rho. J Biol Chem 274: 5868–5879 [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573 [DOI] [PubMed] [Google Scholar]
- Garmendia J, Frankel G, Crepin VF (2005) Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect Immun 73: 2573–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia J, Phillips AD, Carlier MF, Chong Y, Schuller S, Marches O, Dahan S, Oswald E, Shaw RK, Knutton S, Frankel G (2004) TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol 6: 1167–1183 [DOI] [PubMed] [Google Scholar]
- Goosney DL, Celli J, Kenny B, Finlay BB (1999) Enteropathogenic Escherichia coli inhibits phagocytosis. Infect Immun 67: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279: 509–514 [DOI] [PubMed] [Google Scholar]
- Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G (1998) Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Galpha13. Science 280: 2112–2114 [DOI] [PubMed] [Google Scholar]
- Hart MJ, Sharma S, elMasry N, Qiu RG, McCabe P, Polakis P, Bollag G (1996) Identification of a novel guanine nucleotide exchange factor for the Rho GTPase. J Biol Chem 271: 25452–25458 [DOI] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R (1995) The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM (2009) Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol 16: 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizumi Y, Sagara H, Kabe Y, Azuma M, Kume K, Ogawa M, Nagai T, Gillespie PG, Sasakawa C, Handa H (2007) The enteropathogenic E. coli effector EspB facilitates microvillus effacing and antiphagocytosis by inhibiting myosin function. Cell Host Microbe 2: 383–392 [DOI] [PubMed] [Google Scholar]
- Ishizaki T, Naito M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S (1997) p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett 404: 118–124 [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2: 123–140 [DOI] [PubMed] [Google Scholar]
- Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB (1997) Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91: 511–520 [DOI] [PubMed] [Google Scholar]
- Kenny B, Ellis S, Leard AD, Warawa J, Mellor H, Jepson MA (2002) Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol Microbiol 44: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Kourlas PJ, Strout MP, Becknell B, Veronese ML, Croce CM, Theil KS, Krahe R, Ruutu T, Knuutila S, Bloomfield CD, Caligiuri MA (2000) Identification of a gene at 11q23 encoding a guanine nucleotide exchange factor: evidence for its fusion with MLL in acute myeloid leukemia. Proc Natl Acad Sci USA 97: 2145–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC (1998) p115 RhoGEF, a GTPase activating protein for Galpha12 and Galpha13. Science 280: 2109–2111 [DOI] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F (2007) The phosphothreonine lyase activity of a bacterial type III effector family. Science 315: 1000–1003 [DOI] [PubMed] [Google Scholar]
- Luo W, Donnenberg MS (2006) Analysis of the function of enteropathogenic Escherichia coli EspB by random mutagenesis. Infect Immun 74: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Smyth AJ, Gao P, Qin Y, Farrand SK (2003) Mutational analysis of TraR. Correlating function with molecular structure of a quorum-sensing transcriptional activator. J Biol Chem 278: 13173–13182 [DOI] [PubMed] [Google Scholar]
- Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, Tesmer JJ (2007) Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318: 1923–1927 [DOI] [PubMed] [Google Scholar]
- Mao J, Yuan H, Xie W, Wu D (1998) Guanine nucleotide exchange factor GEF115 specifically mediates activation of Rho and serum response factor by the G protein alpha subunit Galpha13. Proc Natl Acad Sci USA 95: 12973–12976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marches O, Covarelli V, Dahan S, Cougoule C, Bhatta P, Frankel G, Caron E (2008) EspJ of enteropathogenic and enterohaemorrhagic Escherichia coli inhibits opsono-phagocytosis. Cell Microbiol 10: 1104–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo S, Lee YM, Dixon JE (2007) Interactions of bacterial effector proteins with host proteins. Curr Opin Immunol 19: 392–401 [DOI] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV (1999) Food-related illness and death in the United States. Emerg Infect Dis 5: 607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R, Petrovska L, Smollett K, Simpson N, Wilson RK, Yu J, Tu X, Rosenshine I, Clare S, Dougan G, Frankel G (2004) Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect Immun 72: 2288–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Koller A, Nordfelth R, Wolf-Watz H, Taylor S, Dixon JE (2007) Identification of a molecular target for the Yersinia protein kinase A. Mol Cell 26: 465–477 [DOI] [PubMed] [Google Scholar]
- Ohlson MB, Huang Z, Alto NM, Blanc MP, Dixon JE, Chai J, Miller SI (2008) Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe 4: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JC, Hall A, Caron E (2002) Vav regulates activation of Rac but not Cdc42 during FcgammaR-mediated phagocytosis. Mol Biol Cell 13: 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitard S, Dean P, Maresca M, Kenny B (2006) The enteropathogenic Escherichia coli EspF effector molecule inhibits PI-3 kinase-mediated uptake independently of mitochondrial targeting. Cell Microbiol 8: 972–981 [DOI] [PubMed] [Google Scholar]
- Ren XD, Schwartz MA (2000) Determination of GTP loading on Rho. Methods Enzymol 325: 264–272 [DOI] [PubMed] [Google Scholar]
- Ritchie JM, Waldor MK (2005) The locus of enterocyte effacement-encoded effector proteins all promote enterohemorrhagic Escherichia coli pathogenicity in infant rabbits. Infect Immun 73: 1466–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee NA, Rivera GM, Dueber JE, Vasilescu D, Mullins RD, Mayer BJ, Lim WA (2008) The pathogen protein EspF(U) hijacks actin polymerization using mimicry and multivalency. Nature 454: 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K (1997) Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387: 725–729 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109: 575–588 [DOI] [PubMed] [Google Scholar]
- Swenson-Fields KI, Sandquist JC, Rossol-Allison J, Blat IC, Wennerberg K, Burridge K, Means AR (2008) MLK3 limits activated Galphaq signaling to Rho by binding to p63RhoGEF. Mol Cell 32: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Nisan I, Yona C, Hanski E, Rosenshine I (2003) EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol Microbiol 47: 595–606 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S (1999) Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol 1: 136–143 [DOI] [PubMed] [Google Scholar]
- Yao Q, Cui J, Zhu Y, Wang G, Hu L, Long C, Cao R, Liu X, Huang N, Chen S, Liu L, Shao F (2009) A bacterial type III effector family uses the papain-like hydrolytic activity to arrest the host cell cycle. Proc Natl Acad Sci USA 106: 3716–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe ME, Rossman KL, Gardner OS, Karnoub AE, Snyder JT, Gershburg S, Graves LM, Der CJ, Sondek J (2007) Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J Biol Chem 282: 13813–13823 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F (2008) Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 15: 1302–1308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.