Abstract
Four strains of anaerobic, Gram-negative bacilli isolated from the human oral cavity were subjected to a comprehensive range of phenotypic and genotypic tests and were found to comprise a homogeneous group distinct from any species with validly published names. 16S rRNA and 23S rRNA gene sequence analyses and DNA–DNA reassociation data revealed that the strains constituted a novel group within the phylum ‘Synergistetes’ and were most closely related to Jonquetella anthropi. Two libraries of randomly cloned DNA were prepared from strain W5455T and were sequenced to provide a genome survey as a resource for metagenomic studies. A new genus and novel species, Pyramidobacter piscolens gen. nov., sp. nov., is proposed to accommodate these strains. The genus Pyramidobacter comprises strains that are anaerobic, non-motile, asaccharolytic bacilli that produce acetic and isovaleric acids and minor to trace amounts of propionic, isobutyric, succinic and phenylacetic acids as end products of metabolism. P. piscolens gen. nov., sp. nov. produced hydrogen sulphide but was otherwise largely biochemically unreactive. Growth was stimulated by the addition of glycine to broth media. The G+C content of the DNA of the type strain was 59 mol%. The type strain of Pyramidobacter piscolens sp. nov. is W5455T (=DSM 21147T=CCUG 55836T).
INTRODUCTION
Synergistes jonesii, a species comprising Gram-negative, anaerobic rods, isolated from goat rumen (Allison et al., 1992) was originally thought to be the sole representative of a novel division, ‘Synergistes’ of the domain Bacteria (Hugenholtz et al., 1998). Subsequently, however, a number of other genera, including Aminobacterium, Aminomonas, Aminiphilus, Anaerobaculum, Dethiosulfovibrio, Jonquetella, Thermanaerovibrio and Thermovirga, were shown to belong to this group (Hugenholtz, 2002; Jumas-Bilak et al., 2007; Vartoukian et al., 2007), although several of these genera were initially misclassified as belonging to the ‘Firmicutes’ (Baena et al., 1999a, b, 2000; Menes & Muxi, 2002; Zavarzina et al., 2000). The neighbouring phylum contains organisms of the genera Deferribacter and Flexistipes, among others, and has therefore been referred to as ‘Deferribacter’ or ‘Flexistipes’. Although ‘Synergistes’ is distinct from this neighbouring phylum, the Bergey's Manual and NCBI taxonomic schemes currently, and incorrectly, lump these groups together as ‘Deferribacter’. No formal phylum name has been proposed for the group currently known as ‘Synergistes’, although the names ‘Aminanaerobia’ (Warnecke et al., 2007) and ‘Synergistetes’ have been suggested (Jumas-Bilak et al., 2007). For the purposes of this article, we will use ‘Synergistetes’, as its relationship with ‘Synergistes’ is obvious and has an appropriate phylum-level construction.
Culture-independent molecular methods based on 16S rRNA gene sequences have been used widely to characterize bacterial communities associated with a wide range of environmental habitats and habitats associated with human and animal diseases. In one such study, a 16S rRNA gene library constructed from subgingival bacterial plaque from a patient with refractory periodontal disease contained a novel clone, BA121, representing a novel taxon in the phylum ‘Synergistetes’ (Paster et al., 2001). Subsequently, we identified 18 clones with essentially an identical sequence in five libraries from individuals with acute ulcerative necrotizing gingivitis, advanced caries or endodontic infections (Munson et al., 2002, 2004; F. E. Dewhirst, unpublished data), suggesting that this novel taxon is a true member of the human oral microbiome. The isolation of four strains with essentially identical 16S rRNA gene sequences from samples associated with oral infections now allows the characterization of a species previously known only as a clone phylotype. Because the only gene sequences currently available for any organism of the ‘Synergistetes’ phylum are 16S rRNA sequences, we undertook a genome survey using shotgun sequencing to get a genomic ‘snapshot’ of this poorly studied phylum to obtain additional sequence data that might be of phylogenetic and taxonomic interest and also provide a resource for metagenomic studies.
METHODS
Bacterial strains and growth conditions.
Strains W5455T and AHN 11662 were isolated from odontogenic abscesses, strain P4G_18 was from an inflamed gingival crevice and strain D031B-08-1 was from a periodontal pocket. Strain AHN 11662 was donated by the late Dr H. Jousimies-Somer of the Anaerobe Reference Laboratory, National Public Health Institute, Helsinki, Finland, and strain D031B-08-1 came from the Virginia Polytechnic Institute Anaerobe Laboratory collection of L. V. H. Moore and the late W. E. C. Moore and was kindly made available by Dr A. Tanner. Jonquetella anthropi ADV 126T was obtained from the CCUG (CCUG 53819T) and J. anthropi E3_33 was isolated from an infected dental root canal. The strains were grown at 37 °C on fastidious anaerobe agar (FAA; LabM, UK) supplemented with 5 % horse blood under anaerobic conditions (N2/H2/CO2, 80 : 10 : 10, by vol.) in an anaerobic workstation (Don Whitley Scientific). Colonial morphologies were determined using a dissecting microscope, after incubation for 7 days. Cellular morphology was recorded after Gram-staining of smears prepared from 2 day FAA cultures. Hanging-drop preparations of 18 h broth cultures were examined, using phase-contrast microscopy, for cellular motility. Transmission electron microscopy was used to examine the cell-wall ultrastructure, as described previously (Downes et al., 2003).
Phenotypic characterization.
Biochemical tests were performed using standard methods (Holdeman et al., 1977; Jousimies-Somer et al., 2002). Susceptibility to special-potency antibiotic discs containing colistin (10 μg), kanamycin (1 mg) and vancomycin (5 μg) was determined on FAA (Jousimies-Somer et al., 2002). Fermentation tests were performed using prereduced, anaerobically sterilized sugars prepared in-house in an anaerobic workstation (Holdeman et al., 1977). Hydrogen sulphide production was determined in a tube containing prereduced, anaerobically sterilized SIM medium (Oxoid), supplemented with 1 μl vitamin K ml−1, 5 μg haemin ml−1, 0.05 % cysteine and 4 % (v/v) salts solution (Holdeman et al., 1977). The strains were grown in peptone-yeast extract broth (PY) and peptone-yeast extract broth with glucose (PYG) (Holdeman et al., 1977) and their short-chain volatile and non-volatile fatty acids were extracted and analysed using GC (Holdeman et al., 1977). Enzyme profiles were obtained with a Rapid ID 32A anaerobe-identification kit (bioMérieux), according to the manufacturer's instructions, with bacteria harvested from Columbia agar plates (LabM) supplemented with 5 % horse blood; the test was performed in triplicate. Susceptibility to amoxicillin (10 μg), ampicillin (2 μg), cefoxitin (10 μg), ciprofloxacin (5 μg), clindamycin (2 μg), gentamicin (10 μg), metronidazole (5 μg), penicillin (1.5 units), tetracycline (5 μg) and trimethoprim (2.5 μg) was investigated using a disc-diffusion method and Iso-Sensitest agar with 5 % horse blood (Oxoid). Zones of inhibition of ≥30 mm in diameter were recorded as representing antibiotic susceptibility, whereas the absence of zones of inhibition around the 7 mm discs was designated as representing antibiotic resistance.
Growth at 25, 30, 37 and 42 °C was assessed using PY medium. Utilization of amino acids was determined by adding alanine, arginine, cysteine, glycine, leucine and lysine (each at 10 mM) to both DP (Magot et al., 1997) and PY media. Growth was measured for up to 10 days by means of spectrophotometric measurement of turbidity at 580 nm. The requirement of NaCl (3 %) for growth and the utilization of thiosulphate, sulphate and sulphite were investigated by adding 20 mM of the sodium salt of each compound to DP medium prior to incubation at 37 °C. The cellular fatty acid composition was determined using acid-catalysed methanolysis to release fatty acid methyl esters (FAMEs) from lyophilized whole cells, as described previously (Sutcliffe, 2000). The FAMEs were extracted into hexane and concentrated under nitrogen gas before being analysed using GC (Sutcliffe, 2000). FAMEs were identified by comparison with authentic FAME standards (Sigma).
Genetic analyses.
The DNA G+C contents of strains W5455T and AHN 11662 were determined using an HPLC method as described previously (Wade et al., 1999). The base composition was also estimated from the genome survey nucleic acid sequence data, by direct counting. A thermal denaturation method (Huß et al., 1983) was used to determine the extent of DNA–DNA relatedness between the strains.
The 16S rRNA genes of the four strains were sequenced as described previously (Downes et al., 2005). Sequences were assembled using the BioEdit program (Hall, 2004) and identified by blast interrogation of the GenBank database (Altschul et al., 1990). Phylogenetic analysis was performed using mega, version 4 (Tamura et al., 2007). Phylogenetic trees were constructed with the neighbour-joining method from distance matrices prepared using the Jukes–Cantor correction (Jukes & Cantor, 1969).
A 16S rRNA gene forward primer and a series of 23S rRNA gene primers, used previously for full sequencing of Helicobacter species (Dewhirst et al., 2005), were used to produce amplicons. The primers used for PCR and sequencing are listed in Supplementary Table S1 (available in IJSEM Online). Amplicon sequencing was performed as described previously (Dewhirst et al., 2005). Genome sequencing contigs containing rRNA genes were identified by means of blastn (Altschul et al., 1990) analysis using 16S rRNA and 23S rRNA gene sequence fragments from manual sequencing. Essentially complete ribosomal operons were assembled manually from the amplicon and genome survey sequences. Contigs containing tRNA were identified using tRNAscan-SE (Lowe & Eddy, 1997).
Construction of DNA libraries.
For library production, strain W5455T was grown on TSBY agar medium (Northeast Laboratory, Waterville, ME, USA) at 37 °C under an N2/CO2/H2 (80 : 10 : 10, by vol.) atmosphere. Cells were lysed using a proteinase K treatment and the DNA was purified by phenol/chloroform extraction (Maniatis et al., 1982). Two strategies were used to fragment the genomic DNA. The first genomic library was produced using RsaI restriction endonuclease digestion (New England Biolabs). The second library was produced using the point-sink shearer method with an Hydroshear DNA-shearing device and a small shearer assembly unit (GeneMachines). To maximize our ability to annotate the survey genome, after separation by gel electrophoresis, an insert size of approximately 1.1–1.6 kb was selected to allow assembly of the paired sequence reads to yield mainly contigs. The DNA was gel-purified using a Qiaquick Spin kit (Qiagen), treated using a DNATerminator End Repair kit and ligated to pSMART-HC Kan (Lucigen, Middleton, WI, USA). After transformation in electrocompetent E. cloni 10G cells (Lucigen), 384 colonies were selected from the first library and 768 from the second. The plasmids were purified using a Perfectprep Plasmid 96 Vac Direct Bind kit (Eppendorf) and sequenced using primers (SL1 and SR2) at each end of the vector. Longer reads were obtained for clones in the second library after addition to the sequencing reaction mix of betaine (Rees et al., 1993).
DNA sequences were assembled using sequencher 4.5 (Gene Codes Corp.) for the first library and celera assembler (J. Craig Venter Institute, Rockville, MD, USA) for the second. The assembled sequences (contigs), together with unassembled single sequence reads (singletons), were subjected to an automatic genome-annotation pipeline (described by Chen et al., 2005). The open reading frame (ORF) identification script, identifying both complete and partial ORFs, used AUG, GUG and UUG as start codons and UAA, UGA and UAG as stop codons. In this study, partial ORFs were defined as reading frames without start or stop codons (or neither) and encoding proteins greater than 50 amino acids in length. For similarity analyses, blast parameters such as word size and substitution matrix were adjusted dynamically on the basis of the lengths of the ORFs, and the expect value (E) was set at 10−5 to screen out non-significant hits. The annotated genome surveys are available at the Human Oral Microbiome Database web resource (http://www.homd.org).
Deposited sequences.
The genome surveys for the first and second libraries of strain W5455T were deposited under GenBank accession numbers DU723013–DU723395 and ET628498–ET629016, respectively. Two ribosomal operons containing 16S rRNA, tRNAs, 23S rRNA and part of the 5S rRNA were deposited under accession numbers EU379932 and EU379933. An Na+/H+ antiporter and IS4-family insertion sequences were deposited under accession numbers EU379934 and EU379935, respectively. The 23S rRNA gene sequences of J. anthropi E3_33 and S. jonesii ATCC 49833T were deposited under accession numbers EU840722 and EU840723, respectively.
RESULTS AND DISCUSSION
Phenotypic and biochemical characteristics
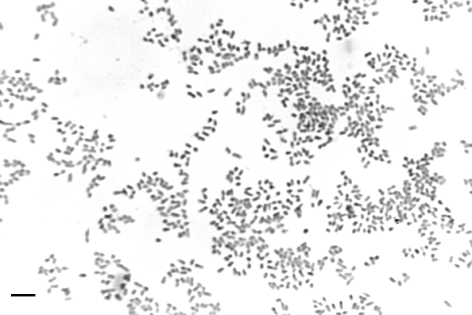
The four novel strains comprised obligately anaerobic, non-motile, non-pigmenting, Gram-negative bacilli (0.7–0.8 μm wide and 0.8–2.2 μm long; Fig. 1). After 7 days incubation on FAA plates, colonies were 0.7–1.1 mm in diameter, circular, entire, high convex to pyramidal and shiny with a consistent opacity and off-white to watery steel-grey in colour when viewed under a plate microscope. The cultures had a distinctive fishy odour. The strains were resistant to special potency discs containing vancomycin and colistin but were sensitive to those containing kanamycin. Growth in PY, PYG and brain heart infusion broth (BHI; LabM) produced a moderately turbid suspension (2 to 3+ on a scale of 0 to 4+). Growth was not enhanced by the addition of carbohydrates to the broth. Growth in DP medium was extremely poor and the addition of NaCl did not stimulate growth. However, glycine, alone among the amino acids tested, did stimulate growth in both DP and PY media. Minimal growth occurred in DP medium supplemented with thiosulphate, sulphate or sulphite. The four strains grew in PY at 25, 30, 37 and 42 °C, with optimal growth occurring at 30 and 37 °C.
Fig. 1.
Micrograph of a Gram-stained smear of strain W5455T. Bar, 5 μm.
The strains were asaccharolytic and major amounts of acetic acid, minor amounts of isovaleric acid and minor to trace amounts of propionic, isobutyric, succinic and phenylacetic acids were produced as end products of metabolism. Hydrogen sulphide was produced but other biochemical tests gave negative results (see the species description). In the Rapid ID 32A identification panel, all four strains were strongly positive for glycine arylamidase, resulting in a profile of 0000 0004 00. J. anthropi strains ADV 126T and E3_33 were strongly positive for both glycine arylamidase and leucyl glycine arylamidase, giving a profile of 0000 0404 00.
Susceptibility testing for all of the antimicrobials investigated revealed that, in each case, strains either grew right up to the disc (showing no zone of inhibition) or a large zone was observed (>30 mm in diameter). In the absence of reference data for these species and antimicrobials, the absence of a zone was regarded as indicating resistance and a zone >30 mm was taken to indicate susceptibility. The four strains had identical antimicrobial susceptibility profiles: they were susceptible to penicillin, ampicillin, amoxicillin, cefoxitin, clindamycin, tetracycline and metronidazole, and resistant to ciprofloxacin, gentamicin and trimethoprim. J. anthropi strains ADV 126T and E3_33 showed the same susceptibility pattern as those for the four novel strains but were also susceptible to ciprofloxacin. The G+C content of the DNA of strains W5455T and AHN 11662 was 59 mol% (by HPLC).
FAME analysis revealed a complex profile that included several unidentifiable fatty acids (Supplementary Table S2, available in IJSEM Online). Most significantly, the FAME profiles are clearly distinct from those of J. anthropi strains (which are dominated by iso-C15 : 0 and C16 : 0 fatty acids) and are also distinct from that of Dethiosulfovibrio peptidovorans (Jumas-Bilak et al., 2007). S. jonesii has previously been reported to exhibit a complex FAME profile, with straight- and branched-chain, saturated and unsaturated FAMEs, along with cyclopropane and 3-hydroxy fatty acids (Allison et al., 1992). Clearly, the FAME profiles of members of the phylum ‘Synergistetes’ can exhibit significant variation and complexity.
Morphological and ultrastructural characteristics
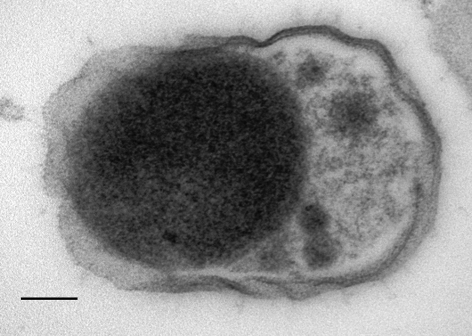
Transmission electron microscopic examination of ultrathin sections of cells of strain W5455T showed the presence of a typical Gram-negative cell wall composed of a thin peptidoglycan layer surrounded by an outer membrane (Fig. 2). The surfaces of the cells of strains W5455T and AHN 11662 had a crenulated appearance (not shown). In the cytoplasm, numerous inclusion bodies (of unknown composition) were observed, as well as the apparent segregation of the DNA into a nuclear area. An internal membrane enclosing the nuclear area could not be visualized with certainty, but may be present. For visualization and three-dimensional positioning of internal structures, cryo-electron microscopy would be the technique of choice (Izard et al., 2008). The general appearance seems to be characteristic of the phylum; transmission electron micrographs of ultrathin sections of cells of S. jonesii, Anaerobaculum mobile, Dethiosulfovibrio russensis and Thermanaerovibrio velox revealed similar cellular morphologies (Allison et al., 1992; Menes & Muxi, 2002; Surkov et al., 2001; Zavarzina et al., 2000).
Fig. 2.
Transmission electron micrograph of an ultrathin section of a cell of strain W5455T. Bar, 100 nm.
Ribosomal rRNA operons
Amplicons of the rRNA operon were successfully produced using the 16S rRNA forward primer Y34 and 23S rRNA reverse primers M83, M85, M86, M88 and M89 (Supplementary Table S1). The amplicons for each primer pair were doublets when examined using agarose gel electrophoresis, indicating that there were at least two distinct rRNA operons of different lengths. The primers listed in Supplementary Table S1 were used for sequencing from the end of the 16S rRNA gene to position 2759 (Escherichia coli numbering) of the 23S rRNA gene. The region between the 16S and 23S rRNA genes contained tRNA Ile (anticodon GAT) and tRNA Ala (anticodon TGC) genes in one version of the operon, and had just a spacer in the other. The partial genome sequencing effort added additional sequence information, allowing us to complete the 23S rRNA gene sequence and extend through half of the 5S rRNA gene sequence. The rRNA operon sequences are available from GenBank under accession numbers EU379932 and EU379933.
Phylogenetic analyses
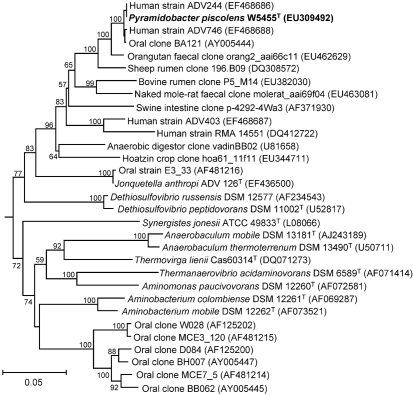
The 16S rRNA gene sequences of the four strains were highly similar, with strains AHN 11662, P4G_18 and D031B-08-1 exhibiting 99.7, 99.7 and 99.6 % sequence similarity, respectively, with strain W5455T. Phylogenetic analysis of the 16S rRNA gene sequence of strain W5455T revealed that the strain belonged to the phylum ‘Synergistetes’ (Fig. 3). The full-length sequence for the 16S rRNA gene was also obtained for strain AHN 11662, and showed 99.7 % sequence similarity with respect to strain W5455T over 1445 unambiguously aligned bases. The most closely related species with a validly published name was J. anthropi (85.7 % sequence similarity). A number of sequences from isolates and clones were more closely related to those of the novel strains than was J. anthropi. These were derived from a variety of sources, including the environment and the rumen and gut of various animal species.
Fig. 3.
Phylogenetic tree based on 16S rRNA gene sequence comparisons over 1175 aligned bases, showing the relationships between strain W5455T and related species. The tree was constructed using the neighbour-joining method, following distance analysis of aligned sequences, and was rooted with Treponema socranskii (not shown; accession no. AF033307). Numbers at branch points are bootstrap percentages (based on data for 100 trees). GenBank accession numbers are shown in parentheses. Bar, 0.05 nucleotide substitutions per site.
Organisms belonging to the phylum ‘Synergistetes’ can be identified quite reliably using blastn analysis with the 16S rRNA sequence Syn_905-981 (5′-TGAAACTCAAAGGAATTGACGGGGGCCCGCACAAGCGGTGGAGCACGTGGTTTAATTCGATGCAAACCGAAGAACCT-3′) targeting the 16S rRNA gene. However, the sequences for some organisms in neighbouring phyla differ by just one or more bases, so any organism with a sequence difference of one or more bases should be checked by constructing a phylogenetic tree. Organisms of the phylum ‘Synergistetes’ can be amplified with the primer pair 9–27 (forward; GAGTTTGATYMTGGCTCAG) and 1483–1501 (reverse; GTTACGACTTCACCCYCCT). This primer pair amplifies organisms in the phyla ‘Synergistetes’ and ‘Spirochaetes’ and the order Coriobacteriales in the phylum ‘Actinobacteria’. This selectivity is due to the presence of a GG in their sequences at Escherichia coli positions 1484 and 1485, where most other bacteria contain a CU. The corresponding CC in the reverse complement primer above is underlined. This specificity was verified in silico for sequences from strains of the phylum ‘Synergistetes’ from GenBank.
A phylogenetic tree based on a comparison of 23S rRNA gene sequences from strain W5455T, J. anthropi E3_33, S. jonesii ATCC 49833T and reference sequences from 12 additional phyla is shown in Fig. 4. As was found in the tree based on 16S rRNA gene sequences, strain W5455T branches with J. anthropi in the phylum ‘Synergistetes’. The 23S rRNA gene sequence data support Hugenholtz's suggestion that ‘Synergistes’ represents a separate branch at the phylum level (Hugenholtz, 2002).
Fig. 4.
Neighbour-joining phylogenetic tree based on 23S rRNA gene sequence comparisons over 2701 positions. Phylogenetic analyses were conducted with mega4. Numbers at branch points are bootstrap percentages (based on 1000 replicates). Bar, 0.05 nucleotide substitutions per site.
The protein translocase subunit SecA has been used as a means of determining deep phylogenetic relationships (Gupta et al., 2003). One of the contigs sequenced contained approximately half of the secA gene (1155 bases, 385 amino acids). A neighbour-joining phylogenetic tree constructed using 26 phylogenetically diverse reference sequences is shown in Fig. 5. Note that, in the secA tree, strain W5455T falls in a deeply branching position with ‘Aquifex aeolicus’ and Thermotoga maritima, as in the tree based on 23S rRNA gene sequences. Thus, analysis of the 16S rRNA and 23S rRNA gene and SecA data supports placement of strain W5455T within the phylum ‘Synergistetes’, and also indicates that ‘Synergistetes’ is a unique phylum that is clearly distinct from the ‘Firmicutes’.
Fig. 5.
Neighbour-joining phylogenetic tree based on secA gene sequences. Numbers at branch points represent bootstrap percentages (based on 1000 replicates). Evolutionary distances were computed using the Poisson correction (Zuckerkandl & Pauling, 1965) method. All positions containing alignment gaps and missing data were eliminated only from pairwise sequence comparisons (pairwise deletion option). In total, there were 1853 positions in the final dataset. Phylogenetic analyses were conducted in mega4. Bar, 0.1 amino acid substitutions per site.
The DNA–DNA reassociation experiments showed that the four strains were closely related, but distinct from J. anthropi. Strain W5455T exhibited 89, 85 and 90 % DNA–DNA relatedness with respect to strains AHN 11662, P4G_18 and D031B-08-1, respectively, but only 11 and 2 % with respect to J. anthropi strains ADV 126T and E3_33.
Genomic survey
Because of the largely biochemically unreactive nature of the novel strains, we expanded the investigation to include genomic data. Low-cost genomic surveys were performed, allowing sequence similarity analysis between gene orthologues. Prior to the current study, the only nucleotide sequence data for any member of the phylum were 16S rRNA gene sequences. This genomic survey provides the first ‘snapshot’ of the genomic content of an organism in the phylum ‘Synergistetes’.
The first library included 385 contigs (up to 2231 bp long) and singletons, comprising a total of 276 552 bp. The second library included 520 contigs (up to 2122 bp long) and singletons, giving a total of 342 989 bp. The overall G+C content of the DNA sequenced was 60.8 mol%, with individual contig G+C contents ranging from 35.3 to 82.9 mol%. Over 1500 ORFs were detected. A total of 1548 blastp hits against the NCBI non-redundant database and 928 hits against Swiss-Prot were found. Some predicted ORFs did not match sequences in the database and may be unique either to this organism or to the phylum. A static annotation of the partial and full-length ORFs is available in Supplementary Tables S3 and S4 (available in IJSEM Online). The Human Oral Microbiome Database website (http://www.homd.org) provides matches for both genome surveys and is automatically updated as new genome sequences are deposited and annotations are refined. The absence of other genomic sequences from members of the phylum ‘Synergistetes’ provides the opportunity to look at orthologous genes in the context of sequence similarities over multiple phyla. The low levels of sequence similarity between strain W5455T and other genomes available in the databases (Supplementary Tables S3 and S4, in IJSEM Online) provide further confirmation of the phylum-level genetic distance between this strain and those sequenced previously.
Although the majority of the blast matches had low similarity scores, some showed high degrees of relatedness. For example, an NhaC-family sodium–proton antiporter from strain W5455T (GenBank accession number EU379934) was found to be highly similar (71–73 % amino acid sequence identity) to sequences from the oral organisms Fusobacterium nucleatum subsp. nucleatum and F. nucleatum subsp. vincentii and Treponema denticola (Kapatral et al., 2002; Seshadri et al., 2004), but was shown to be less than 47 % similar to NhaC from all other organisms in GenBank.
This observation supports the hypothesis that bacteria in the oral cavity readily exchange genetic information (Duncan, 2005). While it is impossible to know the direction of the postulated genetic exchange, it is striking that the three organisms are from different phyla. To explore possible exchange mechanisms, the partial genome sequence was inspected for genes that could participate in genetic exchange. A complete IS4-family insertion sequence element, with inverted repeats and a transposase coding region (GenBank accession no. EU379934), was identified together with a partial sequence coding for an IS200-like transposase. Several partial phage proteins were identified, including a DNA packaging protein B, a phage integrase, a phage tail tape measuring protein and tail-fibre proteins. These genetic elements are common to a wide range of phyla (De Palmenaer et al., 2008; Hambly & Suttle, 2005) and could contribute to genetic exchange.
A notable metabolic characteristic of the named members of the ‘Synergistetes’ phylum is that they are biochemically unreactive, mostly asaccharolytic and possess a proteolytic metabolism (Godon et al., 2005). It is notable that multiple oligopeptide and dipeptide transporters and several proteases were identified but no sugar transporters were found. As expected, a number of proteases of different classes were identified. However, because of the limited information available from these partial genome surveys (approx. 20 % coverage), it is not possible to reconstruct full metabolic pathways. No genes encoding proteins regarded as classical bacterial virulence factors were identified.
In addition to providing the first ‘snapshot’ of the genetic capability of a ‘Synergistetes’ strain, the DNA sequence information generated is useful for the phylogenetic anchoring of sequences obtained in human metagenomic projects. For metagenomic studies to be interpretable, it is essential that genomic information is available for reference organisms at all phylogenetic levels, particularly at the phylum level. Survey sequencing is a low-cost way of obtaining such genetic information for organisms that may not be predominant in a complex environment (Godon et al., 2005).
In our studies to date, we have identified four isolates and 18 clones of this taxon from oral sites (Paster et al., 2001; de Lillo et al., 2006; Munson et al., 2004; F. E. Dewhirst, unpublished observations). In order to identify other examples in the GenBank database, the 16S rRNA gene sequence of strain W5455T was broken down into 500-base segments and used to search GenBank for sequences with ≥99 % identity. Seven clones from two endodontic studies were identified (GenBank accession numbers AY597651, AY597652; DQ643961, DQ643964, EF104597, DQ643963, EF104598; Vianna et al., 2007) along with six from an unpublished study on vaginal flora (EF468690, EF468705, EF468706, EF468707, EF468708 and EF468712). In addition, two human clinical strains, ADV244 (EF468686) and ADV746 (EF468688), were identified (unpublished information from GenBank). Using PCR primer pairs specific for this taxon (based on oral clone BA121), exudates from 7 of 21 untreated endodontic infections produced positive results (Siqueira & Rocas, 2005).
The four strains studied here are highly similar both phenotypically and genotypically and are clearly distinct from any species or genus with validly published names. Strain W5455T shares 86 % 16S rRNA gene sequence similarity with the type strain of J. anthropi, its closest phylogenetic relative. This level of divergence indicates that the strains examined here constitute a novel genus. We therefore propose the name Pyramidobacter piscolens gen. nov., sp. nov. to accommodate this group of strains. Most members of this phylum are asaccharolytic and biochemically unreactive. However, some characteristics that serve to differentiate Pyramidobacter piscolens gen. nov., sp. nov. from J. anthropi have been identified and are shown in Table 1.
Table 1.
Characteristics that serve to differentiate Pyramidobacter piscolens gen. nov., sp. nov. from J. anthropi
Data for Pyramidobacter piscolens gen. nov., sp. nov. are based on the results from the four strains described in this study. Data for J. anthropi are from Jumas-Bilak et al. (2007) and this study.
| Characteristic | Pyramidobacter piscolens | J. anthropi |
|---|---|---|
| Cell size (μm) | 0.7–0.8×0.8–2.2 | 0.8–0.9×1.4–1.7 |
| Leucyl glycine arylamidase | − | + |
| Ciprofloxacin susceptibility | Resistant | Susceptible |
Description of Pyramidobacter gen. nov.
Pyramidobacter (Py.ra.mi.do.bac′ter. Gr. n. pyramis -idos pyramid; L. n. pyramis pyramid; N.L. masc. n. bacter a rod; N.L. masc. n. Pyramidobacter a rod that forms pyramid-like colonies).
Cells are obligately anaerobic, non-motile bacilli. Cells are asaccharolytic and produce major amounts of acetic acid, minor amounts of isovaleric acid and minor to trace amounts of propionic, isobutyric, succinic and phenylacetic acids as end products of metabolism in PYG. The DNA G+C content of the type species is 59 mol%. The type species is Pyramidobacter piscolens.
Description of Pyramidobacter piscolens sp. nov.
Pyramidobacter piscolens (pis.co′lens. L. n. piscis -is fish; L. part. adj. olens smelling; N.L. part. adj. piscolens smelling of fish).
Exhibits the following properties in addition to those given in the genus description. The description is based on the study of the type strain and three other strains. Cells are Gram-negative, 0.7–0.8 μm wide and 0.8–2.2 μm long. After 7 days incubation on FAA plates, colonies are 0.7–1.1 mm in diameter, circular, entire, high convex to pyramidal, smooth, shiny, off-white to grey and opaque. The culture has a distinctive fishy odour. Moderate growth is obtained in PY, PYG and BHI media and growth is not enhanced by the addition of carbohydrates. Growth in DP and PY media is stimulated by the addition of 10 mM glycine. Alanine, arginine, cysteine, leucine and lysine are not utilized. Arabinose, cellobiose, fructose, glucose, lactose, maltose, mannitol, mannose, melezitose, rhamnose, salicin, sucrose, trehalose and xylose are not fermented. Hydrogen sulphide is produced; indole and catalase are not produced. Aesculin, gelatin and urea are not hydrolysed and nitrate is not reduced. Does not grow in 20 % bile. The Rapid ID 32A profile is 0000 0004 00, which corresponds to a strongly positive reaction for glycine arylamidase. The DNA G+C content of the type strain is 59 mol%.
The type strain, W5455T (=DSM 21147T=CCUG 55836T), was isolated from a human odontogenic abscess.
Supplementary Material
Acknowledgments
This research was supported by NIH grants (DE-10374, DE-16937 and DE-17106) from the National Institute for Dental and Craniofacial Research and a grant from the Guy's and St Thomas' Charity (ref. R050724). Professor Hans Trüper is thanked for his expert advice on the construction of the species name. We thank Janet Cheong, Sara Barbuto and Erin Klein for their technical assistance with 23S rRNA gene sequencing, and Derek Spencer for making the second genomic library. We thank Jonathan Eisen for allowing us to use the 23S rRNA gene sequence of S. jonesii. The genome sequencing of S. jonesii was supported by an NSF ‘Assembling the Tree of Life’ grant (0228651) to Jonathan Eisen and Naomi Ward.
Abbreviations
FAME, fatty acid methyl ester
IS, insertion sequence
ORF, open reading frame
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strains W5455T, AHN 11662 and P4G_18 are EU309492, EU309493 and AY207056, respectively.
Tables showing the 16S–23S rRNA gene intervening sequences and 23S rRNA primers, and fatty acid methyl ester profiles of strains of Pyramidobacter piscolens gen. nov., sp. nov., Jonquetella anthropi and Dethiosulfovibrio peptidovorans and static annotations of partial and full-length open reading frames in genomic libraries 1 and 2 are available as supplementary material with the online version of this paper.
References
- Allison, M. J., Mayberry, W. R., McSweeney, C. S. & Stahl, D. A. (1992). Synergistes jonesii gen. nov., sp. nov.: a rumen bacterium that degrades toxic pyridinediols. Syst Appl Microbiol 15, 522–529. [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Baena, S., Fardeau, M. L., Ollivier, B., Labat, M., Thomas, P., Garcia, J. L. & Patel, B. K. (1999a). Aminomonas paucivorans gen. nov., sp. nov., a mesophilic, anaerobic, amino-acid-utilizing bacterium. Int J Syst Bacteriol 49, 975–982. [DOI] [PubMed] [Google Scholar]
- Baena, S., Fardeau, M. L., Woo, T. H., Ollivier, B., Labat, M. & Patel, B. K. (1999b). Phylogenetic relationships of three amino-acid-utilizing anaerobes, Selenomonas acidaminovorans, ‘Selenomonas acidaminophila’ and Eubacterium acidaminophilum, as inferred from partial 16S rDNA nucleotide sequences and proposal of Thermanaerovibrio acidaminovorans gen. nov., comb. nov. and Anaeromusa acidaminophila gen. nov., comb. nov. Int J Syst Bacteriol 49, 969–974. [DOI] [PubMed] [Google Scholar]
- Baena, S., Fardeau, M. L., Labat, M., Ollivier, B., Garcia, J. L. & Patel, B. K. (2000). Aminobacterium mobile sp. nov., a new anaerobic amino-acid-degrading bacterium. Int J Syst Evol Microbiol 50, 259–264. [DOI] [PubMed] [Google Scholar]
- Chen, T., Abbey, K., Deng, W. J. & Cheng, M. C. (2005). The bioinformatics resource for oral pathogens. Nucleic Acids Res 33, W734–W740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lillo, A., Ashley, F. P., Palmer, R. M., Munson, M. A., Kyriacou, L., Weightman, A. J. & Wade, W. G. (2006). Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets. Oral Microbiol Immunol 21, 61–68. [DOI] [PubMed] [Google Scholar]
- De Palmenaer, D., Siguier, P. & Mahillon, J. (2008). IS4 family goes genomic. BMC Evol Biol 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst, F. E., Shen, Z., Scimeca, M. S., Stokes, L. N., Boumenna, T., Chen, T., Paster, B. J. & Fox, J. G. (2005). Discordant 16S and 23S rRNA gene phylogenies for the genus Helicobacter: implications for phylogenetic inference and systematics. J Bacteriol 187, 6106–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes, J., Munson, M. & Wade, W. G. (2003). Dialister invisus sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 53, 1937–1940. [DOI] [PubMed] [Google Scholar]
- Downes, J., Sutcliffe, I. C., Tanner, A. C. R. & Wade, W. G. (2005). Prevotella marshii sp. nov. and Prevotella baroniae sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 55, 1551–1555. [DOI] [PubMed] [Google Scholar]
- Duncan, M. J. (2005). Oral microbiology and genomics. Periodontol 2000 38, 63–71. [DOI] [PubMed] [Google Scholar]
- Godon, J. J., Morinière, J., Moletta, M., Gaillac, M., Bru, V. & Delgènes, J. P. (2005). Rarity associated with specific ecological niches in the bacterial world: the ‘Synergistes’ example. Environ Microbiol 7, 213–224. [DOI] [PubMed] [Google Scholar]
- Gupta, R. S., Pereira, M., Chandrasekera, C. & Johari, V. (2003). Molecular signatures in protein sequences that are characteristic of cyanobacteria and plastid homologues. Int J Syst Evol Microbiol 53, 1833–1842. [DOI] [PubMed] [Google Scholar]
- Hall, T. (2004). BioEdit. Biological sequence alignment editor for Win95/98/NT/2K/XP. Carlsbad, CA: Ibis Biosciences.
- Hambly, E. & Suttle, C. A. (2005). The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol 8, 444–450. [DOI] [PubMed] [Google Scholar]
- Holdeman, L. V. H., Cato, E. P. & Moore, W. E. C. (1977). Anaerobe Laboratory Manual, 4th edn. Blacksburg, VA: Virginia Polytechnic Institute and State University.
- Hugenholtz, P. (2002). Exploring prokaryotic diversity in the genomic era. Genome Biol 3, reviews0003.1–reviews0003.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz, P., Goebel, B. M. & Pace, N. R. (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180, 4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huß, V. A. R., Festl, H. & Schleifer, K. H. (1983). Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4, 184–192. [DOI] [PubMed] [Google Scholar]
- Izard, J., Hsieh, C. E., Limberger, R. J., Mannella, C. A. & Marko, M. (2008). Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Struct Biol 163, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousimies-Somer, H., Summanen, P., Citron, D. M., Baron, E. J., Wexler, H. M. & Finegold, S. M. (2002). Wadsworth Anaerobic Bacteriology Manual, 6th edn. Belmont, CA: Star Publishing.
- Jukes, T. H. & Cantor, C. R. (1969). Evolution of protein molecules. In Mammalian Protein Metabolism, vol. 3, pp. 21–132. Edited by H. N. Munro. New York: Academic Press.
- Jumas-Bilak, E., Carlier, J. P., Jean-Pierre, H., Citron, D., Bernard, K., Damay, A., Gay, B., Teyssier, C., Campos, J. & Marchandin, H. (2007). Jonquetella anthropi gen. nov., sp. nov., the first member of the candidate phylum ‘Synergistetes’ isolated from man. Int J Syst Evol Microbiol 57, 2743–2748. [DOI] [PubMed] [Google Scholar]
- Kapatral, V., Anderson, I., Ivanova, N., Reznik, G., Los, T., Lykidis, A., Bhattacharyya, A., Bartman, A., Gardner, W. & other authors (2002). Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 184, 2005–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T. M. & Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magot, M., Ravot, G., Campaignolle, X., Ollivier, B., Patel, B. K., Fardeau, M. L., Thomas, P., Crolet, J. L. & Garcia, J. L. (1997). Dethiosulfovibrio peptidovorans gen. nov., sp. nov., a new anaerobic, slightly halophilic, thiosulfate-reducing bacterium from corroding offshore oil wells. Int J Syst Bacteriol 47, 818–824. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E. F. & Sambrook, J. (1982). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Menes, R. J. & Muxi, L. (2002). Anaerobaculum mobile sp. nov., a novel anaerobic, moderately thermophilic, peptide-fermenting bacterium that uses crotonate as an electron acceptor, and emended description of the genus Anaerobaculum. Int J Syst Evol Microbiol 52, 157–164. [DOI] [PubMed] [Google Scholar]
- Munson, M. A., Pitt-Ford, T., Chong, B., Weightman, A. J. & Wade, W. G. (2002). Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res 81, 761–766. [DOI] [PubMed] [Google Scholar]
- Munson, M. A., Banerjee, A., Watson, T. F. & Wade, W. G. (2004). Molecular analysis of the microflora associated with dental caries. J Clin Microbiol 42, 3023–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster, B. J., Boches, S. K., Galvin, J. L., Ericson, R. E., Lau, C. N., Levanos, V. A., Sahasrabudhe, A. & Dewhirst, F. E. (2001). Bacterial diversity in human subgingival plaque. J Bacteriol 183, 3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, W. A., Yager, T. D., Korte, J. & von Hippel, P. H. (1993). Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry 32, 137–144. [DOI] [PubMed] [Google Scholar]
- Seshadri, R., Myers, G. S., Tettelin, H., Eisen, J. A., Heidelberg, J. F., Dodson, R. J., Davidsen, T. M., DeBoy, R. T., Fouts, D. E. & other authors (2004). Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A 101, 5646–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira, J. F., Jr & Rocas, I. N. (2005). Uncultivated phylotypes and newly named species associated with primary and persistent endodontic infections. J Clin Microbiol 43, 3314–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkov, A. V., Dubinina, G. A., Lysenko, A. M., Glockner, F. O. & Kuever, J. (2001). Dethiosulfovibrio russensis sp. nov., Dethosulfovibrio marinus sp. nov. and Dethosulfovibrio acidaminovorans sp. nov., novel anaerobic, thiosulfate- and sulfur-reducing bacteria isolated from ‘Thiodendron’ sulfur mats in different saline environments. Int J Syst Evol Microbiol 51, 327–337. [DOI] [PubMed] [Google Scholar]
- Sutcliffe, I. C. (2000). Characterisation of a lipomannan lipoglycan from the mycolic acid containing actinomycete Dietzia maris. Antonie van Leeuwenhoek 78, 195–201. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). mega4: molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Vartoukian, S. R., Palmer, R. M. & Wade, W. G. (2007). The division “Synergistes”. Anaerobe 13, 99–106. [DOI] [PubMed] [Google Scholar]
- Vianna, M. E., Conrads, G., Gomes, B. P. & Horz, H. P. (2007). Quantification and characterization of Synergistes in endodontic infections. Oral Microbiol Immunol 22, 260–265. [DOI] [PubMed] [Google Scholar]
- Wade, W. G., Downes, J., Dymock, D., Hiom, S. J., Weightman, A. J., Dewhirst, F. E., Paster, B. J., Tzellas, N. & Coleman, B. (1999). The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov. and Slackia heliotrinireducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol 49, 595–600. [DOI] [PubMed] [Google Scholar]
- Warnecke, F., Luginbuhl, P., Ivanova, N., Ghassemian, M., Richardson, T. H., Stege, J. T., Cayouette, M., McHardy, A. C., Djordjevic, G. & other authors (2007). Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565. [DOI] [PubMed] [Google Scholar]
- Zavarzina, D. G., Zhilina, T. N., Tourova, T. P., Kuznetsov, B. B., Kostrikina, N. A. & Bonch-Osmolovskaya, E. A. (2000). Thermanaerovibrio velox sp. nov., a new anaerobic, thermophilic, organotrophic bacterium that reduces elemental sulfur, and emended description of the genus Thermanaerovibrio. Int J Syst Evol Microbiol 50, 1287–1295. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl, E. & Pauling, L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, pp. 97–166. Edited by V. Bryson & H.J. Vogel. New York: Academic Press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.