Abstract
During bacterial cytokinesis, hydrolytic enzymes are used to split wall material shared by adjacent daughter cells to promote their separation. Precise control over these enzymes is critical to prevent breaches in wall integrity that can cause cell lysis. How these potentially lethal hydrolases are regulated has remained unknown. Here, we investigate the regulation of cell wall turnover at the Escherichia coli division site. We show that two components of the division machinery with LytM domains (EnvC and NlpD) are direct regulators of the cell wall hydrolases (amidases) responsible for cell separation (AmiA, AmiB and AmiC). Using in vitro cell wall cleavage assays, we show that EnvC activates AmiA and AmiB, whereas NlpD activates AmiC. Consistent with these findings, we show that an unregulated EnvC mutant requires functional AmiA or AmiB but not AmiC to induce cell lysis, and that the loss of NlpD phenocopies an AmiC− defect. Overall, our results suggest that cellular amidase activity is regulated spatially and temporally by coupling their activation to the assembly of the cytokinetic ring.
Keywords: amidase, cell wall, cytokinesis, divisome, peptidoglycan
Introduction
Most bacteria surround themselves with a complex cell envelope that serves as a barrier to insult and a conduit for nutrient uptake. An important component of the envelope is the cell wall or peptidoglycan (PG) layer. This essential layer is constructed from polysaccharide strands that are covalently connected along their length by cross-linked peptides to form a continuous meshwork that encapsulates the cytoplasmic membrane and protects it from osmotic rupture (Vollmer, 2008) (Figure 1). During growth and division, this tough exoskeleton is continuously remodelled by PG synthases called penicillin-binding proteins (PBPs) as well as an array of PG hydrolases capable of cleaving bonds in the PG meshwork (Vollmer et al, 2008). Tight control over these potentially lethal hydrolases must be maintained to prevent them from creating breaches in the cell wall that can lead to cell lysis. Despite the importance of such controls, the cellular mechanisms regulating PG hydrolases have long remained mysterious.
Figure 1.
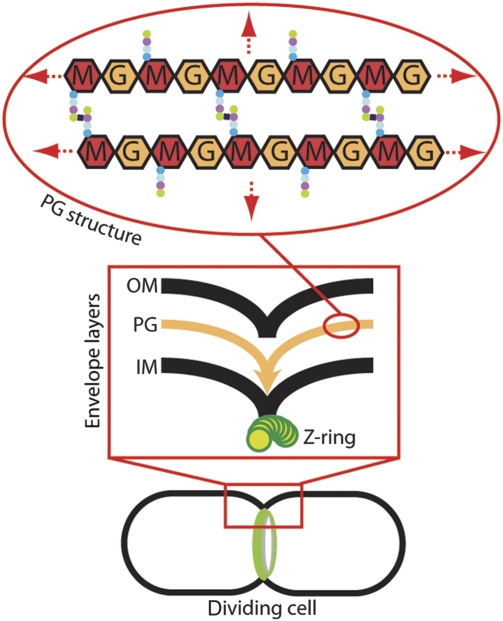
Coordinated envelope constriction in gram-negative bacteria. Diagram of a dividing cell with an assembled cytokinetic ring apparatus (green). The box contains a close-up diagram of the division site highlighting the coordinated constriction of the envelope layers: OM, outer membrane; PG, peptidoglycan; IM, inner membrane; Z-ring, FtsZ cytoskeletal ring. The oval contains a diagram of the PG chemical structure: M, N-acetylmuramic acid; G, N-acetylglucosamine. Coloured dots represent the attached peptides. The PG structure continues in all directions to envelop the cell (red arrows).
The most clearly defined function for cellular PG hydrolases is the splitting of wall material formed between daughter cells during the division process (Vollmer et al, 2008). In Escherichia coli and other gram-negative bacteria, cytokinesis proceeds via the coordinated constriction of all three envelope layers: the inner and outer membranes along with the PG layer sandwiched between them (Figure 1) (den Blaauwen et al, 2008; Uehara et al, 2009). This coordination is achieved by the divisome, a ring-shaped, multiprotein division machine organized by the bacterial tubulin protein, FtsZ (den Blaauwen et al, 2008). One of the primary functions of the divisome is to promote the synthesis of the PG layer that will eventually fortify the new daughter cell poles. This involves several divisome-associated PBPs (den Blaauwen et al, 2008). Although the septal PG produced by these synthases is initially shared by the daughter cells, it must be split shortly after it is formed to allow constriction of the outer membrane to closely follow that of the inner membrane and FtsZ ring (Figure 1). In E. coli, two sets of periplasmic factors are critical for septal PG splitting: LytC-type N-acetylmuramyl-L-alanine amidases (Pfam: Amidase_3) and LytM (lysostaphin/Pfam: Peptidase_M23) factors, both of which are widely distributed among eubacteria (Heidrich et al, 2001; Firczuk and Bochtler, 2007; Vollmer et al, 2008; Uehara et al, 2009). Mutants lacking multiple amidases or LytM factors form long chains of cells that complete inner membrane constriction and fusion, but remain connected by unsplit layers of septal PG that interfere with outer membrane invagination (Heidrich et al, 2001; Priyadarshini et al, 2007; Uehara et al, 2009).
Amidases are PG hydrolases that remove the stem peptide from the glycan strands of PG and can therefore break its cross-links. E. coli encodes three factors with LytC-type amidase domains: AmiA, AmiB and AmiC, all of which are important for cell separation (Heidrich et al, 2001). The founding members of the LytM family of factors, LytM and lysostaphin, are metallo-endopeptidases that cleave pentaglycine cross-bridges in staphylococcal PG (Firczuk and Bochtler, 2007). On the basis of this activity, LytM factors in other bacteria are thought to be PG hydrolases as well, but with altered cleavage specificity because pentaglycine cross-bridges are only found among the staphylococci (Schleifer and Kandler, 1972). E. coli encodes four factors with recognizable LytM domains. Two of them, EnvC and NlpD, have major functions in cell separation (Uehara et al, 2009). The biochemical activities of the E. coli amidases and LytM factors have remained poorly defined. Amidase activity in a purified system has only been shown for AmiA (Lupoli et al, 2009). AmiC amidase activity has only been detected in crude cell extracts, whereas that of AmiB has not been reported so far (Heidrich et al, 2001). Consistent with EnvC possessing PG hydrolase activity, it was able to generate a zone of clearing in a gel-based zymogram assay (Bernhardt and de Boer, 2004). However, its activity has not been investigated in solution, nor has that of NlpD.
Although EnvC, NlpD and all of the amidases except for AmiA are recruited to midcell to participate in the division process (Bernhardt and de Boer, 2003, 2004; Uehara et al, 2009) (T Dinh, unpublished results), it has remained unclear how they work together to specifically promote cell separation during cytokinesis without also promoting cell lysis during interdivisional periods of the cell cycle. Here, we show that the LytM factors, EnvC and NlpD, are not themselves PG hydrolases as previously suspected, but rather function as potent and specific activators of the PG amidases. We infer that the LytM factors have a direct and critical function in the spatiotemporal regulatory mechanism that confines cellular amidase activity to the cytokinetic ring.
Results
The coiled-coil domain of EnvC is a regulatory domain that is necessary and sufficient for recruitment to the divisome
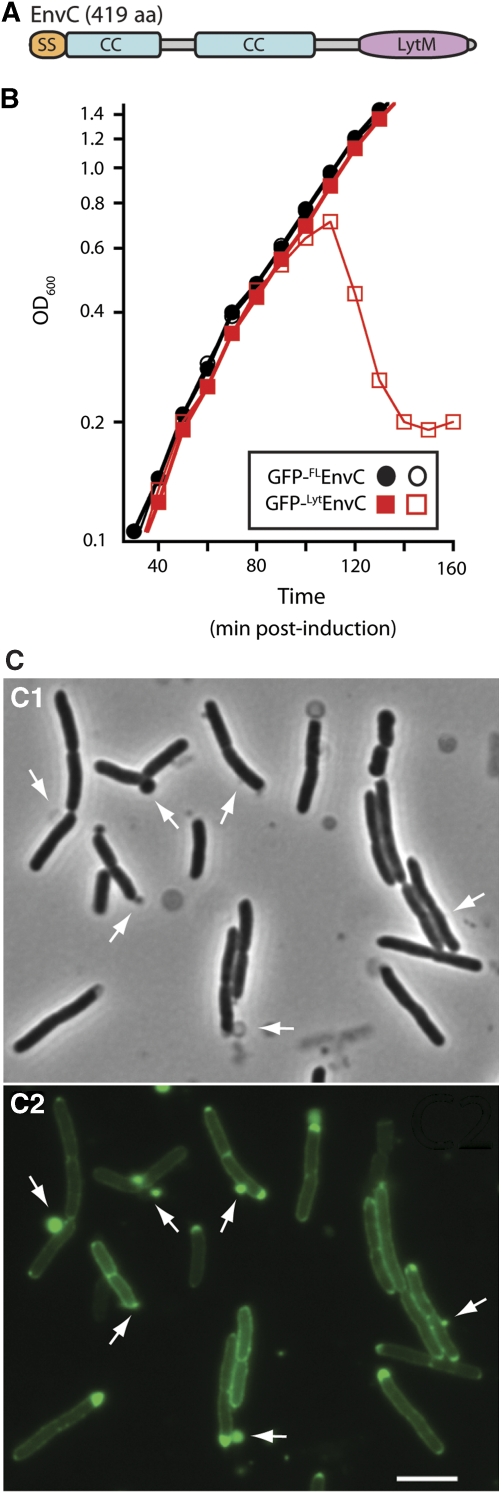
EnvC is a 419 amino-acid protein that contains three identifiable domains: a signal peptide (residues 1–34), a coiled-coil (CC) domain (residues 35–271) and a LytM domain (residues 318–413) (Hara et al, 2002; Ichimura et al, 2002; Bernhardt and de Boer, 2004) (Figure 2A). We hypothesized that the CC domain might serve a regulatory function controlling the putative PG hydrolase activity of EnvC. To explore this possibility, we monitored the growth and morphology of cells producing exported GFP fusions to full length, mature EnvC (35–419) [GFP-FLEnvC] or a truncated version lacking the CC domain, EnvC (278–419) [GFP-LytEnvC]. As expected from earlier results (Bernhardt and de Boer, 2004), GFP-FLEnvC was functional, did not affect growth and localized to the septum (Figure 2B; Supplementary Figure S1A and S1B). Production of GFP-LytEnvC, on the other hand, was extremely toxic and caused cell lysis after low-level induction (Figure 2B). This fusion displayed a largely peripheral (periplasmic) localization pattern and caused the formation of envelope lesions at many locations around the cell body (Figure 2C; Supplementary Figure S1C). Immunoblot analysis revealed that the lytic dose of GFP-LytEnvC was barely detectable relative to the amount of GFP-FLEnvC produced in normally growing cells supplemented with high levels of inducer (Supplementary Figure S2A and S2B). Thus, the CC domain does not control EnvC levels, but instead seems to restrict EnvC activity to the division site by mediating its recruitment to the divisome.
Figure 2.
The LytM domain of EnvC induces lysis. (A) Predicted domain structure of EnvC. SS, signal sequence; CC, coiled coil; LytM, LytM/lysostaphin/peptidase_M23 domain. (B) Wild-type (MG1655) cells harbouring the expression constructs pTU113 [Plac∷ssdsbA-gfp-flenvC] or pTU115 [Plac∷ssdsbA-gfp-lytenvC] integrated at the phage HK022 att site were grown in LB at 37°C to an OD600 of about 0.5. At t=0, the cultures were diluted into fresh LB with (open symbols) or without (closed symbols) 20 μM IPTG and growth was continued at 37°C. Induction of GFP-FLEnvC with much higher IPTG levels (250 μM–1 mM) also failed to affect cell growth (data not shown). Fusions encoded the signal sequence from dsbA for export and the GFP variant used was superfolder GFP (Pédelacq et al, 2006), which we have found is functional in the periplasm following Sec export (T Dinh, unpublished results). (C) Phase contrast (C1) and GFP-fluorescence (C2) micrographs of cells lysing as a result of GFP-LytEnvC production. Arrows highlight cell envelope defects, many of which correspond to regions with an enlarged periplasm as indicated by the accumulation of the exported GFP-LytEnvC fusion. Bar=4 μm. See Supplementary Figure S1C for images of cells before the onset of lysis.
To determine whether the CC domain is sufficient for the recruitment of EnvC to the divisome, we studied the localization of an exported GFP-EnvC (35–277) fusion [GFP-CCEnvC]. To eliminate the possibility that GFP-CCEnvC might be recruited to the septum through EnvC–EnvC interactions rather than EnvC–divisome interactions, we studied its localization in a ΔenvC strain. Periplasmic protein localization studies in this background are complicated by the fact that unfused periplasmic GFP seems to accumulate at the septa of EnvC− cells because the delay in septal PG splitting results in an increased periplasmic volume at these sites (Bernhardt and de Boer, 2004). To account for this, we co-produced exported mCherry and GFP-CCEnvC in the ΔenvC mutant. In these cells, we observed ring or band-like accumulations of GFP-CCEnvC at nascent septa that lacked corresponding accumulations of periplasmic mCherry (Figure 3A). In contrast and as expected, when exported mCherry and unfused GFP were co-produced in ΔenvC cells, their apparent accumulations at septa were always coincident (Figure 3B and C). We conclude that the CC domain is necessary and sufficient for EnvC recruitment to the divisome. Moreover, the failure of GFP-CCEnvC to correct the EnvC− phenotype (Figure 3) indicates that the LytM domain is required for EnvC to promote proper septal PG splitting.
Figure 3.
The CC domain of EnvC is a septal targeting domain. (A–C) Shown are cells of TU176/pTU175 [ΔenvC/Psyn135∷ssdsbA-mCherry] containing the integrated GFP fusion constructs (A) attHKTU128 (Plac∷ ssdsbA-gfp-ccenvC) and (B, C) attHKTB263 (Plac∷ ssdsbA-gfp). Cells were grown to an OD600 of about 0.5 in M9 maltose medium without IPTG (A, B) or with 100 μM IPTG (C), and visualized with GFP (panels A1, B1 and C1), mCherry (panels A2, B2 and C2) or DIC (panels A3, B3 and C3) optics. Arrows highlight GFP-CCEnvC recruitment to nascent division sites that lack a strong periplasmic mCherry signal. Unlike the GFP-CCEnvC construct, which could be visualized in cells grown without IPTG, visualization of periplasmic GFP required the addition of 100 μM IPTG. Psyn135 is a synthetic promoter for constitutive expression. Note that the ΔenvC cells expressing periplasmic GFP-CCEnvC or GFP both display the cell division and cell separation phenotypes typical of EnvC− mutants. (D, E) Shown are cells of TB134 (attHKTB316) [ΔenvC (Plac∷flenvC-mCherry)] containing the integrated GFP fusion constructs (D) attλTU222 (Para∷ ssdsbA-gfp-ccenvC) or (E) attλTU178 (Para∷ ssdsbA-gfp). Cells were grown at 30°C in M9 maltose medium supplemented with 0.2% arabinose but without IPTG, and visualized with mCherry (panels D1 and E1), GFP (panels D2 and E2) or DIC (panels D3 and E3) optics. Bar=4 μm.
To further investigate the function of protein localization in the regulation of EnvC activity, we monitored the effect of GFP-CCEnvC overproduction on the localization and function of FLEnvC–mCherry. When produced as the sole source of full-length EnvC in cells, FLEnvC–mCherry displayed a clear septal localization pattern and promoted proper cell division (Figure 3E) (Uehara et al, 2009). Overproduction of exported GFP-CCEnvC, however, both interfered with FLEnvC–mCherry recruitment to the division site and induced a dominant-negative EnvC− division defect (Figure 3D). Immunoblots indicated that FLEnvC–mCherry remained intact when GFP-CCEnvC was overproduced (Supplementary Figure S2C). We therefore conclude that GFP-CCEnvC effectively competes with FLEnvC–mCherry for a limited number of binding sites at the divisome, and that recruitment to the divisome is critical for EnvC to promote proper septal PG splitting. Interestingly, in contrast to our findings with LytEnvC, delocalized FLEnvC did not cause cell lysis in these experiments (Figure 3D). This implies that proper spatiotemporal regulation of EnvC activity involves more than just the control of its subcellular localization (see Discussion). In this regard, the differential lytic activities of delocalized FLEnvC and LytEnvC suggest that, in addition to controlling EnvC localization, the CC domain may also directly or indirectly regulate the activity of the LytM domain. Notably, although overproduction of GFP-FLEnvC from the integrated attHKTU113 construct (Plac∷ssdsbA-gfp-flenvC) failed to induce lysis at all IPTG levels tested up to 1 mM (Figure 2, data not shown), higher-level GFP-FLEnvC production from a similar construct (attλTU179) with a stronger promoter (Para) was capable of inducing lysis (Supplementary Figure S3). This indicates that the regulatory controls governing FLEnvC activity are saturable and can be overwhelmed by excess protein.
EnvC does not degrade PG in solution
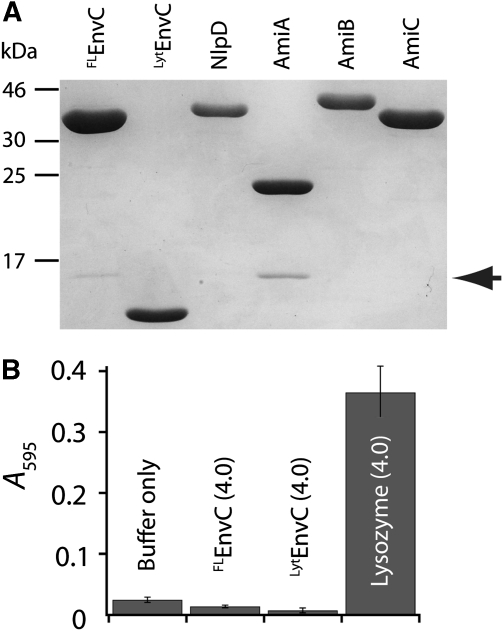
To better define the function of the CC domain in EnvC regulation, we sought to reconstitute EnvC PG hydrolase activity in solution. FLEnvC and LytEnvC were purified (Figure 4A) and tested for their ability to degrade PG using a dye-release assay (Zhou et al, 1988). For this, purified E. coli PG was covalently labelled on its sugar moieties with the dye remazol brilliant blue (RBB). RBB-modified PG was then incubated with purified protein and hydrolytic activity was monitored as dye remaining in the supernatant after the reaction was terminated and centrifuged to pellet intact PG. Control assays readily detected lysozyme-mediated PG cleavage (Figure 4B). In contrast to earlier results using a gel-based zymogram assay (Bernhardt and de Boer, 2004), however, no PG hydrolase activity was observed for FLEnvC or LytEnvC by dye release even after overnight incubation of the reactions (Figure 4B, data not shown).
Figure 4.
EnvC does not degrade PG in solution. (A) Purified proteins were separated by SDS–PAGE (12.5% T) and stained with Coomassie brilliant blue. Proteins used in this study were purified with a 6xHis-SUMO tag (H-SUMO) fused to their N-termini. The tag was removed with purified 6xHis-tagged SUMO protease (H-SP). Cleaved H-SUMO tag and H-SP were depleted from the preparation using fresh Ni-NTA resin. A small amount of H-SUMO remains in the EnvC and AmiA preparations (arrow). (B) Dye-release assays for PG hydrolysis. RBB-PG was incubated with the indicated proteins (4 μM each) for 30 min at 37°C. Undigested PG was pelleted and the absorbance of the supernatant was measured at 595 nm. Longer incubation (>20 h) did not result in significant dye release by either FLEnvC or LytEnvC (data not shown).
LytEnvC requires the amidases, AmiA and AmiB, to induce lysis
The inactivity of both FLEnvC and LytEnvC in vitro prompted us to consider that EnvC may not be a PG hydrolase, a possibility consistent with the fact that EnvC lacks all of the zinc-chelating residues identified in the active site of LytM (Odintsov et al, 2004; Firczuk et al, 2005) (Supplementary Figure S4). Nevertheless, GFP-LytEnvC must cause PG damage in vivo, as it clearly elicited cell lysis. We therefore considered the possibility that rather than being a PG hydrolase itself, EnvC might activate one. To identify the putative EnvC-activated hydrolase(s), we selected for mutants resistant to GFP-LytEnvC production. Interestingly, two resistant mutants with transposon insertions in amiB were isolated, suggesting that EnvC activates AmiB to promote septal PG splitting. To investigate this further, we tested a panel of ami deletion mutants for resistance to GFP-LytEnvC (Figure 5). Deletion of amiB partially restored the plating efficiency of cells producing GFP-LytEnvC, whereas single amiA or amiC deletions had no effect (Figure 5, data not shown). Combining ΔamiA with ΔamiB, but not ΔamiC, fully suppressed the toxicity of LytEnvC (Figure 5). Consistent with the results on solid media, amidase defects delayed or prevented lysis in response to GFP-LytEnvC production when cells were grown in liquid medium (Supplementary Figure S5).
Figure 5.
Mutants with amidase defects are resistant to GFP-LytEnvC production. Cultures of the indicated strains containing the Para∷gfp-lytenvC construct pTU181 integrated at the λ att site were grown overnight at 30°C in LB. Culture densities were normalized and 10-fold serial dilutions were prepared for each. A measure of 10 μl of each dilution was spotted onto the indicated solid media and the plates were incubated overnight at 30°C. The strains used were: TB28 [WT], TU226 [amiB∷Tn4], TU227 [amiB∷Tn6], TB170 [ΔamiB], TB141 [ΔamiA], TU207 [ΔamiA ΔamiB] and TB150 [ΔamiA ΔamiC]. The amiB∷Tn4 and amiB∷Tn6 mutants were isolated in the selection for GFP-LytEnvC resistant mutants.
Amidase activation in vitro
Our results thus far suggested that EnvC may activate AmiA and AmiB, and that GFP-LytEnvC activates them inappropriately to induce cell lysis. To test this directly, we purified the amidases (Figure 4A) and assayed their PG hydrolase activity in the presence and absence of FLEnvC. At high concentrations (4 μM) and long incubation times (4 h to overnight), purified AmiA, AmiB or AmiC alone promoted significant levels of dye release from RBB-PG (Figure 6A). When the amidases were assayed at lower concentrations and/or using shorter reaction times (30 min), minimal dye release was observed (Figure 6B–E). However, consistent with the idea that EnvC activates AmiA and AmiB, the combination of FLEnvC and AmiA or FLEnvC and AmiB using these same conditions resulted in a level of dye release comparable to that of the lysozyme control (Figure 6B–D). FLEnvC did not lead to enhanced PG hydrolysis when it was combined with AmiC as expected from the genetic results (Figures 5 and 6E). Importantly, both end point (Figure 6C and D) and time course assays (Supplementary Figure S6) indicated that LytEnvC and FLEnvC stimulated PG hydrolysis to a similar extent when they were combined with AmiA or AmiB. Thus, the LytM domain is sufficient for enhancing PG hydrolase activity in EnvC–amidase mixtures.
Figure 6.
Enhanced in vitro PG hydrolysis by EnvC–AmiA and EnvC–AmiB combinations. (A) Dye-release assays measuring PG hydrolysis by the amidases in the absence of EnvC. The absorbance of supernatants from the reactions containing the indicated proteins (4 μM) were measured at 595 nm. The reactions were incubated at 37°C for 0.5, 4 or 20 h. Results shown are the average of duplicate reactions. (B) Tubes containing supernatants of dye-release reactions following the incubation of RBB-PG with the indicated proteins (4 μM each) for 30 min at 37°C. (C–E) Same as in (A) except FLEnvC or LytEnvC was combined with the amidases and reactions were all incubated for 30 min at 37°C. Numbers indicate protein concentration in μM. Amidase concentrations were held constant, whereas the amount of FLEnvC or LytEnvC varied. From left to right, a set of amidase reactions starts with no added LytM factor (0) followed by a series of LytM factor additions where the LytM factor concentration was increased by 2 × with each step through the range. Results presented are the average of three independent reactions with the error bars displaying the standard deviation. LZ, lysozyme.
In the in vitro reactions, EnvC–AmiA combinations promoted a greater degree of PG hydrolysis than EnvC–AmiB reactions (Figure 6C and D). AmiB, on the other hand, seemed to have a more dominant function in causing cell lysis when GFP-LytEnvC was produced in vivo. This apparent dichotomy may be explained by the fact that AmiB accumulates at the division site in vivo (T Dinh, unpublished results), whereas AmiA does not (Bernhardt and de Boer, 2003). We suspect that uncontrolled activation of locally concentrated AmiB is more catastrophic than that of delocalized AmiA.
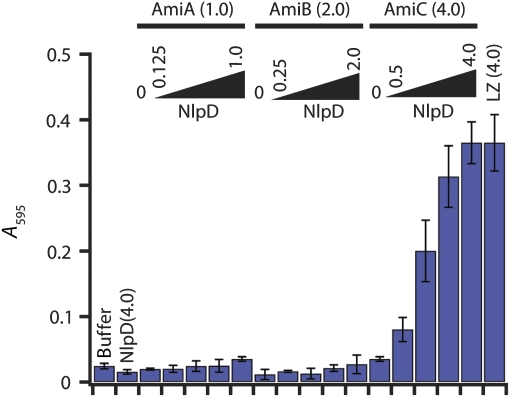
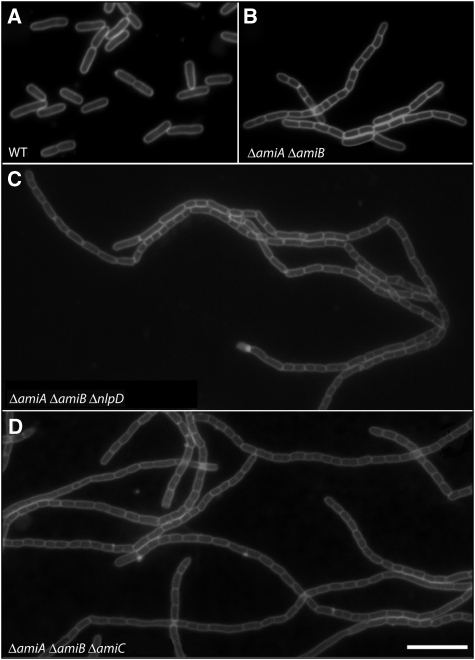
To determine whether other LytM factors are also capable of stimulating PG hydrolysis, we purified a soluble version of NlpD (Figure 4A) and tested its effect on the in vitro activity of the amidases. Interestingly, the results perfectly complemented those with EnvC. The addition of NlpD to AmiC-containing reactions dramatically enhanced PG degradation (Figure 7). In contrast, NlpD had no effect on the activity of reactions containing AmiA or AmiB (Figure 7). Like EnvC, NlpD did not display any PG cleavage activity on its own even though it possesses two of the four LytM catalytic residues (Supplementary Figures S4 and S7). Consistent with the idea that NlpD is needed for AmiC activity in vivo, NlpD inactivation phenocopied the cell separation defect of an amiC deletion. Redundancy of the amidases required that the AmiC− phenotype be visualized in a strain lacking AmiA and AmiB, and loss of AmiC or NlpD dramatically enhanced the cell separation phenotype of this strain (Figure 8).
Figure 7.
Enhanced in vitro PG hydrolysis in reactions containing NlpD and AmiC. Reactions are the same as in Figure 6 except that NlpD replaces EnvC.
Figure 8.
Loss of NlpD phenocopies ΔamiC. Cells of (A) TB28 [WT], (B) TU207 [ΔamiA ΔamiB], (C) TU218 [ΔamiA ΔamiB ΔnlpD] and (D) TB170 [ΔamiA ΔamiB ΔamiC] were grown in LB at 37°C to an OD600 of 0.5. They were then stained with the fixable membrane dye FM1-43-FX, fixed and visualized by fluorescence microscopy as described earlier (Uehara et al, 2009). Bar=8 μm.
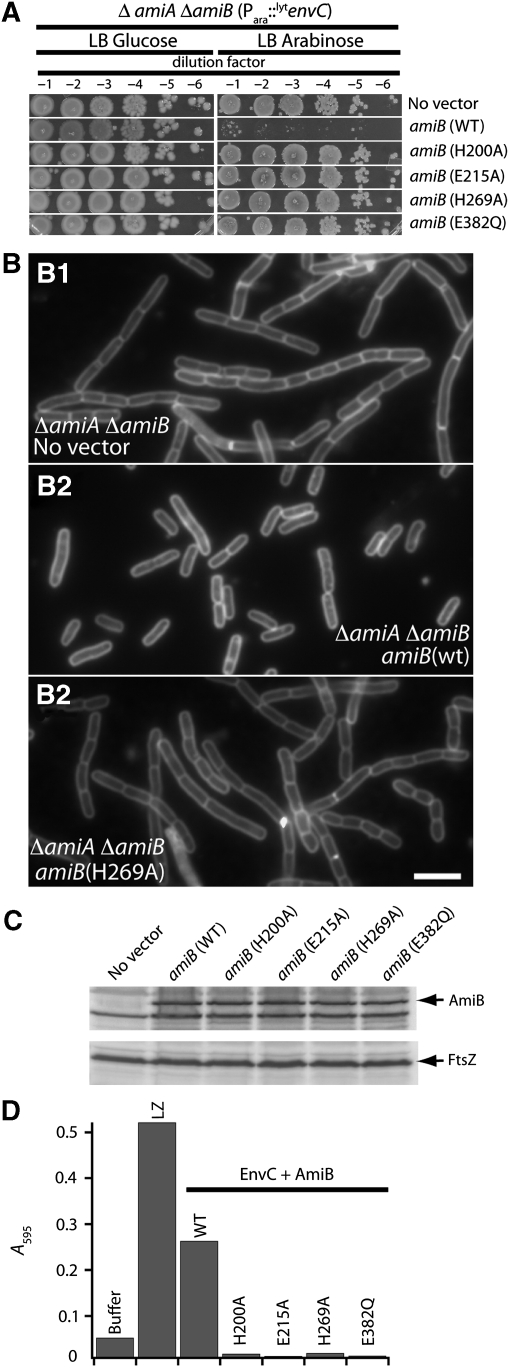
To investigate the function of the amidases in the enhanced PG hydrolysis observed for LytM–amidase reaction mixtures, we analysed the activity of purified AmiB proteins with amino-acid substitutions in predicted active site residues. To guide the design of the mutants, we aligned the AmiB sequence with that of CwlV from Paenibacillus polymyxa and PlyPSA from Listeria monocytogenes phage PSA (Supplementary Figure S7) for which residues important for catalysis have been identified through biochemical and structural analyses (Shida et al, 2001; Korndörfer et al, 2006) (PDB 1jwq). Accordingly, we generated four AmiB mutants (H200A, E215A, H269A and E382Q). All four failed to correct the cell separation and LytEnvC-mediated lysis phenotypes of a ΔamiA ΔamiB double mutant in vivo (Figure 9A–C). Upon purification, all four mutant enzymes also failed to promote PG hydrolysis when they were mixed with EnvC (Figure 9D). Based on this, we conclude that amidase enzymatic activity is indeed necessary for the amidases to promote cell separation during division and for LytEnvC to induce lysis. Furthermore, these results provide strong support for a mechanism in which the amidases provide all the PG hydrolyzing activity in the LytM–amidase reactions.
Figure 9.
AmiB active site residues are required for PG hydrolysis. (A) Cultures of TU207 [ΔamiA ΔamiB] containing chromosomally integrated Para∷gfp-lytenvC and indicated Plac∷ssdsbA-amiB(23–445)-gfp expression constructs were grown overnight at 37°C in LB. Culture densities were normalized and 10-fold serial dilutions were prepared for each. A measure of 10 μl of each dilution was spotted onto the indicated solid media and the plates were incubated overnight at 37°C. (B) Cultures of TU207 [ΔamiA ΔamiB] containing the indicated Plac∷ssdsbA-amiB(23–445)-gfp expression constructs were grown at 37°C in LB to an OD600 between 0.6–0.65. Cells were stained with FM4-64FX, fixed and visualized using fluorescence microscopy. A comparison of AmiB (wt) and AmiB (H269A) is shown, but all other AmiB catalytic mutants tested displayed the same defect in cell separation activity (data not shown). (C) Immunoblots of extracts from the cultures in (B) using anti-GFP antibodies to detect the AmiB-GFP fusions. FtsZ immunoblots are shown as a control for protein loading. (D) Dye-release assays performed as in Figure 6. EnvC and AmiB proteins were present at 2 μM each in the reaction mixtures. Results are the average of duplicate reactions except for AmiB (E215A), which was assayed once.
The biochemical activities of the purified proteins were defined more precisely by incubating them with unlabelled PG and analysing the cleavage products with a combination of HPLC and mass spectrometry. Again, using low levels of the amidases, significant PG cleavage was only observed in reactions with FLEnvC–AmiA, FLEnvC–AmiB and NlpD–AmiC combinations. Importantly, in each of these reactions, the only biochemical activity detected was peptide release by an N-acetylmuramyl-L-alanine amidase (Figure 10). As the odds are small that both EnvC and NlpD themselves possess this exact same activity in latent form, these results provide strong support for the following: (1) AmiA, AmiB and AmiC are indeed N-acetylmuramyl-L-alanine amidases, (2) EnvC and NlpD promote septal PG splitting by activating the amidases at the division site and (3) only specific LytM/amidase combinations yield efficient PG amidase activity.
Figure 10.
Analysis of PG hydrolase activity using HPLC. PG sacculi (unlabelled) were treated with buffer (A), AmiB only (4 μM) (B), FLEnvC only (4 μM) (C) or a mixture of FLEnvC and AmiB (4 μM each) (D) before mutanolysin digestion and HPLC separation of the products. Mutanolysin is an N-acetylmuraminidase that hydrolyzes the M-β-1,4-G bonds in PG. Without prior cleavage by another enzyme, mutanolysin hydrolyzes PG primarily into monomeric and dimeric disaccharide (G-M) units with attached tetrapeptides (Glauner et al, 1988). Enzymatic treatment before mutanolysin addition will alter the product composition. The numbered peaks in the chromatograms were identified using matrix-assisted laser desorption ionization (MALDI) and electrospray mass spectrometry. Peak 1, monomeric disaccharide-tetrapeptide; Peak 2, cross-linked disaccharide-tetrapeptide; Peak 3, free tetrapeptide; Peak 4, free cross-linked tetrapeptide. The profile in (D) is consistent with N-acetylmuramyl-L-alanine amidase activity being active in the reactions before mutanolysin digestion. Results for AmiA and AmiC in mixtures with FLEnvC or NlpD, respectively, were essentially the same as those shown for AmiB and FLEnvC. At the concentrations used, all enzymes were largely inactive in isolation, and only N-acetylmuramyl-L-alanine amidase activity was detected in the mixtures. Results for LytEnvC were also identical to those for FLEnvC. G, N-acetylglucosamine; M, N-acetylmuramic acid; pep4, tetrapeptide.
Discussion
Divisome-activated PG hydrolysis
To properly form the daughter cell poles and drive outer membrane invagination, the divisome of gram-negative bacteria orchestrates a high level of PG hydrolase activity at the division site shortly after it initiates septal PG biogenesis and cell constriction (Heidrich et al, 2001; Priyadarshini et al, 2006; Uehara and Park, 2008; Uehara et al, 2009). While investigating the regulation of a putative PG hydrolase involved in this process, we discovered an essential step in the activation of PG hydrolysis by the cytokinetic ring. We have shown that the E. coli LytM factors, EnvC and NlpD, are divisome-associated activators of the three PG amidases, AmiA, AmiB and AmiC, required for septal PG splitting in E. coli. At high concentrations (4 μM) and long incubation times (⩾4 h), the amidases alone showed some PG cleavage activity in vitro (Figure 6A). This activity was dramatically enhanced when EnvC or NlpD were included in the reaction (Figures 6 and 7). EnvC specifically enhanced PG hydrolysis in combination with AmiA and AmiB, and NlpD specifically enhanced PG hydrolysis when combined with AmiC (Figures 6 and 7). Although the possibility that the LytM factors possess some latent ability to hydrolyze PG directly is difficult to rule out conclusively, all of our evidence indicates instead that the predominant function of EnvC and NlpD is to activate PG hydrolysis by their cognate amidases. In support of this interpretation: (1) EnvC and NlpD did not show any PG cleavage activity in solution even when they were incubated with PG at high concentrations for long periods (>20 h) (Figure 4, data not shown), (2) EnvC and NlpD lack residues known to be important for catalysis by Staphylococcus aureus LytM (Supplementary Figure S4), (3) amidase activity was the only PG hydrolase activity detected following HPLC analysis of reaction products produced by LytM–amidase mixtures (Figure 10) and (4) substitution of AmiB residues implicated in catalytic activity abolished the hydrolytic activity of EnvC–AmiB mixtures (Figure 9). One significant implication of these results is that they help explain why cell separation requires both the amidases and the LytM factors (Heidrich et al, 2001; Priyadarshini et al, 2006; Uehara et al, 2009). Apparently, the basal PG hydrolase activity we and others observe for the amidases in vitro (Figure 6A) (Heidrich et al, 2001; Lupoli et al, 2009) is insufficient to promote cell separation in vivo even though both AmiB and AmiC accumulate at the division site (Bernhardt and de Boer, 2003) (T Dinh, unpublished results). Rather, efficient cell separation also requires that the amidases be activated by LytM factors. Though E. coli produces two different activators and three different amidases, it is clear that EnvC–AmiA and EnvC–AmiB systems have the predominant functions in cell separation. The cell separation defect of EnvC− cells is far more pronounced than that of NlpD− mutants (Uehara et al, 2009), and of all the double-amidase mutants, those lacking AmiA and AmiB have the most severe separation phenotype (Chung et al, 2009). Interestingly, although the phenotypes of EnvC− and AmiA− AmiB− mutants are similar, they are not identical. In addition to cell separation defects, the loss of EnvC activity causes FtsZ-ring assembly defects at high temperatures and is synthetically lethal with defects in the Min system (Hara et al, 2002; Ichimura et al, 2002; Bernhardt and de Boer, 2004). EnvC therefore seems to have a function in stabilizing the divisome in addition to activating the amidases to promote cell separation, and it will be interesting to elucidate how these functions are connected in future efforts.
The process of septal PG splitting is delicate surgery. To avoid catastrophe, the divisome must be equipped with ‘fail-safe' mechanisms that prevent activation of the PG splitting enzymes until the rest of the machinery is properly assembled and PG synthesis can be precisely coordinated with hydrolysis. One or more divisomal components may therefore serve as modulators of the LytM–amidase system. Excellent candidates for additional regulatory factors would be the as yet unidentified factors that recruit the amidases and LytM factors to the divisome (protein or PG structure), and/or the synthetic PBPs. Another intriguing candidate is FtsN, as the binding of its SPOR domain to peptide-free glycan strands generated by amidase activity promotes the localization of its essential domain to the divisome (Ursinus et al, 2004; Gerding et al, 2009). This is proposed to set up a self-enhancing cycle in which the essential domain of FtsN promotes cell constriction and amidase activation, which in turn recruits more FtsN to the divisome via its SPOR domain to further stimulate constriction (Gerding et al, 2009). Thus, although our discovery and reconstitution of the LytM–amidase activation systems has revealed an important step in the regulation of PG biogenesis at the septum, much additional work will be needed to fully understand how the divisome balances PG synthesis and degradation at the division site to drive cell constriction without inadvertently causing lysis. We expect that further mechanistic explorations in this area will benefit significantly from the biochemical and molecular tools established in this study.
Spatiotemporal control of PG hydrolysis
In addition to coordinating amidase activation with PG synthesis at the divisome, regulatory mechanisms must also be in place to keep the levels of unwanted PG cleavage throughout the envelope to a minimum at times in the cell cycle before divisome assembly. Failure to do so would also risk the formation of lysis-inducing breaches in the PG network. EnvC, NlpD, AmiB and AmiC are all recruited to the division site (Bernhardt and de Boer, 2003, 2004; Uehara et al, 2009) (T Dinh, unpublished results), and this is likely to contribute significantly to the spatial control over their activities. Precise amidase localization is not always essential for proper septal PG splitting; however, as in the absence of AmiB or AmiC, AmiA effectively catalyses cell separation without accumulating at sites of constriction (Chung et al, 2009). Our current results suggest that confinement of the AmiA activator (EnvC) may be sufficient to restrict amidase activity to septal PG. We showed that the CC domain of EnvC is necessary and sufficient to target it to the division site (Figures 2 and 3). In addition, an EnvC variant lacking the CC domain (LytEnvC) efficiently caused cell lysis (Figure 2B; Supplementary Figure S2). GFP-LytEnvC also failed to be recruited to the division site (Figure 2C), suggesting that protein localization is important in the control of EnvC activity. However, preventing the localization of FLEnvC to the division site by the overproduction of GFP-CCEnvC or by blocking FtsZ assembly did not result in rapid cell lysis (Figure 3D) (Bernhardt and de Boer, 2004). Thus, in addition to its function in protein localization, the CC domain also seems to have an important function in coupling the activation of EnvC, and therefore the amidases, to divisome recruitment. The observation that FLEnvC and LytEnvC have equivalent amidase activation activities in vitro (Figure 6; Supplementary Figure S6) suggests that the CC domain does not have an autoregulatory function. Rather, it seems that at least one additional cellular factor is required to prevent EnvC from activating the amidases before the cytokinetic ring is assembled. An attractive possibility is that the CC domain interacts with an inhibitory factor that blocks EnvC activity throughout the periplasm and that the inhibitor is released when EnvC associates with the divisome via its CC domain (Figure 3) thus allowing localized amidase activation there. As GFP-CCEnvC overproduction did not induce cell lysis in our experiments (Figure 3D), however, such putative inhibitor(s) would have to be relatively abundant, or require both the CC and LytM domains of EnvC for a stable interaction.
The mechanism of amidase activation?
The detailed mechanism of amidase activation by the LytM factors EnvC and NlpD is not yet known, but we consider several possibilities: (1) efficient cleavage of PG by the amidases requires some prior deformation or hydrolysis of bonds in the PG substrate by the LytM factors, and/or vice versa, (2) the amidases enter the periplasm in an inactive state and require some covalent modification (e.g. proteolytic processing) by the LytM factors for activation, (3) PG-bound LytM proteins recruit the amidases to the PG via specific LytM–Ami interactions facilitating the association of the amidases with their substrate or (4) LytM–Ami interactions allosterically activate the amidases. These scenarios are not exclusive, and more complicated ones are possible. So far, we have not detected any obvious protein modifications in our in vitro cell wall cleavage assays, and all our data argue against the possibility that EnvC or NlpD possess PG hydrolytic activity. The observed specificity of the LytM factors in the activation of their cognate amidases also argues against a general deformation of the PG substrate by EnvC or NlpD that might render bonds more accessible for amidase cleavage. Hence, we favour one of the latter two possibilities for amidase activation or some combination of them.
Diverse activities for PG hydrolase folds?
Bacterial genomes typically encode numerous proteins with domains predicted to possess PG hydrolase/cleavage activity (Firczuk and Bochtler, 2007; Vollmer, 2008). Although many of these factors are likely to indeed be PG hydrolases, our results suggest that at least a portion of them may have acquired the ability to regulate other PG hydrolases in addition to, or in lieu of, an intrinsic ability to cleave PG. We thus anticipate that the hydrolase-activator regulation described here will prove to be part of a general strategy used by bacteria to control when and where PG hydrolysis occurs. Given the importance of this decision for their survival, however, we anticipate that additional studies will reveal that bacterial cells superimpose a diverse array of regulatory layers on this core activation system.
A regulatory function for EnvC and NlpD was unexpected considering their relatedness to LytM and lysostaphin endopeptidases as well as the inference from zymography that EnvC has PG hydrolase activity (Bernhardt and de Boer, 2004). For zymography, proteins are separated in an SDS–polyacrylamide gel containing purified PG. The gel is then incubated in buffer to promote protein refolding, and PG in the gel is stained with methylene blue. A zone of clearing at the position where a protein of interest migrates is typically interpreted as evidence for PG hydrolase activity. Purified EnvC produced such a zone of clearing (Bernhardt and de Boer, 2004), but our current results suggest that this is probably not due to hydrolytic activity. PG degradation by EnvC was not detected in solution (Figure 4B), and its LytM domain lacks residues thought to be critical for catalysis (Supplementary Figure S4). Although we did not detect any EnvC-mediated PG cleavage, the protein did readily bind PG in co-sedimentation assays (Supplementary Figure S8). We therefore posit that clearing in a zymogram can result from either genuine PG degradation or from PG binding and dye exclusion. In the case of EnvC, the signal likely stems from its PG-binding activity. A similar observation was recently made for the SpoIID protein involved in cell wall remodelling during sporulation in Bacillus subtilis (Morlot et al, 2010). Thus, our results emphasize the need to interpret results based solely on zymography with caution.
Daughter cell separation and rapid β-lactam-induced lysis
E. coli mutants lacking multiple LytM factors or amidases not only fail to complete division, they display a fundamentally different response to β-lactam antibiotics (Heidrich et al, 2002; Chung et al, 2009; Uehara et al, 2009). Although wild-type cells lyse rapidly through lesions at their division sites, lysis of these mutants is slower and accompanied by a progressive loss in cell shape (Chung et al, 2009; Uehara et al, 2009). β-lactams, therefore, seem to induce a lethal divisome malfunction by tricking the machinery into activating the LytM–amidase splitting system when PG synthesis is compromised. The clinical success of these drugs highlights the therapeutic potential of molecules capable of interfering with the proper regulation of septal PG splitting. Our findings suggest that PG synthesis inhibition is not the only avenue by which potential antibiotics might tip the cellular scales in favour of PG hydrolysis and lysis. Molecules that promote the untimely activation of the amidases should be equally effective.
Materials and methods
Media, bacterial strains and plasmids
Cells were grown in LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or minimal M9 medium (Miller, 1972) supplemented with 0.2% casamino acids and 0.2% sugar. Unless otherwise indicated, antibiotics were used at 10 (chloramphenicol; Cam), 15 (ampicillin; Amp) or 20 (kanamycin; Kan) μg/ml.
The bacterial strains and plasmids used in this study are listed in Supplementary Tables S1 and S2, respectively. A detailed description of their construction is given in Supplementary data. All strains used for in vivo experiments are derivatives of MG1655 (Guyer et al, 1981), and all deletion alleles were either sourced from the Keio knockout collection (Baba et al, 2006) or constructed to resemble those in the collection. All plasmids used for in vivo experiments except pTU175 are derivatives of the CRIM plasmids developed by Haldimann and Wanner (2001). They were integrated into phage attachment sites (HK022 or λ) using the helper vectors pTB102 (Bernhardt and de Boer, 2005) or pInt-ts (Haldimann and Wanner, 2001), respectively, as described earlier (Haldimann and Wanner, 2001). pTU175 is a low-copy vector with a pSC101 origin. It constitutively produces periplasmic mCherry (Shaner et al, 2004).
Fluorescence microscopy and growth curves
Fluorescence microscopy was performed essentially as described earlier (Uehara et al, 2009). Please see figure legends for details about growth conditions and sample preparation methods used for specific experiments.
Selection for mutants resistant to LytEnvC
The parental strain MG1655 (attHKTU115) (attλTU181) [(Plac∷ssdsbA-gfp-lytenvC) (Para∷ssdsbA-gfp-lytenvC)] was mutagenized with a KanR mariner transposon (Chiang and Rubin, 2002) delivered by conjugation from the donor strain BW19851/pSC189. LytEnvC-resistant mutants from the resulting library were selected by growing cells in the presence of arabinose and IPTG. The positions of the transposon insertions were mapped by arbitrarily primed PCR essentially as described (Bernhardt and de Boer, 2004). See Supplementary data for a detailed description of the methods used for the mutagenesis and selection.
Protein purification
All proteins were overexpressed and purified with a 6xHis-SUMO (H-SUMO) tag fused to their N-termini (Mossessova and Lima, 2000; Marblestone et al, 2006). The sequence of the affinity tag in all cases was MRGSHHHHHHMASG. The SUMO sequence was amplified from the Saccharomyces cerevisiae genome (gene Smt3) as described earlier (Bendezú et al, 2009). After purification of an H-SUMO fusion protein by metal-affinity chromatography, the H-SUMO tag was removed using 6xHis-tagged SUMO protease (H-SP) (Bendezú et al, 2009). Cleavage reactions were passed through Ni-NTA resin to remove free H-SUMO and H-SP, yielding a pure preparation of the desired protein without added non-native residues. The only exceptions were AmiC, and the AmiB protein preparations used for the experiments in Figure 9. For AmiC, the sequence RRAEL was left on its N-terminus. The AmiB proteins had an additional serine at their N-termini and the dipeptide LE appended to their C-termini. Specific purification procedures for each protein preparation used in this study are given in Supplementary data.
Preparation of sacculi labelled with RBB
Sacculi were prepared from strain TU163 [Δlpp] as described by Uehara et al (2009). Isolated sacculi were treated with 200 μg/ml amylase (Sigma, St Louis, MO) at 37°C for 2 h in 1 × PBS and washed with water. The amylase-treated sacculi were then incubated with 20 mM RBB (Sigma) in 0.25 M NaOH overnight at 37°C. The preparation was neutralized with HCl, and RBB-labelled sacculi were pelleted by centrifugation (21 000 × g, 20 min, room temperature). The sacculi were then repeatedly resuspended in water and pelleted by centrifugation until the supernatant was clear. The final pellet was resuspended in water containing 0.02% azide and stored at 4°C.
Dye-release assay for PG hydrolysis
A measure of 10 μl of RBB-labelled sacculi were incubated at 37°C for various times with purified amidases and/or LytM factors in 100 μl of PBS buffer (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl and 2.7 mM KCl, pH 7.4). Final protein concentrations and reaction times are indicated in Figures 4, 6, 7 and 9. Reactions in Figures 4, 6, 7 and 9 were terminated by incubating them at 95°C for 5 min. Reactions in Supplementary Figure S6 (60 μl total) were terminated by the addition of 30 μl of ethanol. Following termination, all reactions were centrifuged at 21 000 × g for 20 min at room temperature. Supernatants were removed and their absorbance was measured at 595 nm. The time course experiment in Supplementary Figure S6 indicates that most of the cleavage reactions in Figures 4, 6, 7 and 9 were performed to completion.
HPLC analysis of products from PG cleavage reactions
Wild-type sacculi from TB28 were prepared for this assay as described earlier (Uehara et al, 2009). The sacculi were incubated with amidases and/or LytM factors (EnvC or NlpD) at 37°C overnight in PBS. Following heat inactivation, the samples were digested with 5 U Mutanolysin (Sigma) at 37°C overnight. The digests were centrifuged (21 000 × g, 10 min, room temperature) and the muropeptides in the supernatants were prepared for and analysed by HPLC as described earlier (Uehara et al, 2009). To identify the compound that eluted at 26 min in Figure 10D (Peak 4), the fraction was analysed with a Voyager-DE MALDI-TOF mass spectrometer (Perseptive Biosystems). The molecular weight of the compound was m/z 927.21, which represents free cross-linked tetrapeptide (calculated MW=904) plus Na+. Identification of the compound that eluted at 11 min in Figure 10D (Peak 3) was carried out using a 6520 Accurate-Mass quadrupole time-of-flight liquid chromatography/mass spectrometry system (Agilent) with a Jupiter Proteo C12 column (250 × 4.6 mm, 4 μm, 90 Å) from Phenomenex. The samples were injected onto the column and eluted at a flow rate of 0.5 ml/min with 1% formic acid in water for 5 min followed by a linear gradient of 1–10% methanol over 10 min. The compound eluted at 6.1 min under these conditions with an m/z of 462.2, which represents free tetrapeptide (calculated MW=461) plus H+.
Supplementary Material
Acknowledgments
We thank Piet de Boer, David Rudner, Ry Young, James T Park and members of the laboratory for critical reading of the manuscript. We also thank Brad L Pentelute, Adam Barker and John Collier for help with protein purification and MALDI-TOF, and Emma Doud and Suzanne Walker for help with LC/MS. This work was supported by the Massachusetts Life Science Center, the Burroughs Wellcome Fund and the National Institutes of Health (R01 AI083365-01A1). TGB holds a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Hale CA, Bernhardt TG, de Boer PA (2009) RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2003) The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48: 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2004) Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol 52: 1255–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SL, Rubin EJ (2002) Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296: 179–185 [DOI] [PubMed] [Google Scholar]
- Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D (2009) Rapid {beta}-lactam-induced lysis requires successful assembly of the cell division. Proc Natl Acad Sci USA 106: 21872–21877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA (2008) Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev 32: 321–344 [DOI] [PubMed] [Google Scholar]
- Firczuk M, Bochtler M (2007) Folds and activities of peptidoglycan amidases. FEMS Microbiol Rev 31: 676–691 [DOI] [PubMed] [Google Scholar]
- Firczuk M, Mucha A, Bochtler M (2005) Crystal structures of active LytM. J Mol Biol 354: 578–590 [DOI] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezú FO, Hale CA, Bernhardt TG, de Boer PA (2009) Self-enhanced accumulation of FtsN at division sites, and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol 191: 7383–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B, Höltje JV, Schwarz U (1988) The composition of the murein of Escherichia coli. J Biol Chem 263: 10088–10095 [PubMed] [Google Scholar]
- Guyer MS, Reed RR, Steitz JA, Low KB (1981) Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol 45: 135–140 [DOI] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183: 6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Narita S, Karibian D, Park JT, Yamamoto Y, Nishimura Y (2002) Identification and characterization of the Escherichia coli envC gene encoding a periplasmic coiled-coil protein with putative peptidase activity. FEMS Microbiol Lett 212: 229–236 [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Höltje JV (2001) Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol 41: 167–178 [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H, Höltje JV (2002) Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol 184: 6093–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Yamazoe M, Maeda M, Wada C, Hiraga S (2002) Proteolytic activity of YibP protein in Escherichia coli. J Bacteriol 184: 2595–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korndörfer IP, Danzer J, Schmelcher M, Zimmer M, Skerra A, Loessner MJ (2006) The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J Mol Biol 364: 678–689 [DOI] [PubMed] [Google Scholar]
- Lupoli TJ, Taniguchi T, Wang TS, Perlstein DL, Walker S, Kahne DE (2009) Studying a cell division amidase using defined peptidoglycan substrates. J Am Chem Soc 131: 18230–18231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marblestone JG, Edavettal SC, Lim Y, Lim P, Zuo X, Butt TR (2006) Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci 15: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ (2010) A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev 24: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876 [DOI] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak M, Bochtler M (2004) Latent LytM at 1.3A resolution. J Mol Biol 335: 775–785 [DOI] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24: 79–88 [DOI] [PubMed] [Google Scholar]
- Priyadarshini R, de Pedro MA, Young KD (2007) Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol 189: 5334–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, Popham DL, Young KD (2006) Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol 188: 5345–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36: 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shida T, Hattori H, Ise F, Sekiguchi J (2001) Mutational analysis of catalytic sites of the cell wall lytic N-acetylmuramoyl-L-alanine amidases CwlC and CwlV. J Biol Chem 276: 28140–28146 [DOI] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG (2009) LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191: 5094–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Park JT (2008) Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J Bacteriol 190: 3914–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursinus A, van den Ent F, Brechtel S, de Pedro M, Höltje JV, Löwe J, Vollmer W (2004) Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J Bacteriol 186: 6728–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W (2008) Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev 32: 287–306 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P, Foster S (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32: 259–286 [DOI] [PubMed] [Google Scholar]
- Zhou R, Chen S, Recsei P (1988) A dye release assay for determination of lysostaphin activity. Anal Biochem 171: 141–144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.