Abstract
Background
Bacterial targeting of tumours is an important anti-cancer strategy. We previously showed that strain SL7838 of Salmonella typhimurium targets and kills cancer cells. Whether NO generation by the bacteria has a role in SL7838 lethality to cancer cells is explored. This bacterium has the mechanism for generating NO, but also for decomposing it.
Methods
Mechanism underlying Salmonella typhimurium tumour therapy was investigated through in vitro and in vivo studies. NO measurements were conducted either by chemical assays (in vitro) or using Biosensors (in vivo). Cancer cells cytotoxic assay were done by using MTS. Bacterial cell survival and tumour burden were determined using molecular imaging techniques.
Results
SL7838 generated nitric oxide (NO) in anaerobic cell suspensions, inside infected cancer cells in vitro and in implanted 4T1 tumours in live mice, the last, as measured using microsensors. Thus, under these conditions, the NO generating pathway is more active than the decomposition pathway. The latter was eliminated, in strain SL7842, by the deletion of hmp- and norV genes, making SL7842 more proficient at generating NO than SL7838. SL7842 killed cancer cells more effectively than SL7838 in vitro, and this was dependent on nitrate availability. This strain was also ca. 100% more effective in treating implanted 4T1 mouse tumours than SL7838.
Conclusions
NO generation capability is important in the killing of cancer cells by Salmonella strains.
Background
Strains of Salmonella typhimurium and other bacteria tend to target tumours and have been used for cancer therapy and as vehicles of gene delivery in enzyme prodrug therapy (GDEPT) [1-3]. In our previous GDEPT work with a number of reductive prodrugs, including our newly discovered 6-chloro-9-nitro-5-oxo-5H-benzo [a]phenoxazine (CNOB), we employed S. typhimurium strain SL7838 (ΔsopE and ΔaroA) to deliver the chrR6 gene to tumours. SL7838 is attenuated with enhanced preference for tumour localization, and the chrR6-encoded enzyme (ChrR6) possesses superior capacity to activate reductive prodrugs [4,5]. The treatments generated a dual killing effect of tumour cells, one due to the SL7838 bacteria themselves, and the other to the activated prodrugs [4,5].
Here we investigated the mechanism of cancer cell killing by SL7838. It is thought that competition for nutrients by bacteria plays a role. However, bacteria may also be able to produce substances that are toxic to cancer cells. One such candidate is the nitric oxide free radical molecule (NO). That this radical has anticancer activity is shown by the fact that high expression levels of mammalian nitric oxide synthase (iNOS), which generates NO, or the external provision of this molecule (e.g. by nitro-aspirin) arrest tumour proliferation [6-9]. Like other S. typhimurium strains, SL7838 possesses nitrate and nitrite reductases and is thus able to convert nitrate and nitrite to NO under oxygen limitation [10]. As nitrate and nitrite are present in mammalian body (30-150 and 0.3-20.0 μM, respectively [11]), and tumours contain hypoxic regions [4,12-15], SL7838 has the potential to generate NO in tumours. However, this bacterium also possesses the Hmp and NorV enzymes that decompose NO in order to detoxify it. Thus, its ability to generate NO would depend on the relative activity of these opposing pathways under given conditions. We examined the capacity of SL7838 to generate NO in a variety of settings and report that NO generation is likely to play a major role in its lethality to cancer cells.
Methods
Bacterial strains, plasmids primers and chemicals
See Table S1. Salmonella typhimurium strain SL7838 was transformed by electroporation [16] with plasmids pCGSL1, encoding the Lux operon, and pFVP25.1, encoding ChrR6. Deletion of the norV, hmP genes (to generate strain SL7842) was carried out as described [17], using the specified primers and plasmids (Additional file 1); deletions were confirmed by three independent PCR assays. The NO donor, 2-(N,N-Diethylamino)-diazenolate 2-oxide (DEA NONOate); nitrate, nitrite hemoglobin and sodium dithionite were purchased from Sigma Inc. MO; Griess reagent (for NO measurement), 4-amino-5-methylamino-2',7' difluorofluorescein diacetate (DAF-FM di-acetate) from Molecular Probe Inc. CA; and SensoLyte™ caspase assay kit were from AnaSpec Inc. CA.
Tumour cell lines and viability assays
JC (murine mammary cancer) cells were obtained from Cancer Research UK; 4T1 (murine mammary cancer) and A-2780 (human ovarian cancer) and 293T (human kidney cancer) from ATCC. MDA-MDB-435 (human breast cancer) was kindly supplied by Dr. Gambhir. All cells were grown as adherent cultures in DMEM medium supplemented with 10% FBS and 1% pen/strep. Cell viability was measured by MTS assay, as described [4].
Cancer cell infection and chemical exposure
Confluent cancer cells were infected with the appropriate bacterial strain [MOI, 10; 4], and after incubation (1 h, 37°C) were washed twice with preheated DMEM and further incubated in DMEM plus gentamycin (1 h, 37°C; 100 μg/ml). Following another wash, the cells were suspended in DMEM-gentamycin (10 μg/ml) and exposed to NO3- (150 μM), NO2- (2 μM), or the NO donor DEA NONOate (final NO concentration, 0.2 μM).
Probing intracellular NO
Washed cell were suspended in PBS buffer (50 μl) supplemented with DAF-FM-diacetate (5 μM) and incubated in the dark for 15 min. 5 μl were spotted on a microscope slide, air dried in the dark, and after the addition of 5 μl Vectashield® fluorescence anti-quenching medium, were covered by a glass cover slip. DAF-FM reacts quantitatively with NO to form a fluorescent benzotriazole derivative. Cells were visualized at 1000× magnification (Olympus B×60 upright fluorescence microscope), imaged (Hamamatsu Orca1 CCD), and pseudocolored, with identical settings for each image (Image Pro Plus 5.0).
Oxygen and nitric oxide quantification in implanted tumours in live mice
O2 and NO profiles of tumours were determined in groups of two mice, one with uninfected, the other infected with SL7838; measurements were made at three individual regions of each tumour (which had an average depth of 14 mm). Clark-type NO and O2 microsensors were used [18,19]. Microsensors were connected to a picoammeter and polarized at +750 and -800 mV for NO and O2 measurements, respectively. Amperometric responses were recorded with DAQCard-AI-16XE-50 (National Instruments, Austin, TX) data acquisition system, using the μ-profiler (Lubos Ploerecky, MPI Bremen, Germany) software at 1s intervals. Each recording represented an average of 2,000 signals at 10 kHz frequency. The microsensors were mounted on a 3-axis motorized micromanipulator (MM 33; Märzhäuser, Wetzlar, Germany) controlled by μ-profiler software (3564-K-024-BC; Faulhaber Group, Schönaich, Germany) for μ-positioning. Tumours were overlaid with low melt agarose (4%, Sigma, Inc.), using a transparent plastic ring. The microsensor tip was placed at the surface of the tumour manually using a stereo microscope (Omano, Inc). The profiles were measured beginning from the inside of the tumour. The NO sensor has a sensitivity range of ~2 - 5 pA μM-1 and a spatial resolution of 60-80 μm; it was calibrated as described [18].
Treatment of implanted tumours
4T1 tumours were implanted in immunocompetent BALB/c mice as before [5] - s.c.; 106 cell inoculum. The resulting tumours (10-14 day post inoculation) were injected with PBS, SL7838, or SL7842 (105CFU). Tumour burden (by caliper) and the animal weight were determined at appropriate intervals. The tumour and the bacterial cells were visualized as before [5] via luminescence due to firefly luciferase and Lux expression, respectively. Imaging of bacterial Lux activity in anesthetized (2% isoflurane) mice was done in an IVIS Spectrum system (Caliper, CA). In addition, imaging was performed 10 min after intraperitoneal injection of luciferin (150 μL of 30 mg/ml solution). The signal generated after luciferin addition (Luc expression) includes the signal due to the bacterial Lux expression, but since the former was more than 50 fold greater, the latter is negligible. All animal studies were performed according to approved Stanford's IACUC and biosafety committee protocols.
Statistical Analyses
Student T tests were performed in all statistical analysis, except for the Kaplan-Meier survival curves, where Logrank tests were used. P-values are indicated; values of less than 0.05 were considered significant.
Results
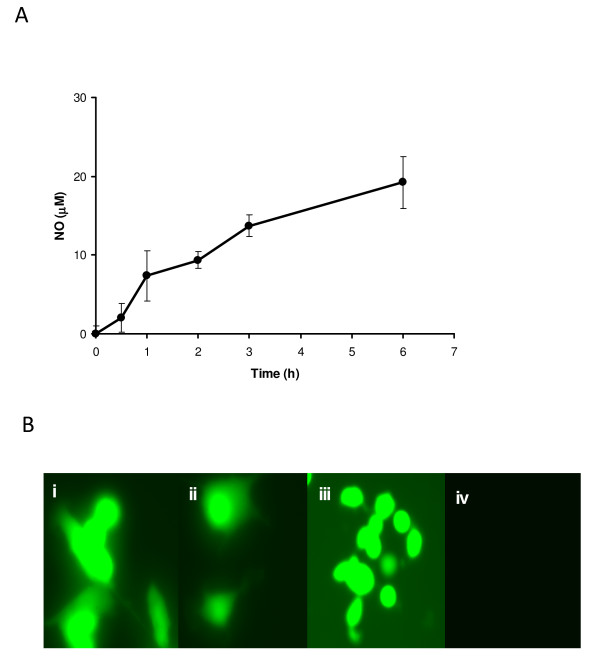
We determined whether SL7838 cell suspensions can generate NO. When supplied nitrate (500 μM) under partial anaerobiosis, the suspensions generated NO for several hours (Figure 1A). We next determined if SL7838 can generate NO after infecting JC murine cancer cells. L-NAME (20 μM) was used to inhibit the indigenous NO generating activity of JC cells (due to iNOS), and NO formation was detected by DAF-FM diacetate; the latter fluoresces in the presence of NO, as we confirmed experimentally (Figure. 1BiA). Addition of nitrate or nitrite (at serum concentrations) to SL7838-infected JC cells generated the fluorescence signal (Figure. 1Bii and 1Biii). Similar results were obtained with infected human MDA-MB-435 breast, or A2780 ovarian cancer cells (not shown).
Figure 1.
A. NO production by SL7838 cell suspensions (A660, 1.5; glucose-M9 medium) incubated without shaking in the presence of nitrate. NO concentration was determined by the oxy-haemoglobin method [24]; B. NO (green fluorescence) production in SL7838-infected JC cells as measured by DAF-FM diacetate. The mammalian NO synthase inhibitor, L-NAME (20 μM), was added to JC cell growth medium 24 h prior to bacterial infection. SL7838-infected JC cells were supplemented with 0.2 μM NO donor (DEA NONOate) positive control (i), 150 μM NO3- (ii), or 2 μM NO2- (iii), or none of these compounds (iv). Uninfected JC cells receiving 150 μM NO3- also did not generate the fluorescence, as in (iv). The experiment was run in triplicate. Error bars represent STDEV.
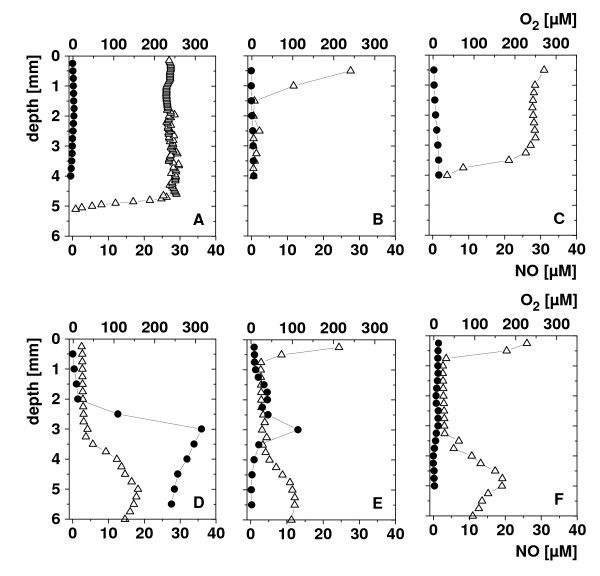
We then explored NO generation directly in implanted s.c. 4T1 tumours in living mice after intratumoural administration of SL7838. The tumours were impaled with a microelectrode for NO measurement [18,19]; an oxygen microelectrode measured oxygen concentration in parallel. The tumours were examined at three separate locations. In the uninfected control tumours, no NO generation was seen, and the oxygenation levels changed with tumour depth as well as location (Figure. 2, panels A-C) revealing, as expected, several regions of anoxia (<~1 μM oxygen). In the infected tumours (Figure. 2, panels D-F), NO generation of up to 300 μM was seen (region D); the amount varied in different regions (e.g., lower levels in region E; none in F). Anaerobiosis tended to be more marked in the infected tumours, but there was no obvious correlation between the tumour oxygenation status and NO production.
Figure 2.
In vivo measurement of NO, and O2 in dorsal 14-day old 4T1 subcutaneous tumours in BALB/c mice (50-100 mm3-). Symbols: Panels A-C: vertical NO (black circle) and oxygen (white triangle) concentration profiles in 3 different spots of an uninfected tumour; Panels D-F: such profiles in a tumour that was infected intratumourally with the bacteria 24 h prior to the measurement. Amperometric microsensors were used; representative results are presented.
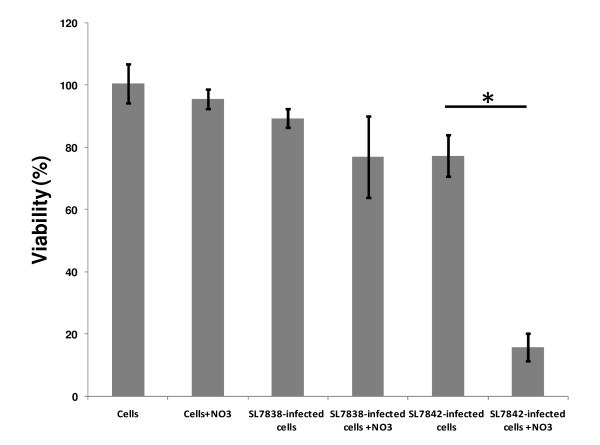
In order to test if NO generation by SL7838 contributes to its lethality to cancer cells, we generated strain SL7842, which was rendered more active in NO generation by deleting the norV and hmp genes that encode the pathways for decomposing this radical; while SL7838 cell suspensions caused complete disappearance of added NO (90 μM) within ca. 200 min (as measured by Greiss assay), those of SL7842 generated no decomposition; the NO donor, DEA NONOate, was used in these experiments. SL7848-infected 4T1 cells exhibited much greater loss of viability than those infected with SL7838 (Figure. 3; compare bars 4 and 6 from the left). The killing of the cancer cells was dependent both on nitrate availability and bacterial infection. Invasion/replication assays showed that both the strains were equally competent in invading and replicating in cancer cells (not shown). Similar results were obtained with JC, A2780, or 293T cancer cells (not shown).
Figure 3.
Loss of viability at 48 h of 4T1 breast cancer cells in vitro. Viability loss at 48 h of 4T1 breast cancer cells in vitro with no additions ("cells") and as a result of NO3- (150 μM) addition with or without infection by strain SL7838, or SL7842, as indicated (n = 3; error bars indicate standard deviation, *p <0.0002).
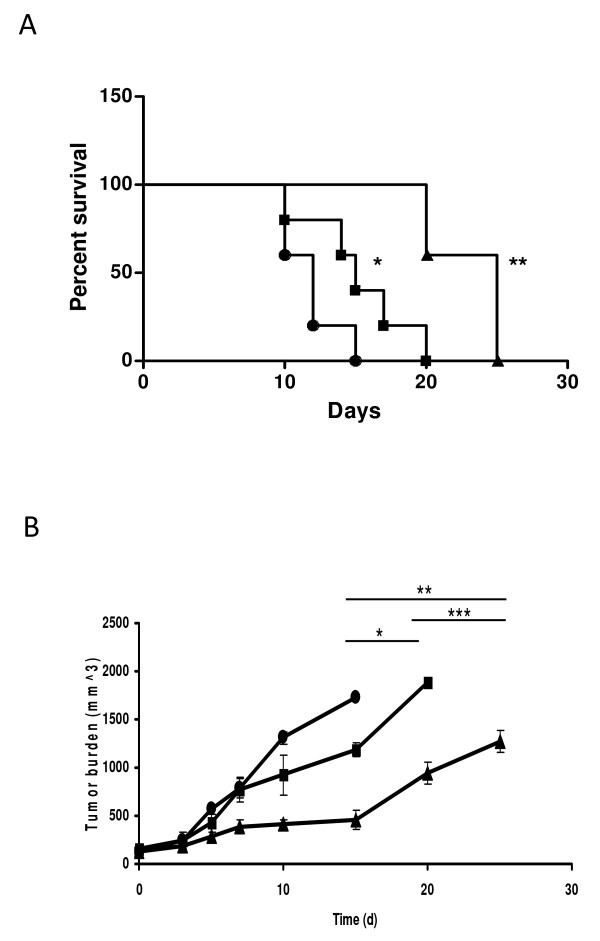
Strain SL7842 proved more effective also in treating implanted 4T1 tumours in mice. Following the development of subcutaneous (s.c.) tumours (10 - 14 days post inoculation of 106 4T1 cells [5]), separate groups of mice were injected intratumourally with saline (control), strain SL7838, or SL7842. While treatment with SL7838 did increase survival (Figure. 4A; Log Rank Test, *p <0.01) as reported before, treatment with SL7842 generated ~100% greater survival (Log Rank Test, **p <0.003). Similar outcome is evident from tumour burden measurements (Figure. 4B). The bacteria remained localized to the tumour (not shown), as revealed by bioluminescence imaging as we previously reported ([5]; see Methods); and the mice exhibited no significant change in weight, suggesting non-toxicity of the treatment.
Figure 4.
Efficacy of bacterial treatment of implanted cancer in mice. A. Survival (Kaplan-Meier) curves of BALB/c mice bearing s.c. 4T1 tumours (50-100 mm3). Symbols: untreated mice administered saline (control; black circle); SL7838-treated mice (black square); SL7842-treated mice (black triangle) (n = 5 animals/group) (*p <0.015, **p < 0.003). Bacteria (105 CFU) were administered intratumourally at the start of the experiment. B. Tumour burden measured at indicated time points using caliper. Symbols as above. T-tests *p < 0.064, **p < 0.022, ***p < 0.035
Discussion
Salmonella bacteria are known to kill infected cancer cells [5,21,22]. These bacteria have the capacity to generate NO, but also possess a pathway to decompose it. Since NO is a known anti carcinogen [6-9], this investigation addressed two questions, whether Salmonella strain SL7838 can generate NO under conditions found inside the cells and within the tumours; and whether this potential capacity has a role in cancer treatment by these bacteria. Cell suspensions of this bacterium under partial anaerobiosis generated NO, indicating that the pathway for the formation of this radical is more active than that involved in its decomposition. A similar result was seen within several cancer cell lines infected with this bacterium. NO was generated in the presence of LNAME, ensuring that the mammalian iNOS was rendered inactive, and no NO was generated by nitrate-supplemented uninfected cancer cells, or the infected cancer cells not exposed to nitrate (Figure. 1Biv). Thus, the intracellular NO seen in these experiments was of bacterial origin and contingent on nitrate availability. This result also shows that Salmonella infecting these cells experience anaerobiosis, since oxygen is their preferred electron acceptor, and they generate NO from nitrate (or nitrite) only under anaerobic conditions.
We further investigated whether NO generation occurred within the implanted 4T1 tumours in mice using micro sensors for both NO and oxygen. The electrodes were impaled in the tumours and measurements were made in several locations. NO generation can occur within the tumours regardless of bacterial infection because of mammalian nitric oxide synthases. Of these three such enzymes, nNOS and eNOS are constitutive but generate very low levels of NO. The enzyme responsible for most NO generation is iNOS, which requires induction by cytokines [20]. However, no NO was detected in the uninfected tumours, indicating lack of activity of these enzymes. Substantial NO was seen within the tumours infected by the bacteria, showing once again that the bacterial infection was the cause of NO generation. Both the distribution of NO as well as of oxygen within the tumours was highly uneven as might be expected form previous findings of tumour heterogeneity and the fact that intratumourally injected SL7838 remain confined to localized tumour regions [5].
The above results affirmed the first query of this investigation, i.e., that Salmonella SL7838 does indeed generate NO under conditions found within cancer cells and tumours. The second question concerning the role of NO in lethality was addressed by generating strain SL7848 with deactivated NO decomposing pathway. This strain was more active in killing cancer cells in vitro. Furthermore, it had a vastly greater capacity to treat implanted 4T1 tumours in mice compared to strain SL7838 which because it retains the NO decomposition pathway has lower capacity to generate NO. We conclude that NO generation plays an important role in anticancer activity of Salmonella bacteria, and that enhancement of this capacity can benefit cancer therapy utilizing these agents by themselves or as vehicles of gene delivery.
Conclusions
Salmonella stains with enhanced capacity for NO generation are likely to prove beneficial in anti-cancer strategies aimed at using these bacteria for gene delivery and cancer treatment. Since many other bacteria can also reduce NO3- and NO2- [23], this approach may prove to be generally beneficial in anti-cancer measures involving bacteria.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YB constructed the mutants, designed and conducted many of the experiments, and wrote along with FS first draft of the manuscript. FS & DdB performed NO and oxygen measurements in implanted tumours. SHT & CHC carried out molecular imaging and statistical analysis. AM, along with YB, conceived the study, participated in its design and coordination and put together the final manuscript version. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Bacterial strains, plasmids, and primers. All bacterial strains and molecular tools used in this study.
Contributor Information
Yoram Barak, Email: Yoram.Barak@codexis.com.
Frank Schreiber, Email: fschreib@mpi-bremen.de.
Steve H Thorne, Email: thornesh@upmc.edu.
Christopher H Contag, Email: ccontag@stanford.edu.
Dirk deBeer, Email: dbeer@mpi-bremen.de.
A Matin, Email: amatin@stanford.edu.
Acknowledgements
We thank Drs. Laura McLaughlin, Michael Benoit and Malika Larabi for contributing ideas and assistance in experimental work. Drs. Trevor Lawley provided plasmids pKD3/4 and Ferric Fang the Δhmp Salmonella typhi strain. This work was supported by grant 1 RO1 CA125074-01A1 from National Cancer Institute (to AM), the National Cancer Institute In vivo Molecular Imaging Centre at Stanford grant (P50 CA114747) and the John A and Cynthia Fry Gunn Research Fund and Stanford Office of Technology Licensing (1105626-100-WOAAA). YB was supported, in part, by a Lady Davis postdoctoral fellowship. FS was supported by Max-Planck-Society.
References
- Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B. Combination bacteriolytic therapy for the treatment of experimental tumours. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15155–15160. doi: 10.1073/pnas.251543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek JM, Low KB, Bermudes D. Tumour-targeted Salmonella as a Novel Anticancer Vector. Cancer Res. 1997;57(20):4537–4544. [PubMed] [Google Scholar]
- Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fischer J. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumour-targeting in vivo. Nat Biotechnol. 1999;17(1):37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- Barak Y, Thorne SH, Ackerley DF, Lynch SV, Contag CH, Matin A. New enzyme for reductive cancer chemotherapy, YieF, and its improvement by directed evolution. Mol Cancer Ther. 2006;5(1):97–103. doi: 10.1158/1535-7163.MCT-05-0365. [DOI] [PubMed] [Google Scholar]
- Thorne SH, Barak Y, Liang W, Bachmann MH, Rao J, Contag CH, Matin A. CNOB/ChrR6, a new prodrug enzyme cancer chemotherapy. Mol Cancer Ther. 2009;8(2):333–341. doi: 10.1158/1535-7163.MCT-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emma L, Williams MBAD. Nitric oxide and metastatic cell behaviour. BioEssays. 2005;27(12):1228–1238. doi: 10.1002/bies.20324. [DOI] [PubMed] [Google Scholar]
- Gao J, Liu X, Rigas B. Nitric oxide-donating aspirin induces apoptosis in human colon cancer cells through induction of oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(47):17207–17212. doi: 10.1073/pnas.0506893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashfi K, Rigas B. Molecular targets of nitric-oxide-donating aspirin in cancer. Biochem Soc Trans. 2005;33(Pt 4):701–704. doi: 10.1042/BST0330701. [DOI] [PubMed] [Google Scholar]
- McLaughlin LM, Demple B. Nitric Oxide-Induced Apoptosis in Lymphoblastoid and Fibroblast Cells Dependent on the Phosphorylation and Activation of p53. Cancer Res. 2005;65(14):6097–6104. doi: 10.1158/0008-5472.CAN-04-4254. [DOI] [PubMed] [Google Scholar]
- Spector MP, Garcia del Portillo F, Bearson SMD, Mahmud A, Magut M, Finlay BB, Dougan G, Foster JW, Pallen MJ. The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology. 1999;145(11):3035–3045. doi: 10.1099/00221287-145-11-3035. [DOI] [PubMed] [Google Scholar]
- Schulz R, Schmidt D, Blum A. Decreased plasma levels of nitric oxide derivatives in obstructive sleep apnoea: response to CPAP therapy. Thorax. 2000;55:1046–1051. doi: 10.1136/thorax.55.12.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. Exploiting the hypoxic cancer cell: mechanisms and therapeutic strategies. Molecular Medicine Today. 2000;6:157–162. doi: 10.1016/S1357-4310(00)01677-4. [DOI] [PubMed] [Google Scholar]
- Brown JM. The Hypoxic Cell: A Target for Selective Cancer Therapy--Eighteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1999;59(23):5863–5870. [PubMed] [Google Scholar]
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4(6):437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6(8):583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- O'Callaghan D, Charbit A. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol Gen Genet. 1990;223(1):156–158. doi: 10.1007/BF00315809. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber F, Polerecky L, de Beer D. Nitric Oxide Microsensor for High Spatial Resolution Measurements in Biofilms and Sediments. Analytical Chemistry. 2008;80(4):1152–1158. doi: 10.1021/ac071563x. [DOI] [PubMed] [Google Scholar]
- Gieseke A, Nielsen JL, Amann R, Nielsen PH, de Beer D. In situ substrate conversion and assimilation by nitrifying bacteria in a model biofilm. Environmental Microbiology. 2005;7(9):1392–1404. doi: 10.1111/j.1462-2920.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- Kitasato A, Tajima Y, Kuroki T, Tsutsumi R, Adachi T, Mishima T, Kanematsu T. Inflammatory cytokines promote inducible nitric oxide synthase-mediated DNA damage in hamster gallbladder epithelial cells. World J Gastroenterol. 2007;13(47):6379–6384. doi: 10.3748/wjg.13.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Thorne SH, Ackerley DF, Lynch SV, Contag CH, Matin A. New enzyme for reductive cancer chemotherapy, YieF, and its improvement by directed evolution. Molecular Cancer Therapeutics. 2006;5(1):97–103. doi: 10.1158/1535-7163.MCT-05-0365. [DOI] [PubMed] [Google Scholar]
- Jain KK. Use of bacteria as anticancer agents. Expert Opinion on Biological Therapy. 2001;1(2):291–300. doi: 10.1517/14712598.1.2.291. [DOI] [PubMed] [Google Scholar]
- Zumft W. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61(4):533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Noack E. Nitric oxide assay using hemoglobin method. Methods Enzymol. 1994;233:240–250. doi: 10.1016/s0076-6879(94)33027-1. full_text. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacterial strains, plasmids, and primers. All bacterial strains and molecular tools used in this study.