Abstract
Polycomb (PcG) and Trithorax (TrxG) group proteins act antagonistically to establish tissue-specific patterns of gene expression. The PcG protein Ezh2 facilitates repression by catalysing histone H3-Lys27 trimethylation (H3K27me3). For expression, H3K27me3 marks are removed and replaced by TrxG protein catalysed histone H3-Lys4 trimethylation (H3K4me3). Although H3K27 demethylases have been identified, the mechanism by which these enzymes are targeted to specific genomic regions to remove H3K27me3 marks has not been established. Here, we demonstrate a two-step mechanism for UTX-mediated demethylation at muscle-specific genes during myogenesis. Although the transactivator Six4 initially recruits UTX to the regulatory region of muscle genes, the resulting loss of H3K27me3 marks is limited to the region upstream of the transcriptional start site. Removal of the repressive H3K27me3 mark within the coding region then requires RNA Polymerase II (Pol II) elongation. Interestingly, blocking Pol II elongation on transcribed genes leads to increased H3K27me3 within the coding region, and formation of bivalent (H3K27me3/H3K4me3) chromatin domains. Thus, removal of repressive H3K27me3 marks by UTX occurs through targeted recruitment followed by spreading across the gene.
Keywords: chromatin, histone demethylase, myogenesis, transcription, UTX
Introduction
Tissue-specific patterns of gene expression are established through the antagonistic functions of Polycomb group (PcG) and Trithorax group (TrxG) proteins (Ringrose and Paro, 2004; Brock and Fisher, 2005; Muller and Verrijzer, 2009). Although several enzymatic activities have been identified in both the PcG and TrxG groups of proteins (Ringrose and Paro, 2004; Brock and Fisher, 2005), a hallmark of their function seems to be the ability to methylate histones at specific genomic loci—the PcG protein Ezh2 (KMT6) marks repressive chromatin by trimethylation of histone H3 Lys 27 (H3K27me3), whereas the KMT2 family of TrxG proteins mark transcriptionally active chromatin by trimethylation of histone H3 Lys 4 (H3K4me3). In addition, genome-wide studies of chromatin states have revealed the existence of bivalent—H3K4me3 and H3K27me3—chromatin domains that mark developmentally regulated genes that are not actively transcribed (Bernstein et al, 2006). Although initially identified in pluripotent cells, bivalent chromatin domains have also been shown to be present at developmentally regulated genes in cells of restricted potency, including terminally differentiated cells (Mohn et al, 2008). The fact that bivalently marked genes are not expressed suggests that the repressive effects of H3K27me3 dominate over the activating effects of H3K4me3 when both marks are present (Barski et al, 2007). As the repressive H3K27me3 mark can be heritably transmitted to daughter cells to maintain specific gene expression programs (Hansen et al, 2008), expression of developmentally regulated genes would require the removal of the H3K27me3 mark to permit the activation of gene expression. The identification of UTX (KDM6A) and JMJD3 (KDM6B) as proteins that mediate demethylation of H3K27me3 provided insight into the mechanism by which promoters could transition from a transcriptionally repressed to a transcriptionally active state (Agger et al, 2007; De Santa et al, 2007; Hong et al, 2007; Lan et al, 2007; Lee et al, 2007; Chaturvedi et al, 2009). Proteomics analysis has shown that UTX complexes with MLL3/HALR(KMT2C), MLL4/ALR(KMT2D), and PTIP (Cho et al, 2007; Issaeva et al, 2007; Lee et al, 2007). However, the mechanism by which the H3K27 demethylases are targeted to subsets of genes in a tissue-specific manner has not been established.
During development, expression of myogenin (Myog) is restricted to cells of the skeletal muscle lineage, whereas muscle creatine kinase (CKm) is restricted to cells of the skeletal and cardiac muscle lineages. These two genes represent an excellent model for studying tissue-specific gene expression as their activation is temporally distinct during myogenesis (Bergstrom et al, 2002). As would be expected of genes that demonstrate muscle-specific gene expression, both of these loci have polycomb-associated H3K27me3 marking of nucleosomes in human embryonic fibroblasts (Bracken et al, 2006). Similarly, it has been shown that the CKm gene is targeted by the PcG protein Ezh2 in muscle progenitor cells leading to an enrichment of the H3K27me3 repressive chromatin mark at this locus (Caretti et al, 2004). Upon myogenic differentiation, H3K27me3 is lost at the CKm gene (Caretti et al, 2004) and both the Myog and CKm genes become marked by TrxG-mediated H3K4me3 permitting gene expression (Rampalli et al, 2007). In this study, we sought to decipher the mechanism by which demethylation of H3K27me3 is mediated at the muscle-specific Myog and CKm genes during myogenesis.
Results
Demethylation of muscle genes occurs through a two-step process
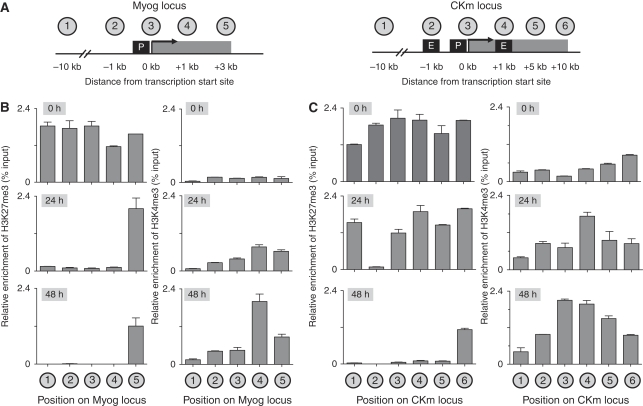
Native chromatin immunoprecipitation (ChIP) (Brand et al, 2008) was used to profile the distribution of PcG-mediated H3K27me3 and TrxG-mediated H3K4me3 across the Myog and CKm genes (Figure 1) during serum withdrawal-induced differentiation of C2C12 myoblasts (Yaffe and Saxel, 1977). To compare enrichment of histone modifications at different genomic positions, qPCR amplifications with each primer set were normalized using genomic DNA. Paralleling genome-wide studies that have demonstrated H3K27me3 marks chromatin over extended chromatin domains (Barski et al, 2007), we observe extensive enrichment of H3K27me3 throughout the 5′ regulatory region, and the coding region of the CKm and Myog genes in proliferating (0 h) myoblasts, whereas H3K4me3 levels remain relatively low at these repressed loci (Figure 1B and C, top panel). By 24 h of differentiation, the Myog gene is expressed (Rampalli et al, 2007; see Figure 6B) and demonstrates extensive loss of H3K27me3 across the 5′ regulatory region and portions of the coding region (but not at the 3′ UTR), whereas the 5′ end of the coding region becomes enriched for H3K4me3 (Figure 1B, middle panel). In contrast, the CKm gene (which is transcriptionally silent at 24 h (Rampalli et al, 2007; see Figure 6B) remains enriched for H3K27me3 across the loci with the exception of Region 2, which represents its upstream muscle-specific enhancer (Figure 1C, middle panel). This loss of H3K27me3 at the enhancer position is not caused by reduced nucleosome occupancy (Schones et al, 2008) or nucleosome instability (Jin et al, 2009) as histone H3 remains associated with the CKm enhancer throughout differentiation (Supplementary Figure 1B). By 48 h of differentiation, both Myog and CKm are expressed at high levels (Rampalli et al, 2007; see Figure 6B), and display extensive loss of H3K27me3 both within the coding region and as far upstream as −10 kb of the gene (Figure 1B and C, bottom panels). This is accompanied by enrichment of H3K4me3 within the gene (Figure 1B and C, bottom panels). Interestingly, we see reduced but continued enrichment of the repressive H3K27me3 mark in the 3′ end of both the expressing myog and CKm genes. The importance of retaining the H3K27me3 mark in this region is not clear but its position suggests a possible function in transcript processing. Thus, expression of both the Myog and CKm genes are regulated through the antagonistic methylation activities of polycomb (H3K27me3) and trithorax (H3K4me3) group proteins. In addition, demethylation at the CKm gene seems to occur in two temporally distinct steps.
Figure 1.
Methylation of histone H3 at the Myog and CKm genes during muscle differentiation. (A) Schematic representation of the Myog and CKm genes. P represents gene promoters, whereas E in the CKm gene represents enhancer (distal and intronic) elements. The encircled numbers above the schematic indicate the location on the gene where each of the different probe sets localize for analysis of ChIP studies. (B, C) Native ChIP analysis of the temporal (0, 24, and 48 h) changes in H3K27me3 or H3K4me3 enrichment across the Myog and CKm genes. Chromatin isolated from C2C12 cells at various stages of differentiation was subjected to immunopurification using either an anti-H3K27me3 or an anti-H3K4me3 antibody. After deproteination, immunopurified DNA was quantitated by qPCR using probes that recognize the different regions indicated in the schematic in Figure 1A.
The demethylase UTX mediates removal of H3K27me3 at the Myog and CKm genes
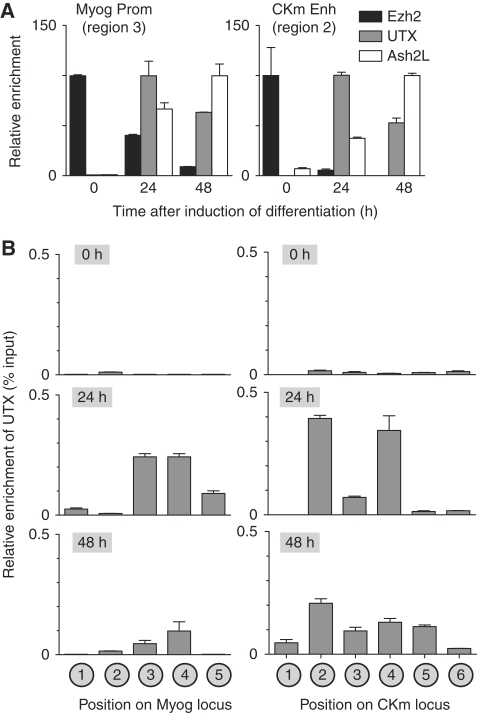
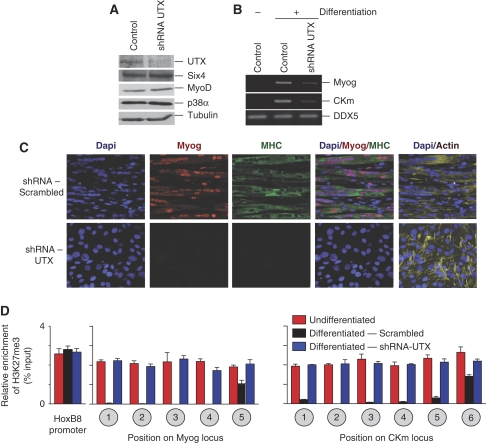
Demethylation of H3K27me3 in mammalian cells has been shown to be mediated through the activity of the enzymes JMJD3 and UTX (Agger et al, 2007; De Santa et al, 2007; Hong et al, 2007; Lan et al, 2007; Lee et al, 2007). Although JMJD3 seems to have a global function in modulating H3K27me3 levels in the cell, ectopic expression of UTX has only modest effects on cellular levels of this repressive histone mark (Agger et al, 2007; Lan et al, 2007), suggesting it could be acting to demethylate-specific genomic loci. ChIP analysis demonstrates recruitment of UTX to the transcriptional regulatory region of both the Myog and CKm genes at 24 h (Figure 2A and B, Myog Region 3 and Ckm Region 2). This recruitment of UTX coincides with a loss of the antagonistic Ezh2 H3K27 methyltransferase enzyme (Figure 2A) and removal of the repressive H3K27me3 mark at both genes (Figure 1B and C). Ash2L1 (the primary Ash2L isoform expressed in differentiating C2C12 cells; see Supplementary Figure 2) is maximally enriched at both genes by 48 h (Figure 2A), coinciding with the increased enrichment of H3K4me3 within their respective coding regions (Figure 1B and C). This differential temporal recruitment of Ash2L1 and UTX to muscle-specific genes is consistent with earlier results examining the binding of these two proteins to the HoxA1-3 and HoxB1-3 genes in differentiating NTERA2 cells (Lee et al, 2007), and supports our earlier findings that Ash2L1/MLL2(KMT2B) establishes this transcriptionally permissive H3K4me3 mark at muscle-specific genes (Rampalli et al, 2007). Interestingly, we see the recruitment of UTX at 24 h (Figure 2B, Ckm Region 4) to the previously described intronic enhancer of the CKm gene (Johnson et al, 1989), though we do not see demethylation of H3K27me3 within this region. It is unclear why UTX is unable to demethylate H3K27me3 at this location, but suggests that the activity of this enzyme could be modulated through other epigenetic markings of chromatin. This observation is consistent with studies of the Hox locus demonstrating UTX is enriched in multiple regions that remain marked by H3K27me3 (Lan et al, 2007). By 48 h of differentiation, UTX is enriched across the coding region of both the Myog and CKm genes (Figure 2B) and can be detected at regions of the gene that demonstrate loss of the repressive H3K27me3 mark (Figure 1B and C). To confirm a function for UTX in mediating the demethylation of these two muscle-specific genes, we performed shRNA-mediated knockdown of this protein using lentiviral constructs (Figure 3A). Knockdown of UTX in differentiating myoblasts leads to an marked decrease in both Myog and CKm expression while having no effect on expression levels of the ubiquitously expressed DDX5 gene (Figure 3B and data not shown). Furthermore, immunocytochemistry experiments demonstrate that knockdown of UTX in C2C12 cells prevents the formation of multinucleated, myosin heavy chain (MHC) positive myotubes (Figure 3C). Concomitant with the loss of expression, knockdown of UTX in differentiating myoblasts inhibits demethylation of H3K27me3 at both the CKm and Myog genes (Figure 3D), but has no effect on the transcriptionally silent HOXB8 promoter that is bivalently marked in C2C12 cells (Bernstein et al, 2006). Thus, UTX is responsible for removing the repressive H3K27me3 mark at the Myog and CKm genes during myogenesis, though we note that the relative distribution of the H3K27 demethylase across the Myog and CKm loci (Figure 2B; 48 h differentiation) suggests that different mechanisms may exist for establishing demethylation of the distinct regions across the genes.
Figure 2.
UTX becomes associated with the Myogenin and CKm loci during C2C12 myogenesis. (A) Cross-linked chromatin from C2C12 cells (growing (0 h) or differentiated (24 or 48 h)) was subject to ChIP analysis using antibodies directed against UTX, Ezh2, or Ash2L. Immunopurified DNA was quantitated by qPCR using probes that recognize either the Myog promoter (left panel) or the distal enhancer of the CKm gene (right panel). To account for variability in IP efficiency of the three different antibodies, relative enrichment is plotted as a percentage of the maximal enrichment value observed for each factor individually during the 48 h time course after first calculating the absolute enrichment as a percentage of input chromatin. (B) X-ChIP analysis of the temporal (0, 24, and 48 h) changes in UTX enrichment across the Myog and CKm genes. Chromatin isolated from C2C12 cells at various stages of differentiation was subjected to immunopurification using an anti-UTX antibody. After deproteination, immunopurified DNA was quantitated by qPCR using probes that recognize the different regions indicated in the schematic in Figure 1A.
Figure 3.
UTX mediates removal of the H3K27me3 mark at the Myog and CKm genes during myogenesis. (A) Western blot analysis of UTX knockdown in differentiating C2C12 cells. Nuclear extracts prepared from cells infected with lentivirus expressing shRNA that target UTX or control (scrambled) were separated by SDS–PAGE and analysed by western blot using the antibodies indicated. Each panel represent two lanes run on the same gel that have been cropped due to space constraints. Uncropped blots are shown in Supplementary Figure 7. (B) Gene expression in C2C12 cells after UTX knockdown. Primers specific for Myog, CKm, or the ubiquitously expressed DDX5 mRNA were used to amplify cDNA prepared from C2C12 cells under growth (−) or differentiation (+) conditions. Cells were infected with the non-targeted scrambled shRNA (control) or an UTX-targeted shRNA as indicated. (C) C2C12 cells infected with control (scrambled) or UTX-targeted shRNA were differentiated for 72 h and then examined by immunocytochemistry for actin (phalloidin stain), Myog, or MHC, whereas the DNA was stained with DAPI. Images were generated on a Zeiss 510 Confocal microscope using a × 40 lens. (D) Native ChIP analysis measured relative H3K27me3 levels at various regions within the Myog, CKm, or HoxB8 loci. C2C12 cells were infected with control (shRNA-scrambled) or UTX (shRNA-UTX) targeting shRNA, selected for infection using puromycin and then differentiated for 72 h. Positions on the locus correspond to regions indicated in the schematic in Figure 1A.
Six4 targets UTX to the Myog and CKm genes during myogenesis
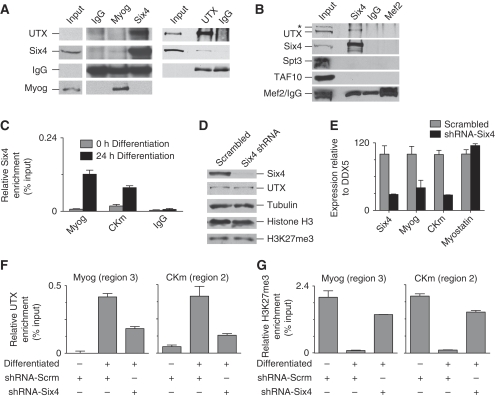
The localized demethylation of the CKm enhancer at 24 h of differentiation suggests that UTX is targeted to specific genes through an interaction with transactivators. Among the transcriptional regulators known to associate with the Myog promoter, we observed an association between UTX and the homeobox protein Six4 in reciprocal immunoprecipitations using nuclear extracts prepared from C2C12 cells that had been differentiated for 24 h (Figure 4A). Further confirming this interaction, UTX and Six4 co-immunoprecipitate from nuclear extracts prepared from human erythroleukemia (K562) cells that also express both proteins endogenously (Figure 4B). ChIP experiments demonstrate that Six4 is recruited to the regulatory region of both Myog and CKm at 24 h of differentiation (Figure 4C; Supplementary Figure 3), when UTX is strongly associated with these genes (Figure 2). Consistent with its previously described function in regulating transcription of the Myog and CKm genes (Spitz et al, 1998; Himeda et al, 2004), we observe that a 70% knockdown of Six4 in differentiating myoblasts results in a similar decrease in both Myog and Ckm gene expression, without affecting UTX, Myostatin, or global H3K27me3 levels (Figure 4D and E). This decreased expression of muscle-specific genes in the absence of Six4 coincides with a decrease in UTX recruitment to the regulatory regions of both the Myog and CKm (Figure 4F), and results in an increased enrichment of the repressive H3K27me3 mark at these genes (Figure 4G). Taken together, these results demonstrate that Six4 facilitates expression of muscle-specific genes by recruiting UTX to their regulatory region.
Figure 4.
Six4 targets UTX to muscle-specific genes. (A) Proteins were immunoprecipitated from C2C12 nuclear extracts (24 h differentiation) using antibodies directed against UTX, Myog, Six4, or control IgG. Immunoprecipitated proteins, and 1/50th of input, were examined by western blot using the antibodies indicated. The left panel corresponds to an immunoprecipitation experiment, where the input and elution samples were processed together. The blot was cropped to remove lanes corresponding to flow-through fractions, and thus the input is separated from the elution fractions by a white bar but was run on the same gel and has the same exposure. Uncropped versions of the UTX and Six 4 analysis are shown in Supplementary Figure 6A. (B) Proteins were immunoprecipitated from nuclear extracts prepared from erythroleukemia cell line K562 using antibodies directed against Six4, Mef2, or control IgG. Immunoprecipitated proteins, and 1/10th of input, were examined by western blot using the antibodies indicated. The asterisk (*) marks a non-specific band observed from the UTX antibody in K562 cells. Mef2/IgG indicates that the two Mef2 bands observed in K562 cells co-migrated with IgG. To provide evidence that the stronger band observed in the Mef2 IP lane represents immunoprecipitated Mef2, we provide a shorter exposure of the blot in Supplementary Figure 6B where the slower migrating Mef2 band is distinguishable from the IgG. Note that all elution bands in this experiment migrate slightly higher than their counterpart in the input because the samples were eluted in a buffer containing urea. (C) Cross-linked chromatin from C2C12 cells (growing (0 h) or differentiated (24 h)) was subjected to ChIP analysis of Six4 binding. Immunopurified DNA was quantitated by qPCR using hydrolysis probes that recognize Myog (Region 3), CKm (Region 2), or IgH. (D) Western blot analysis of protein extracts isolated from cells infected with lentivirus expressing shRNA targeting Six4 or a scrambled control. Nuclear extracts prepared from cells infected with lentivirus expressing shRNA that target UTX or control (scrambled) were separated by SDS–PAGE and analysed by western blot using the antibodies indicated. Each panel represents two lanes run on the same gel that have been cropped due to space constraints. Uncropped blots are shown in Supplementary Figure 8. (E) RNA isolated from cells infected with lentivirus expressing Six4 targeting (or control) shRNA was reverse transcribed and subjected to qPCR analysis. Expression levels are expressed relative to DDX5. (F) Cross-linked chromatin from growing (−), or 24 h differentiated (+) C2C12 cells was subjected to ChIP analysis using antibodies directed against UTX. Immunopurified DNA was quantitated by qPCR using probes that recognize the promoter (Region 3) of Myog, or the upstream enhancer (Region 2) of CKm. (G) Native chromatin from growing (−), or 24 h differentiated (+) C2C12 cells was subject to ChIP analysis using antibodies directed against H3K27me3. Immunopurified DNA was quantitated by qPCR using probes that recognize the promoter (Region 3) of the Myog gene, or the upstream enhancer (Region 2) of CKm.
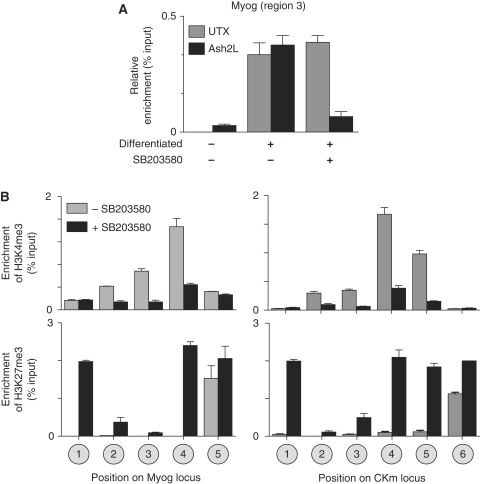
Recruitment of Ash2L1/MLL2(KMT2B)-containing H3K4 methyltransferase complexes to muscle-specific promoters by Mef2d requires phosphorylation of the transactivator by p38 MAPK (Rampalli et al, 2007). Here, we used the pharmacological inhibitor SB203850 to determine whether Six4-mediated recruitment of UTX to the Myog and CKm genes is also p38 MAPK dependent. ChIP analysis demonstrated that, in contrast to Ash2L1, UTX recruitment to the Myog promoter does not require p38 MAPK activity (Figure 5A). Consistent with this observation, inhibition of p38 MAPK signalling did not prevent the demethylation of H3K27me3 within the transcriptional regulatory region of either Myog (Figure 5B, Regions 2 and 3) or CKm (Figure 5B, Regions 2 and 3) at 48 h of differentiation. In contrast, spreading of the demethylase activity into the coding region of the gene was impaired at both Myog and CKm in the presence of SB203580 (Figure 5B, Myog Region 4 and CKm Regions 4 and 5). As we have observed earlier (Rampalli et al, 2007), SB203580 prevented Ash2L1 recruitment (Figure 5A) and H3K4me3 accumulation (Figure 5B) on the Myog and Ckm genes. Thus, we examined the effect of Ash2L knockdown on demethylation of the Myog gene. Interestingly, the knockdown of Ash2L in differentiating C2C12 cells (Supplementary Figure 4A) leads to a similar distribution of H3K27me3 at the Myog gene, where the coding region of the gene remains enriched for the repressive histone mark (Supplementary Figure 4B). This demonstrates that an event downstream of Ash2L1 recruitment is necessary for spreading of the UTX demethylase activity into the coding region of these muscle genes.
Figure 5.
UTX recruitment to the Myog and CKm genes is p38 MAPK independent. (A) Cross-linked chromatin from growing (differentiation −), or 48 h differentiated (differentiation +) C2C12 cells that had been treated with or without the pharmacological inhibitor of p38 MAPK (SB203580) was subjected to ChIP analysis using antibodies recognizing either Ash2L or UTX. (B) Native ChIP analysis was performed on C2C12 cells that had been induced to differentiate for 48 h in the presence (+) or absence (−) of the p38 MAPK inhibitor SB203580. H3K4me3 (top panels) or H3K27me3 (bottom panels) ChIPs were analysed by qPCR using probes specific for position on the Myog or CKm locus as outlined in Figure 1A.
Blocking of RNA Pol II elongation leads to the formation of bivalent H3K27me3/H3K4me3 marks
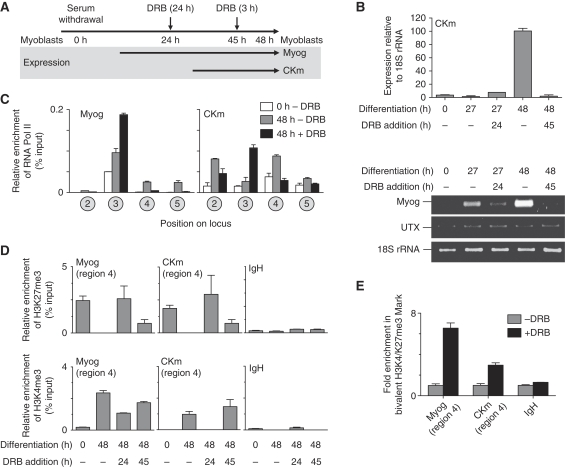
Inhibition of p38 MAPK activity in differentiating myoblasts leads to the formation of a transcriptionally poised promoter that is enriched for RNA Polymerase II (Pol II) (Simone et al, 2004; Rampalli et al, 2007). Furthermore, UTX has been shown to associate with Rpb1 and to co-localize with elongating Pol II (Rpb1 phosphorylated at Ser2) on polytene chromosomes (Smith et al, 2008). We therefore used the pharmacological inhibitor of transcriptional elongation DRB (Figure 6A) to examine whether UTX-mediated demethylation within the gene may require the elongating Pol II. As expected, DRB treatment blocked expression of CKm and Myog (Figure 6B), resulting in the accumulation of Pol II within the 5′ end of the genes (Figure 6C, Region 3). To determine the effect of Pol II elongation on H3K27 demethylase activity, H3K27me3 native ChIP was performed on chromatin isolated from 48 h differentiated C2C12 cells that had been treated with DRB for 3 h (added at time point 45 h of differentiation) or 24 h (added at time point 24 h of differentiation) (Figure 6D). Under both conditions, DRB treatment leads to a marked increase in H3K27me3 enrichment within the Myog and CKm coding regions (Region 4) compared with the control-treated cells. Remarkably, the increased levels of H3K27me3 observed after DRB treatment are accompanied by a loss of UTX within the coding region, but not the regulatory regions, of the genes (Supplementary Figure 5A) and an increased association of Ezh2 and Suz12 (Supplementary Figure 5B) within the genes demonstrating the dynamic relationship between PcG and TrxG proteins at developmentally regulated genes. Taken together, this data strongly suggests that the demethylase activity of UTX migrates into the coding region of the Myog and CKm genes with elongating Pol II.
Figure 6.
Induced stalling of Pol II at actively transcribed genes establishes bivalent chromatin domains. (A) Timeline for the addition of DRB during C2C12 differentiation. Arrows in the shaded box provide approximate times at which the Myog and CKm genes become expressed during myogenesis. (B) Differentiating C2C12 cells were treated with DRB as indicated. Random-primed cDNA was prepared from C2C12 cells and subjected to qPCR (top panel) using Taqman primer/probes sets that amplify CKm, or 18S rRNA or semi-quantitative PCR (lower panel) using primers specific for Myog, UTX, or 18S rRNA. (C) Cross-linked chromatin from C2C12 cells (growing (0 h), −DRB), or differentiated for 48 h in the presence (48 h, +DRB) or absence (48 h, −DRB) of DRB was subject to ChIP analysis using an antibody directed against Pol II (Rpb1). Immunopurified DNA was quantitated by qPCR using hydrolysis probes that recognize the regions indicated in the schematic in Figure 1A. (D) Chromatin was isolated from C2C12 cells in growth (0 h) or differentiation conditions (48 h) that had been treated with 100 μM DRB as indicated in the timeline in Figure 6A. (E) Native re-ChIP analysis was performed on the chromatin isolated from differentiating C2C12 cells (48 h) that have been incubated in the presence (+DRB) or absence (−DRB) of the inhibitor of transcriptional elongation DRB for 3 h. Chromatin immunoprecipitated with an anti-H3K4me3 antibody was purified, and re-immunoprecipitated using an anti-H3K27me3 antibody. Immunopurified DNA was quantitated by qPCR using hydrolysis probes that recognize the IgH gene or the +1000 region (Region 4) of the Myog or CKm genes.
Interestingly, the enrichment of H3K27me3 at the Myog and CKm genes after induced Pol II stalling (treatment with DRB at time point 45 h) did not coincide with a loss of the active H3K4me3 mark from the same loci (Figure 6D), leading to the formation of an apparent bivalent (H3K27me3/H3K4me3) chromatin domain. However, when DRB treatment was initiated at 24 h of differentiation, the formation of the bivalent mark was restricted to the Myog gene (Figure 6D, DRB addition 24 h). The differential marking of chromatin at Myog and CKm at this time point is remarkable because the Myog gene is already transcribed before adding DRB at 24 h of differentiation, whereas the CKm gene has not yet begun to be expressed at this stage of differentiation (Figure 6B). Only after the CKm gene has begun transcribing (DRB at 45 h) can the bivalent mark be formed on this gene, suggesting that an initial round of transcription is necessary for the formation of bivalent marks. Finally, to determine whether the co-localization of H3K27me3 and H3K4me3 marks reflect true bivalent domains (as opposed to a mixed population of cells that were differentially marked with one of the two methylations), we performed native ChIP-ReChIP analysis. Analysis of chromatin from cells treated with or without DRB show that genes that harbour a stalled Pol II can be marked by both H3K27me3 and H3K4me3 at the same allele (Figure 6E). These results suggest that the formation of bivalent domains occur through the stalling of Pol II at transcribed genes.
Discussion
During myogenesis, the muscle-specific genes Myog and CKm transition from a transcriptionally repressive state of H3K27me3 marked chromatin to the transcriptionally permissive state of H3K4me3 marked chromatin. In this study, we demonstrate that the histone demethylase UTX is recruited to these two muscle genes to remove the repressive H3K27me3 mark. Although the H3K4 methyltransferases MLL3(KMT2C) and MLL4(KMT2D) are core components of the UTX complex (Cho et al, 2007; Issaeva et al, 2007; Lee et al, 2007), we find that Ash2L1/MLL2(KMT2B) is the major methyltransferase mediating H3K4me3 at the Myog and CKm genes (Figure 5; Rampalli et al, 2007). This is consistent with recent results showing that MLL1(KMT2A)/MLL2(KMT2B), but not MLL3(KMT2C)/MLL4(KMT2D), is responsible for mediating H3K4 trimethylation at the Hox locus, though both MLL family subsets affect gene expression within the cluster (Wang et al, 2009). Importantly, we observe that UTX associates with the enhancer regions of the CKm gene before gene expression, leading to a localized demethylation of the upstream regulatory element. Furthermore, extensive loss of H3K27me3 is observed within the promoter region of the Myog gene under condition where expression of this gene is blocked in C2C12 cells using a p38 MAPK inhibitor (Simone et al, 2004; Rampalli et al, 2007), or Ash2L knockdown (Rampalli et al, 2007). This, combined with studies showing that UTX associates with regions of the Hox gene cluster in the absence of RNA Pol II (Lan et al, 2007), demonstrates that the initial recruitment of UTX to muscle-specific genes is not through direct association with the elongating polymerase. Instead, we find that removal of H3K27me3 from the Myog and CKm genes occurs through a two-step process. In a first step, UTX is targeted to muscle-specific genes by the transcriptional regulator Six4 resulting in a localized demethylation of the transcriptional regulatory region upstream of the transcription start site. Once associated with a specific locus, UTX spreads into the coding region of the genes via an elongating polymerase-mediated mechanism. Taken together, these results provide novel insight into the mechanism by which UTX acts to remove the H3K27me3 mark over several kilobases of sequence at specific loci.
We have demonstrated that the DNA-bound transcriptional regulator Six4 is responsible for targeting UTX to the Myog and CKm gene regulatory regions. The strength of the association suggests that the interaction may not be direct, and could be mediated by TLE1, which was identified by yeast two-hybrid as an interacting partner of UTX (Grbavec et al, 1999), and interacts with Six family of proteins to co-regulate gene expression (Lopez-Rios et al, 2003). Alternatively, the interaction could be modulated through a post-translational modification of Six4, as we have previously observed for the association between Ash2L1/MLL2(KMT2B) and phosphorylated Mef2d (Rampalli et al, 2007). An essential function for Six4 in the activation of both the Myog (Spitz et al, 1998; Grifone et al, 2005) and CKm (Himeda et al, 2004) genes has previously been established. Deletion of the Six4-binding element in the promoter of the Myog gene abolished correct expression of a transgenic reporter (Spitz et al, 1998), whereas similar studies with a mutant Six4-binding site in a CKm transgenic reporter demonstrated significant loss of expression in muscle (Nguyen et al, 2003). In addition, the Six1/Six4 double knockout mice demonstrate general muscle hypoplasia and a loss of Myog expression (Grifone et al, 2005). However, the mechanism by which Six4 participates in the activation of transcription has not previously been established. Our study suggests that the function of Six4 is to target the UTX/KMT2D demethylase complex to muscle-specific promoters to activate gene expression. This function for Six4 in establishing tissue-specific gene expression is surprising, as expression of this homeobox protein is not limited to muscle tissue (Ohto et al, 1998). Interestingly, Mef2d (which is also expressed in multiple tissues) recruits the Ash2L1/KMT2B complex to muscle-specific genes in a p38-MAPK-dependent manner during myogenesis (Rampalli et al, 2007), whereas the muscle-specific transcriptional activator MyoD establishes a transcriptionally poised promoter at the Myog gene (Pol II, p300, and acetylated histone H3 are all present) in the absence of p38-MAPK activity (Simone et al, 2004; Rampalli et al, 2007). MyoD has also been shown to recruit P-TEFb to the Myog promoter to mediate phosphorylation of Pol II at Ser2 (Giacinti et al, 2006). This co-operation between the three families of transcription factors is consistent with the fact that binding sites for Six4 and Mef2d are enriched at MRF bound genes that become activated during myogenesis (Blais et al, 2005). Thus, the tissue-specific transcriptional regulator MyoD seems to establish a transcriptionally poised promoter at specific genes, whereas transcriptional regulators Six4 and Mef2d that are expressed in multiple tissues have a function in recruiting TrxG proteins to overcome the repressive effects of PcG proteins to permit gene expression. The fact that Six4 and Mef2d expression is not limited to muscle suggests that these two proteins could collaborate with other tissue-specific transactivators to overcome the repressive effects of PcG in other cell types.
Once targeted to muscle-specific genes by Six4, UTX seems to migrate into the coding region of the genes with the elongating Pol II. Examination of H3K27me3 enrichment across the CKm locus at 24 h (prior to expression of this gene) clearly demonstrates a localized demethylation that does not spread into the gene. Upon expression, the coding region of this gene becomes extensively demethylated. In addition, we observe that either pharmacological inhibition of p38 MAPK activity, or a knockdown of Ash2L prevent demethylation of the coding region (but not the regulatory elements) of both Myog and CKm genes. The fact that these conditions block Pol II from engaging in transcription at these two genes (Simone et al, 2004; Rampalli et al, 2007) supports the notion that UTX moves across their coding regions with the elongating polymerase. Indeed, Smith et al (2008) have previously shown an interaction between UTX and Pol II in cell extracts, while also demonstrating that UTX co-localizes with the elongating Pol II on polytene chromosomes in Drosophila. This argument is strengthened by experiments showing that DRB-induced stalling of Pol II at the Myog and CKm genes leads to a loss of UTX-mediated H3K27me3 demethylation activity within the coding region of the gene. There are many cases of transcription-related factors spreading across the coding region of genes with Pol II, the RNA processing machinery being a prime example (Pandit et al, 2008; Brookes and Pombo, 2009). In addition, it has been shown that TrxG proteins (both Ash1 and Kismet) that stimulate transcriptional elongation indirectly reduce levels of H3K27 methylation within target genes (Papp and Muller, 2006; Srinivasan et al, 2008). Interestingly, we observe that the 3′ end of both the Myog and CKm genes are demethylated to a lesser extent than the rest of the coding region of the gene. As UTX associates with Pol II phosphorylated at Ser 2 (Smith et al, 2008), it is possible that the demethylase is lost in this region as the elongating Pol II becomes dephosphorylated to permit transcriptional termination (Ahn et al, 2004; Kim et al, 2004). Consistent with this possibility, we observe reduced association of UTX with the 3′ end of the Myog and CKm genes (Figure 2). Taken together, these results suggest that the demethylase activity of UTX moves into the coding region of the Myog and CKm genes with elongating Pol II. We have also observed that UTX demethylates the region located 10 kb upstream of the CKm and Myog genes. Although it is possible that UTX could spread away from the coding region using intergenic transcription (Dye et al, 2006), divergent transcription (Seila et al, 2008), or self-polymerization (Li et al, 2009), the molecular mechanism through which UTX mediates removal of the repressive H3K27me3 mark in this distal region remains to be determined.
Bivalent (H3K4me3/H3K27me3) chromatin domains mark developmentally regulated genes in both pluripotent (Bernstein et al, 2006) and terminally differentiated cells (Mohn et al, 2008). Interestingly, the use of DRB to block elongation of Pol II at the transcribing Myog and CKm genes leads to the formation of bivalent (H3K4me3/H3K27me3) chromatin marks. This relationship between the formation of bivalent marks and the presence of stalled Pol II is interesting in the light of recent findings. Although the importance of stalled Pol II in regulating gene expression has been long appreciated (Bentley and Groudine, 1986; Rougvie and Lis, 1988; Krumm et al, 1992, 1995), recent studies in Drosophila (Muse et al, 2007; Zeitlinger et al, 2007) have suggested that this process is more prevalent than previously believed. In mesodermal tissue derived from Drosophila embryos, 12% of genes have a stalled polymerase (Zeitlinger et al, 2007). Among loci possessing a stalled Pol II, the number of developmentally regulated genes is disproportionately high (Zeitlinger et al, 2007). Consistent with this finding, inactive CpG-rich promoters that tend to be marked by bivalent methylation at H3K4 and H3K27 marks have been shown to be bound by Pol II (Mohn and Schubeler, 2009). Surprisingly, in our studies, the formation of the bivalent mark was restricted to genes that have previously been transcribed. This need for gene expression to establish bivalent marks is consistent with expression arrays studies, suggesting that tissue-specific genes are sporadically expressed in pluripotent embryonic stem cells (Efroni et al, 2008). Furthermore, studies using global run-on sequencing demonstrate that genes that have stalled Pol II express low, but significant levels of full-length transcripts (Core et al, 2008). On the basis of these findings, we propose that bivalent chromatin domains are established through stalling of Pol II at transcribed genes whose activity is modulated by the antagonistic functions of PcG and TrxG proteins.
In conclusion, we have demonstrated that the histone demethylase UTX is targeted to muscle-specific genes by the transcriptional activator Six4 to mediate removal of the repressive H3K27me3 mark during myogenesis. After initial targeting of muscle-specific genes, we show that spreading of H3K27me3 demethylase activity of UTX across the gene requires an actively elongating polymerase. Thus, this work provides novel insight into the mechanism by which UTX mediates the removal of H3K27me3 marks over extended distances at developmentally regulated genes.
Materials and methods
Antibodies
Commercial antibodies used in these studies include H3K4me3 (Abcam ab8580), H3K27me3 (Abcam ab6002), H3 antibody (Millipore 06-755), Myog (Santa Cruz SC-576), Suz12 (Abcam ab12073), Mef2 (Santa Cruz sc-17785, sc-13917), RPB1 (Abcam ab5408), and Ezh2 (Zymed 36-6300). Antibodies directed against Ash2L (Demers et al, 2007), TAF10 (Wieczorek et al, 1998), and Spt3 (Brand et al, 1999) have been described earlier. The Six4 antibody against the full-length protein was generated in rabbits as described earlier (Spitz et al, 1998) and purified by affinity to the cognate recombinant proteins (A.B. unpublished reagent). The rabbit anti-UTX antibody was generated against a recombinant His-tagged protein corresponding to amino acids 550–728 of human UTX protein that was expressed in bacteria.
Cell culture
The mouse myoblast cell line C2C12 was maintained at <80% confluency in DMEM containing 10% FBS, and differentiated in DMEM containing 2% horse serum, 10 μg/ml insulin, and 10 μg/ml transferrin. Lentivirus expressing shRNA targeting UTX or Six4 (Sigma) were used to infect C2C12 cells as described earlier (Yoon and Chen, 2008). Briefly, C2C12 cells were infected with lentivirus at 20% confluency in growth media. Infected cells were then re-infected 24 h later with fresh lentivirus, and selection with puromycin began 36 h after the initial infection. Differentiation was induced 48 h after the start of the infection, and continued for an additional 24–72 h. Stalling of Pol II was induced by the addition of 5,6-dichloro-1-β-D-ribobenzimidazole (Sehgal et al, 1976) (DRB) at a final concentration of 100 μM to C2C12 cells. For p38 inhibition studies, SB203580 was added to the differentiation media at a final concentration of 10 μM as described earlier (Zetser et al, 1999).
Chromatin immunoprecipitation
Analysis of histone modifications was performed using native ChIP as described earlier (Rampalli et al, 2007; Brand et al, 2008). Association of transcriptional regulators with specific regions of the genome was performed using X-ChIP as described earlier (Brand et al, 2004). Chromatin from C2C12 cells (cross-linked using 1% formaldehyde) was sheared using a Bioruptor (Diagenode) to obtain a resolution of ∼400 bp. Immunoprecipitated DNA was subjected to qPCR analysis using hydrolysis probes (see Supplementary data for Primer/Probe sequences). To determine relative enrichment (Brand et al, 2008), qPCR amplification of immunoprecipitated samples was normalized to genomic DNA for each primer set individually. Normalized values obtained from a mock IP (IgG) were subtracted from those observed in the specific IP, and then corrected for sample variation through division by normalized values observed from 1/50th of the input chromatin. Average values of an experiment represent biological replicates that are displayed with error bars corresponding to ±s.d. Each experiment was performed at least twice, and yielded similar results.
Double chromatin immunoprecipitation (native ChIP-ReChIP)
Analysis of bivalent marks was performed using a native ChIP-ReChIP method developed based on a previously established native ChIP protocol (Rampalli et al, 2007; Brand et al, 2008). Nuclei were isolated from 4 × 107 C2C12 cells treated with DRB (or vehicle), and their chromatin was fragmented with MNase (Supplementary Figure 5C). Nucleosomes were then isolated from the nuclei by hydroxyapatite chromatography before immunopurification with antibodies recognizing H3K4me3 or rabbit IgG. After washing, immunoprecipitated chromatin was eluted from the antibodies with a buffer containing 5 mM NaPO4 (pH 7.2), 600 mM KCl, 0.5 mM EDTA, and 25 mM DTT—note that a high concentration of DTT (up to 100 mM) does not disrupt the integrity of the nucleosomes (Dorigo et al, 2004). The eluate was then subjected to hydroxyapatite chromatography, and immunoprecipitated with antibodies recognizing H3K27me3 or mouse IgG. DNA recovered from the second immunoprecipitation was subjected to qPCR analysis as described earlier. Average values of an experiment represent biological replicates that are displayed with error bars corresponding to ±s.d. Each experiment was performed at least twice, and yielded similar results.
Reverse-transcription qPCR assays
Random-primed cDNA prepared from total RNA was subjected to duplex qPCR. Probes specific for Myog, CKm, or Acta1 were labelled with 5′ fluorescein phosphoramidite and 3′ black hole quencher-1, whereas the control DDX5 (Su et al, 2007) specific probe was labelled with 5′ Joe NHS Ester and 3′ Iowa Black FQ. In experiments where DRB was used to block transcriptional elongation, duplex qPCR was performed with a 5′Yakima Yellow/3′Eclipse Dark Quencher-labelled 18S rRNA primer/probe set (Eurogentec). Relative expression was calculated using Δ(ΔCt) of the gene-specific probes compared with the control (DDX5) gene-specific probe.
Co-immunoprecipitations
Nuclear extracts were prepared from differentiating C2C12 cells at 24 h time point as described earlier (Dignam et al, 1983). Rabbit polyclonal antibodies were attached to Protein A Dynabeads (Invitrogen), and then incubated with 5 mg of nuclear extract for 16 h. Immunoprecipitated material was washed three times with buffer D (20 mM Hepes pH 7.6, 300 mM KCl, 0.5 mM EDTA, 10% glycerol and 0.1% Nonidet-40), eluted in either 1 × SDS loading dye or a urea containing buffer (50 mM Tris pH 8.3, 6 M Urea, 5 mM EDTA, and 0.05% SDS), and then subjected to western blot analysis using the indicated antibodies.
Supplementary Material
Acknowledgments
We thank LiFang Li and Esther Mak for technical assistance, and Yubing Liu for Six4 antibody purification. This work was supported by grants from the Canadian Institutes of Health Research (to FJD and MB), the Muscular Dystrophy Association (to AB); and National Institutes of Health (to KG). FJD holds a Canadian Research Chair in Epigenetic Regulation of Transcription. MB holds a Canadian Research Chair in Regulation of Gene Expression. SS is a fellow of the Fonds de la Recherche en Sante Quebec.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bentley DL, Groudine M (1986) A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature 321: 702–706 [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ (2002) Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 9: 587–600 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ (2008) Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation (N-ChIP) of nucleosomes isolated using hydroxyapatite chromatography. Nature Protocols 3: 398–409 [DOI] [PubMed] [Google Scholar]
- Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M (2004) Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol 11: 73–80 [DOI] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L (1999) Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem 274: 18285–18289 [DOI] [PubMed] [Google Scholar]
- Brock HW, Fisher CL (2005) Maintenance of gene expression patterns. Dev Dyn 232: 633–655 [DOI] [PubMed] [Google Scholar]
- Brookes E, Pombo A (2009) Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep 10: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V (2004) The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi CP, Hosey AM, Palii C, Perez-Iratxeta C, Nakatani Y, Ranish JA, Dilworth FJ, Brand M (2009) Dual role for the methyltransferase G9a in the maintenance of {beta}-globin gene transcription in adult erythroid cells. Proc Natl Acad Sci USA 106: 18303–18308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K (2007) PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M (2007) Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell 27: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306: 1571–1573 [DOI] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Haussecker D, West S, Proudfoot NJ (2006) Turnover and function of noncoding RNA polymerase II transcripts. Cold Spring Harb Symp Quant Biol 71: 275–284 [DOI] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E (2008) Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C, Bagella L, Puri PL, Giordano A, Simone C (2006) MyoD recruits the cdk9/cyclin T2 complex on myogenic-genes regulatory regions. J Cell Physiol 206: 807–813 [DOI] [PubMed] [Google Scholar]
- Grbavec D, Lo R, Liu Y, Greenfield A, Stifani S (1999) Groucho/transducin-like enhancer of split (TLE) family members interact with the yeast transcriptional co-repressor SSN6 and mammalian SSN6-related proteins: implications for evolutionary conservation of transcription repression mechanisms. Biochem J 337 (Part 1): 13–17 [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P (2005) Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 132: 2235–2249 [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300 [DOI] [PubMed] [Google Scholar]
- Himeda CL, Ranish JA, Angello JC, Maire P, Aebersold R, Hauschka SD (2004) Quantitative proteomic identification of Six4 as the Trex-binding factor in the muscle creatine kinase enhancer. Mol Cell Biol 24: 2132–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA 104: 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E (2007) Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G (2009) H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Wold BJ, Hauschka SD (1989) Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Mol Cell Biol 9: 3393–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Hickey LB, Groudine M (1995) Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev 9: 559–572 [DOI] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M (1992) The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev 6: 2201–2213 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Li H, Motamedi MR, Yip CK, Wang Z, Walz T, Patel DJ, Moazed D (2009) An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing. Mol Cell 34: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J, Tessmar K, Loosli F, Wittbrodt J, Bovolenta P (2003) Six3 and Six6 activity is modulated by members of the groucho family. Development 130: 185–195 [DOI] [PubMed] [Google Scholar]
- Mohn F, Schubeler D (2009) Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet 25: 129–136 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D (2008) Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Muller J, Verrijzer P (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K (2007) RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen QG, Buskin JN, Himeda CL, Fabre-Suver C, Hauschka SD (2003) Transgenic and tissue culture analyses of the muscle creatine kinase enhancer Trex control element in skeletal and cardiac muscle indicate differences in gene expression between muscle types. Transgenic Res 12: 337–349 [DOI] [PubMed] [Google Scholar]
- Ohto H, Takizawa T, Saito T, Kobayashi M, Ikeda K, Kawakami K (1998) Tissue and developmental distribution of Six family gene products. Int J Dev Biol 42: 141–148 [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD (2008) Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol 20: 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Muller J (2006) Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ (2007) p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol 14: 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R (2004) Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT (1988) The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal PB, Darnell JE Jr, Tamm I (1976) The inhibition by DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell 9: 473–480 [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA (2008) Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL (2004) p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36: 738–743 [DOI] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A (2008) Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, Maire P (1998) Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci USA 95: 14220–14225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Dorighi KM, Tamkun JW (2008) Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet 4: e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY (2007) Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 8: 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A (2009) Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek E, Brand M, Jacq X, Tora L (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393: 187–191 [DOI] [PubMed] [Google Scholar]
- Yaffe D, Saxel O (1977) Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270: 725–727 [DOI] [PubMed] [Google Scholar]
- Yoon MS, Chen J (2008) PLD regulates myoblast differentiation through the mTOR-IGF2 pathway. J Cell Sci 121: 282–289 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetser A, Gredinger E, Bengal E (1999) p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J Biol Chem 274: 5193–5200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.