Abstract
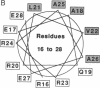
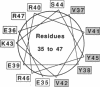
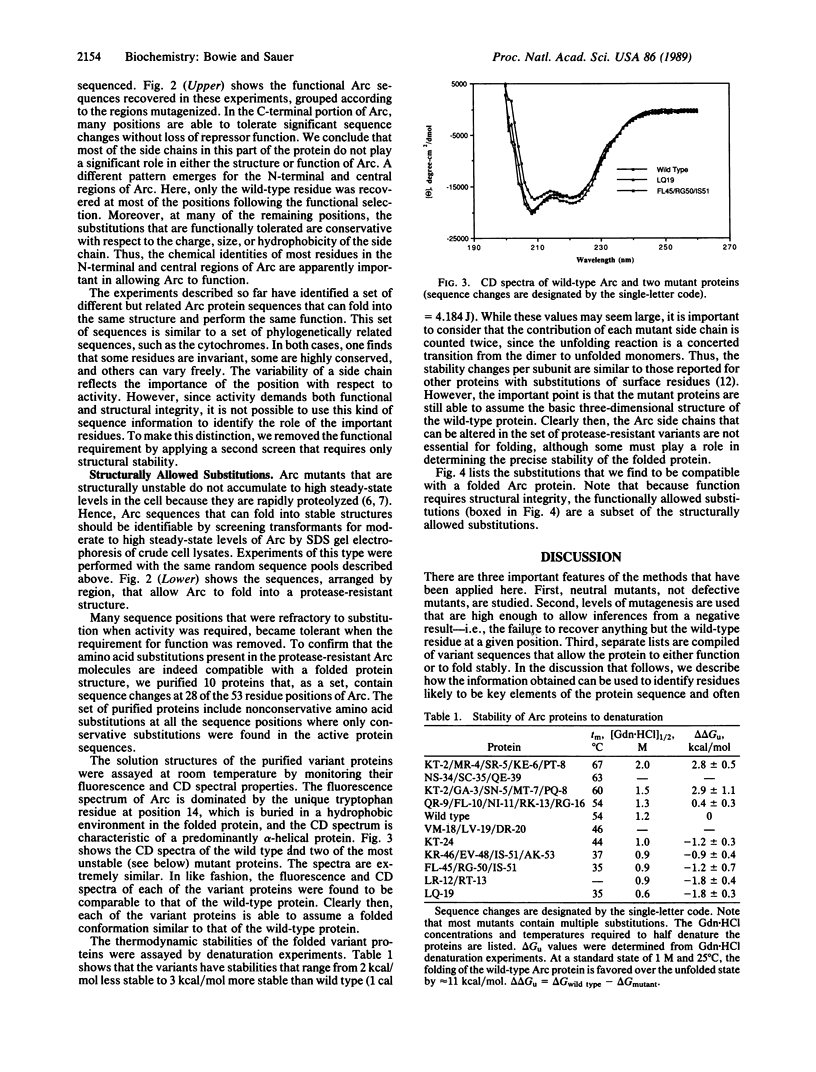
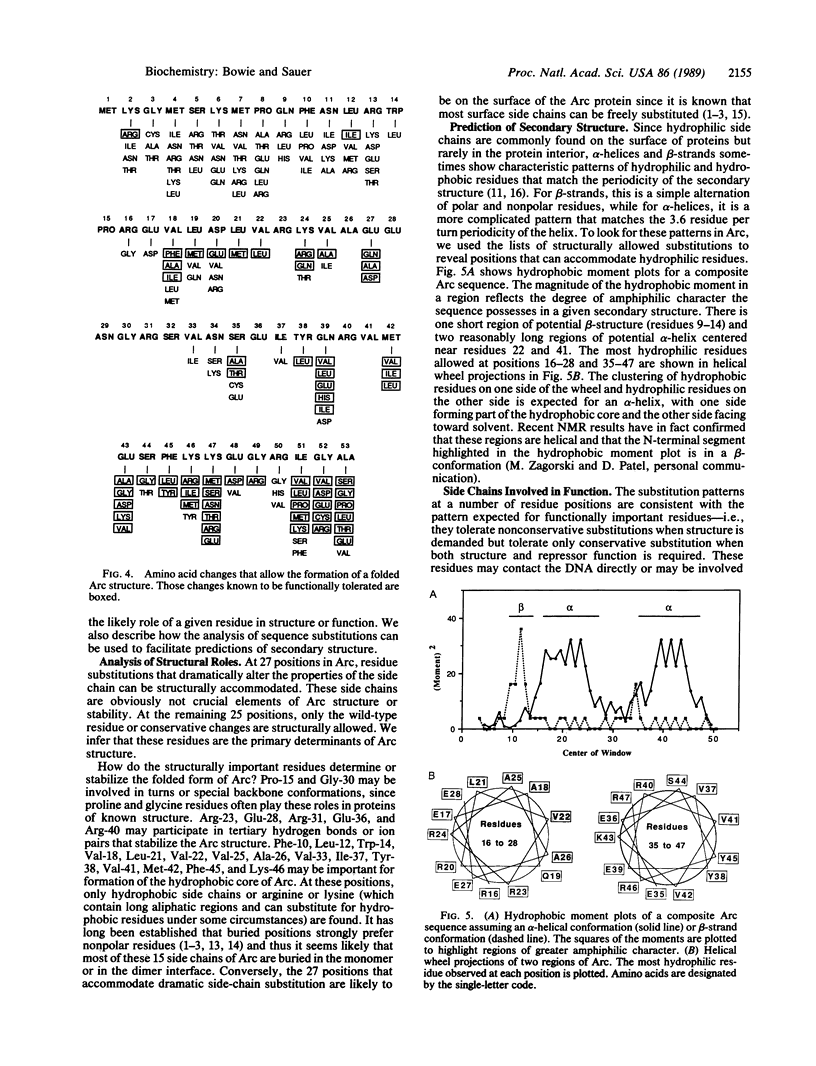
We have generated an extensive genetic map of functionally allowed and/or structurally allowed amino acid substitutions in Arc repressor, a DNA binding protein of unknown structure. Analysis of the allowed substitution patterns identifies residues that are likely to be involved in protein function and identifies side chains that play important structural roles, including residues likely to form the hydrophobic core. The identities of approximately one-third of the residues in Arc repressor are functionally important, about one-half are structurally important, and the remainder are unimportant for either structure or function. The patterns of obligatory hydrophobic positions permit strong predictions of secondary structure.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Sun D. P., Wilson K., Wozniak J. A., Cook S. P., Matthews B. W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987 Nov 5;330(6143):41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- Bashford D., Chothia C., Lesk A. M. Determinants of a protein fold. Unique features of the globin amino acid sequences. J Mol Biol. 1987 Jul 5;196(1):199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Sturtevant J. M., Sauer R. T. Effect of single amino acid replacements on the thermal stability of the NH2-terminal domain of phage lambda repressor. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5685–5689. doi: 10.1073/pnas.81.18.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight K. L., Sauer R. T. DNA binding specificity of the Arc and Mnt repressors is determined by a short region of N-terminal residues. Proc Natl Acad Sci U S A. 1989 Feb;86(3):797–801. doi: 10.1073/pnas.86.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lesk A. M., Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J Mol Biol. 1980 Jan 25;136(3):225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- Miller J. H. Genetic studies of the lac repressor. XI. On aspects of lac repressor structure suggested by genetic experiments. J Mol Biol. 1979 Jun 25;131(2):249–258. doi: 10.1016/0022-2836(79)90075-5. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Nussbaum A. L., Struhl K. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986;44(2-3):177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- Reidhaar-Olson J. F., Sauer R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science. 1988 Jul 1;241(4861):53–57. doi: 10.1126/science.3388019. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susskind M. M. A new gene of bacteriophage P22 which regulates synthesis of antirepressor. J Mol Biol. 1980 Apr 25;138(4):685–713. doi: 10.1016/0022-2836(80)90060-1. [DOI] [PubMed] [Google Scholar]
- Sweet R. M., Eisenberg D. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol. 1983 Dec 25;171(4):479–488. doi: 10.1016/0022-2836(83)90041-4. [DOI] [PubMed] [Google Scholar]
- Vershon A. K., Bowie J. U., Karplus T. M., Sauer R. T. Isolation and analysis of arc repressor mutants: evidence for an unusual mechanism of DNA binding. Proteins. 1986 Dec;1(4):302–311. doi: 10.1002/prot.340010404. [DOI] [PubMed] [Google Scholar]
- Vershon A. K., Youderian P., Susskind M. M., Sauer R. T. The bacteriophage P22 arc and mnt repressors. Overproduction, purification, and properties. J Biol Chem. 1985 Oct 5;260(22):12124–12129. [PubMed] [Google Scholar]