Summary
We report a detailed restriction map of the bacteriophage T4 genome and the alignment of this map with the genetic map. The sites cut by the enzymes BglII, XhoI, KpnI, SalI, PstI, EcoRI and HindIII have been localized. Several novel approaches including two-dimensional (double restriction) electrophoretic separations were used.
Introduction
Detailed restriction maps of a number of viral genomes (Roberts 1976) have played an important role in the recent analyses of these systems. However, a detailed physical map has not yet been determined for one of the most intensively investigated bacterial viruses, bacteriophage T4. Two features of the T4 genome have hampered attempts to define the restriction map. First, because the DNA contains glucosylated 5-hydroxymethyl cytosine (HMC) rather than cytosine, it is resistant to restriction-enzyme cleavage. Second, the genome is very large (about 166 kb, Wood and Revel 1976). We have circumvented the first problem by using a multiple mutant of T4 that incorporates cytosine rather than HMC into its DNA (Snyder et al. 1975; Wilson et al. 1977). To cope with the large genome size we have used a new two-dimensional method for separation of the numerous restriction fragments (O’Farrell, in preparation), and have applied some novel approaches to the detection and ordering of these restriction fragments.
The positions of 231 restriction enzyme cutting sites, including all the sites cut by the restriction enzymes BglII, XhoI, KpnI, SalI, and PstI, and most of the sites cut by EcoRI and HindIII are reported here. The restriction map has been aligned with the genetic map by localizing, on the restriction map, the positions of a number of cloned fragments of T4 DNA. The positions of these cloned fragments on the genetic map had previously been determined by marker rescue procedures (Mattson et al. 1977). An additional reference point is a unique BamHI restriction site (Takahashi et al. 1979), whose location on the T4 genetic map has been determined (Wilson et al. 1980).
Two independent published efforts have identified positions of SalI, SmaI and KpnI restriction sites and some of the BglII sites but did not include alignment to the genetic map (Kiko et al. 1979; Rüger et al. 1979; Carlson and Nicolaisen 1979). Additionally, extensive analysis of DNA clones localized a number of EcoRI sites (Wilson et al. 1977). As will be discussed below, our results substantially agree with these reports, as well as with other recent data.
Materials and Methods
a) Bacteria and Phage
A T4 phage carrying four mutations, [g56amE51, g42amC87, ΔdenB NB5060, alcW7] was propagated on a permissive bacterial host QD Su3. Growth of this bacteriophage, designated alcW7, on a non-suppressing host ED8689 (sup°, trpR−,hsdR− derivative of W3350) gave a high titer of phage containing cytosine. Unless specified otherwise, T4 DNA refers to the cytosine-containing DNA isolated from this phage. E. coli 5K(R1) was used to check the approximate extent of substitution of C for HMC (Wilson et al. 1977). The C containing phage had a plating efficiency of 10−5 on this strain and appears to have no residual HMC. Bacterial strains and T4 alcW7 where obtained from G.G. Wilson (Wilson et al. 1977).
b) Enzymes and Chemicals
Endonuclease EcoRI was generously provided by Pat Bedinger and Janet Natzle. All other restriction endonucleases were obtained from New England Biolabs. The T4 DNA polymerase was a kind gift from Ursula Hibner. The polymerase was prepared according to Morris et al. (1979), had a specific activity of about 18,000 units per mg and was free of measurable nuclease contamination. Most notably no 5′ exonuclease was detected in this polymerase preparation. Agarose was Seakem ME grade, α 32P dATP was purchased from New England Nuclear.
c) Restriction Enzyme Cleavage and DNA Labeling Reaction
All the restriction enzymes we have examined work well in a single buffer (designated TA buffer) having the following composition: 33 mM Tris acetate pH 7.9, 66 mM potassium acetate, 10 mM magnesium acetate, 0.5 mM dithiothreitol and 100 µg/ml bovine serum albumin (nuclease free). No EcoRI* activity or other “star” activities were observed even after extensive over-digestions of ØX174 DNA in TA buffer. However, one site in T4 DNA was cut very slowly by BglII although it may represent a “star site” (a sequence related to that preferred by the restriction enzyme), there is no evidence that the specificity of the enzyme is decreased in the buffer used here. Direct tests showed that EcoRI was more stable and more active in TA buffer than in one of the frequently used EcoRI buffers (100 mM Tris C1 pH 7.2, 100 mM NaC1 5 mM MgCl2). Other enzymes gave levels of activity roughly equal to the nominal activities quoted by the supplier.
Use of T4 DNA polymerase, which is also active in TA buffer, provides an extremely convenient method for labeling restriction fragments. In the absence of deoxyribonucleoside triphosphate, the 3′ exonuclease activity of the polymerase removes nucleotides from 3′ ends of the DNA fragments and these are replaced with labeled nucleotides in a polymerization reaction initated by addition of radioactive substrates (Englund 1972). In a typical reaction 100 ng of T4 DNA in 10 µl was cut with a restriction enzyme; 100 ng of T4 DNA polymerase (in 1 µl of 0.2 M KPO4, pH 7.5, 50% glycerol, 1% mercaptoethanol) was then added and the reaction incubated for about 2 min at 37 °. Finally, α32P dATP was added (10 µM) along with the other three unlabeled deoxyribonucleotide triphosphates (100 µM) and incubation was continued for about 4 min to replace about 100 terminal nucleotides. The labeling reaction was terminated by heating to 70 ° for 5 min. The detailed characteristics of this labeling reaction will be reported separately (O’Farrell, in preparation). It is important to note that the number of nucleotides replaced by labeled nucleoitdes can be regulated. Here, about 100 nucleotides at the termini of each restriction fragment have been replaced and, unless a subsequent restriction enzyme cut is made within this short labeled region, the label behaves as an end-specific label.
Restriction fragments labeled by the above procedure can be redigested with a second restriction enzyme immediately after the heat step. In this case, only the termini generated by the first digestion will be labeled. Such a digest with, for example, XhoI and PstI will be designated XhoI (32P)/ PstI, indicating that the termini generated by XhoI cleavage are labeled while the termini generated by PstI are not labeled.
To obtain partial digests, T4 DNA was treated with different levels of restriction enzyme and, after each digest was end labeled, it was analyzed on one dimensional gels. Selected levels of digestion were then analyzed by a two-dimensional scheme (described in Results and Discussion). Generally, on the basis of the one-dimensional separations, several digests were mixed to produce a fairly equal distribution among higher order partials and the smaller partials. Even though some enzymes, such as PstI, produced digests in which the starting material and/or the limit restriction fragments were vastly more abundant than the intermediates, the high sensitivity of autoradiography allowed adequate detection of the partials.
d) Electrophoresis
Restriction fragments were separated electrophoretically on flat bed gels of a suitable agarose concentration in TAE buffer (40 mM Tris acetate pH 8.1, 20 mM sodium acetate, 2 mM EDTA), at a voltage gradient of 2 V/cm (Sharp et al. 1973; Helling et al. 1974). Gels were stained with ethidium bromide and the fluorescence observed under ultraviolet illumination. For autoradiography, gels were dried thoroughly under vacuum and exposed directly to X-Omat (Kodak) X-ray film, with or without an intensifying screen (Kodak Lighting Plus).
Two-dimensional restriction analysis was performed using a new method which will be described in detail in a separate publication (O’Farrell, in preparation) but is outlined here. As described by Potter et al. (1977), restriction fragments separated according to size by agarose gel electrophoresis can be recovered, redigested with a second restriction enzyme and each fraction again separated according to size by agarose gel electrophoresis to generate a two-dimensional separation. To simplify this procedure, some investigators, after separating restriction fragments in the first dimension, have digested the DNA fragments with a second restriction enzyme within the agarose gel (Rosenvold and Honigman 1977; Kovacic and Wang 1979). The new two-dimensional method used here incorporates an electrophoresis step which brings together the second restriction enzyme and the separated fragments in a narrow zone stacked at the edge of the gel and thereby greatly improves both the in situ digestion and the subsequent separation. This method produces sharp spots, and allows the use of numerous restriction enzyme combinations which were previously impractical. When combined with end labeling and analysis of partial digests (Smith and Birnstiel 1976; Kovacic and Wang 1979), it greatly extends our capacity to determine restriction maps of complex genomes. Its application is illustrated more thoroughly under “Results” below.
e) Hybridization of Cloned T4 DNA Fragments to Separated Restriction Fragments of T4 DNA
Chemically activated paper (diazophenylthioether paper) was prepared according to a method described by Brian Seed (personal communication). T4 DNA was either cut with a single enzyme (BglII, XhoI, EcoRI, HindIII or PstI), with various pairwise combinations of these enzymes, or with a combination of all five enzymes. Each digest was separated on both a 0.5% agarose gel and a 1.5% agarose gel and the separated DNA fragments transferred to the activated paper according to the methods of Alwin et al. (1977), as subsequently modified (Wahl et al. 1979). As described by the latter authors, the same filters were hybridized in sequence with a series of labeled probes. Total T4 DNA and 6 fragments of T4 DNA cloned in pBR322 were labeled and used as probes.
The DNA fragments were labeled using T4 DNA polymerase, rather than the more common nick-translation method described by Rigby et al. (1977). For these reactions, DNAs to be labeled were first restricted, then, using the T4 polymerase reaction, about 800 bases at the termini were replaced with α32P deoxynucleotide triphosphates.
Results and Discussion
Strain Consideration
The substitution of cytosine by dHMC in T4 DNA is achieved by two phage encoded enzymes: The product of gene 42, a hydroxymethylase (HMase), generates dHMC from dCMP, while the product of gene 56, dCTPase, reduces the level of the more usual substrate for DNA synthesis. Bacteriophage T4 encoded nucleases endoII and endoIV are involved in host DNA breakdown, but will degrade T4 DNA containing cytosine (Kutter and Wiberg 1968). A variety of multiply mutant T4 strains capable of incorporating cytosine into viable phage have been constructed (Snyder et al. 1976; Morton et al. 1978; Wilson et al. 1977) and are referred to as C-T4 strains. The combination of mutations most effective in eliminating HMC are deficient in dCTPase, endoIV and HMase (Morton etal. 1978); an additional mutation, alc, is required to overcome a block affecting growth of cytosine containing T4 (Kutter et al. 1975; Snyder et al. 1976).
Because of the impact on restriction enzyme analysis, the choice of alternative alleles used for C-T4 strain construction must be considered. In particular, mutations in the denB locus, which encodes endoIV, confer no distinctive phenotype. Usually, to simplify identification, deletions which remove not only denB but a nearby selectable marker are used. For the map construction reported here, we chose a strain described by Wilson et al. (1977) [am E51 (dCTPase), am C87 (HMase), deletion denB rlI NB5060, alc W7]. This C-T4 strain grows unusually well, and its DNA can be cleaved to completion with restriction enzymes. As shown in Fig. 1, the deletion in this strain, rIIB NB5060, begins approximately 0.3 kb in from the N terminus of the rIIB gene and extends through the entire D region into ac--a total of approximately 3.2kb (Bautz and Bautz 1967; Depew etal. 1975; D. Pribnow, personal communication). Other endoIV deletions commonly used in C-T4 strains are also illustrated in Fig. 1.
Fig. 1.
A map of the regions of the T4 genome deleted in some of the den B mutations used for the construction of C-T4 strains. The mutation labeled Bruner’s “rIIH23” was used by Kiko etal. (1979), Rüger et al., (1979) and Marsh (1980). As discussed by Kutter et al. (1975), this deletion was confused with the original rIIH23. The C-T4 strain used here contains the rIINB5060 deletion. Carlson and Nicolaisen (1979) have used a strain carrying saΔ9 for restriction mapping
The Map
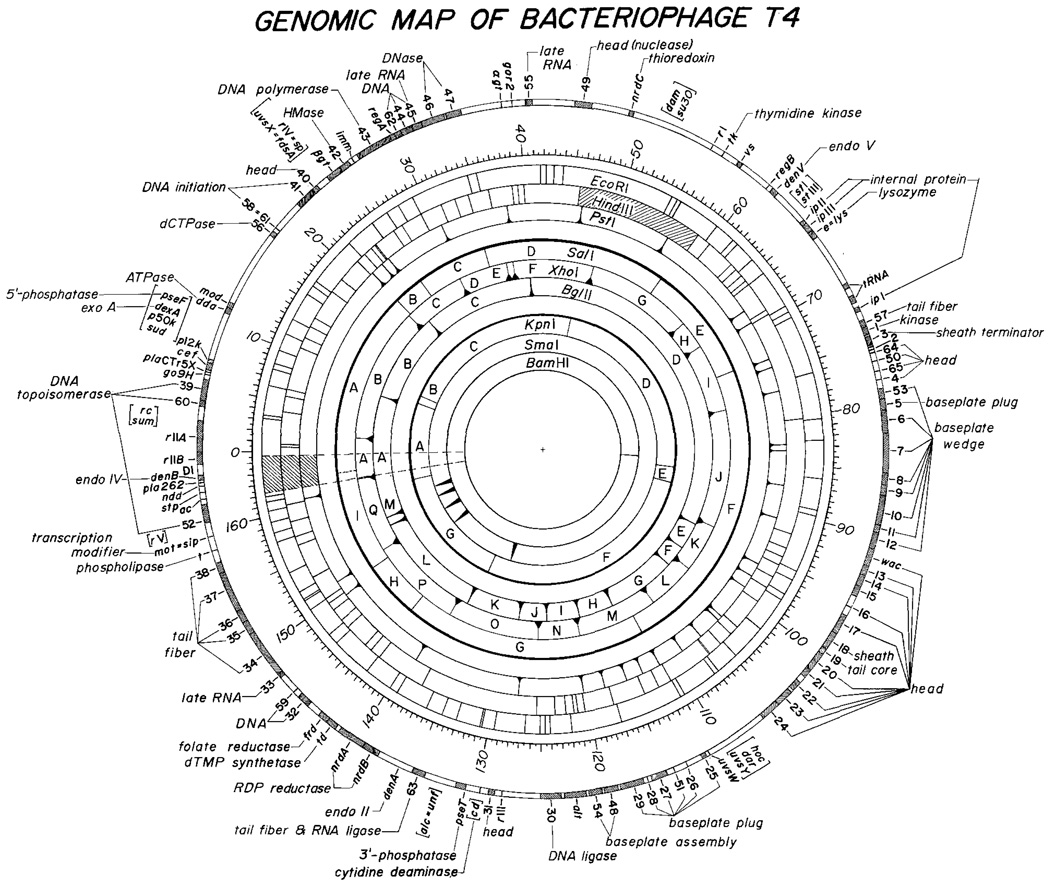
Figure 2 and Tables 1–5 present a detailed physical map of the T4 genome. To facilitate comparison we have set the origin of the physical map at the origin of the genetic map. Methods used in map construction include: (1) one- and two-dimensional agarose gel analysis of multiple restriction-enzyme digests; (2) analysis of partial digestion products using a new two-dimensional technique; and (3) hybridization of identified cloned fragments to blots of one-dimensional gel analyses of various multiple enzyme digests. The following description is designed to illustrate the capacity, limitations and accuracy of the methods; more precise procedural details will be presented in a subsequent paper (O’Farrell, in preperation).
Fig. 2.
The restriction map of bacteriophage T4 and its alignment with the genetic map. The positions of BglII, XhoI, PstI, HindIII and EcoRI sites are taken from our own data. The order of most of the SalI, and KpnI fragments is as determined by Carlson (1979). Our data confirmed her order, identified an additional fragment, and defined the alignment of the SalI and KpnI sites with other restriction sites and with the genetic map. The SmaI positions were defined by aligning of the map derived by Kiko et al. (1979) with our map. The genetic map was modified from the map of Wood and Revel (1976) primarily according to data summarized by Mosig (1980). The positions of the genetic markers have been further adjusted to correspond to the physical map according to data from cloned DNA fragments as summarized in Table 5
Table 1.
Positions of restriction enzyme sites in bacteriophage T4 for KpnI, SalI, BglII, XhoI and PstI measured as distance in kb from the rIIA/rlIB junction
| Map position |
Band number |
Fragment size |
Distal terminus |
|
|---|---|---|---|---|
| KpnI | A | 5 | 12.4 | 9.6 |
| B | 7 | 1.4 | 11.0 | |
| C | 3 | 36.6 | 47.6 | |
| D | 2 | 39.25 | 86.85 | |
| E | 6 | 3.05 | 89.9 | |
| F | 1 | 46.1 | 136.0 | |
| G | 4 | 24.25 | 160.25 | |
| SalI | A | 2 | 33.7 | 21.2 |
| B | 8 | 3.9 | 25.1 | |
| C | 6 | 9.2 | 34.3 | |
| D | 4 | 23.7 | 58.0 | |
| E | 5 | 15.15 | 73.15 | |
| F | 1 | 38.4 | 111.5 | |
| G | 3 | 31.55 | 143.1 | |
| H | 7 | 7.15 | 150.3 | |
| BglII | A | 3 | 15.9 | 8.55 |
| B | 4 | 13.95 | 22.5 | |
| C | 2a | 17.4 | 39.9 | |
| D | 1 | 55.35 | 95.25 | |
| E | 8b | 4.2 | 99.45 | |
| F | 9 | 3.25 | 102.7 | |
| G | 5 | 10.75 | 113.45 | |
| H | 7a | 5.45 | 118.9 | |
| I | 7b | 5.3 | 124.2 | |
| J | 8a | 4.35 | 128.55 | |
| K | 6 | 8.25 | 136.8 | |
| L | 2b | 17.4 | 154.2 | |
| M | 10 | 1.2 | 155.4 | |
| XhoI | A | 12 | 2.2 | 1.7 |
| B | 1 | 19.1 | 20.8 | |
| C | 8 | 8.9 | 29.7 | |
| D | 9a | 7.0 | 36.7 | |
| E | 13 | 0.9 | 37.6 | |
| F | 5a | 11.6 | 49.2 | |
| G | 3 | 15.05 | 64.2 | |
| H | 11 | 4.0 | 68.2 | |
| I | 7 | 9.6 | 77.8 | |
| J | 2 | 16.6 | 94.4 | |
| K | 9b | 6.75 | 101.15 | |
| L | 9c | 6.75 | 107.9 | |
| M | 5b | 11.6 | 119.5 | |
| N | 10 | 5.9 | 125.4 | |
| O | 4b | 13.0 | 138.4 | |
| P | 4a | 13.2 | 151.6 | |
| Q | 6 | 10.8 | 162.35 | |
| PstIa | A | 1 | 23.25 | 21.30 |
| B | 1.45 | 22.75 | ||
| C | 0.95 | 23.70 | ||
| D | 8 | 6.8 | 30.5 | |
| E | 7a | 7.45 | 37.95 | |
| F | 6 | 8.2 | 46.15 | |
| G | 4 | 9.95 | 56.1 | |
| H | 2b | 11.5 | 67.6 | |
| I | 1.5 | 69.1 | ||
| J | 11 | 4.3 | 73.4 | |
| K | 0.9 | 74.3 | ||
| PstIa | L | 15 b | 2.7 | 77.0 |
| M | 9 | 5.8 | 82.8 | |
| N | 0.6 | 83.4 | ||
| O | 13 | 3.8 | 87.2 | |
| P | 14 | 3.55 | 90.75 | |
| Q | 10 | 5.3 | 96.05 | |
| R | 1.5 | 97.55 | ||
| S | 1.35 | 98.9 | ||
| T | 12 | 4.0 | 102.9 | |
| U | 0.6 | 103.5 | ||
| V | 15d | 2.55 | 106.05 | |
| W | 15c | 2.6 | 108.65 | |
| X | 15a | 2.8 | 111.45 | |
| Y | 16 | 2.1 | 113.55 | |
| Z | 1.9 | 115.45 | ||
| AA | 15 e | 2.45 | 117.9 | |
| BB | 15f | 2.35 | 120.25 | |
| CC | 7b | 7.15 | 127.4 | |
| DD | 2a | 11.8 | 139.2 | |
| EE | 3 | 10.5 | 149.7 | |
| FF | 1.55 | 151.25 | ||
| GG | 5 | 9.65 | 160.9 |
The PstI sites were mapped by a new method involving partial digestions. The sizes listed were derived from this analysis rather than a measurement of fragments generated by a PstI digest. Because of limited accuracy and resolution the smaller bands seen in a PstI digest could not be unambiguously assigned to predicted fragments
Table 5.
Correspondence between physical and genetic map
| Genetic locus | Physical map a | Genetic map b |
|---|---|---|
| rII A/B junction | 0 | 0 |
| 39 | 4.0–5.8 | 3.4–5.2 |
| 42 | 25.2–26.0 | 23.0–23.8 |
| 43 | 26.7–29.9 | 25.0–28.2 |
| 45 | 31.7–32.4 | 32.7–33.3 |
| 47 | 34.4–35.5 | 37.2–38.1 |
| e(A.A. 71/72) | 65.6 | 66.2 |
| tRNA cluster | 70.1–71.7 | 69.9–72.3 |
| 1 | 73.9–74.35 | 73.3–74.2 |
| 50 | 76.7 | 77.7 |
| 7/8 | 85.l | 87.0 |
| 15 | 94.0–94.8 | 96.5–97.3 |
| 21 | 101.3–102.0 | 103.0–103.7 |
| 24 | 105.0–106.4 | 106.1–107.5 |
| 29 | 116.4–118.7 | 116.0–118.2 |
| 30 | 123–125 | 121.8–123.3 |
| 63 | 133.9–135.1 | 132.4–133.7 |
| td | 141.8–142.7 | 140.2–141.0 |
| 32 | 144.4–145.5. | 144.2–145.7 |
| 33 | 146.7–147.0 | 146.8–147.1 |
| 34/35 junction | 151.6 | 151.6 |
| 37/38 junction | 156.7 | 156.7 |
| rII A/B junction | 166.0 | 166.0 |
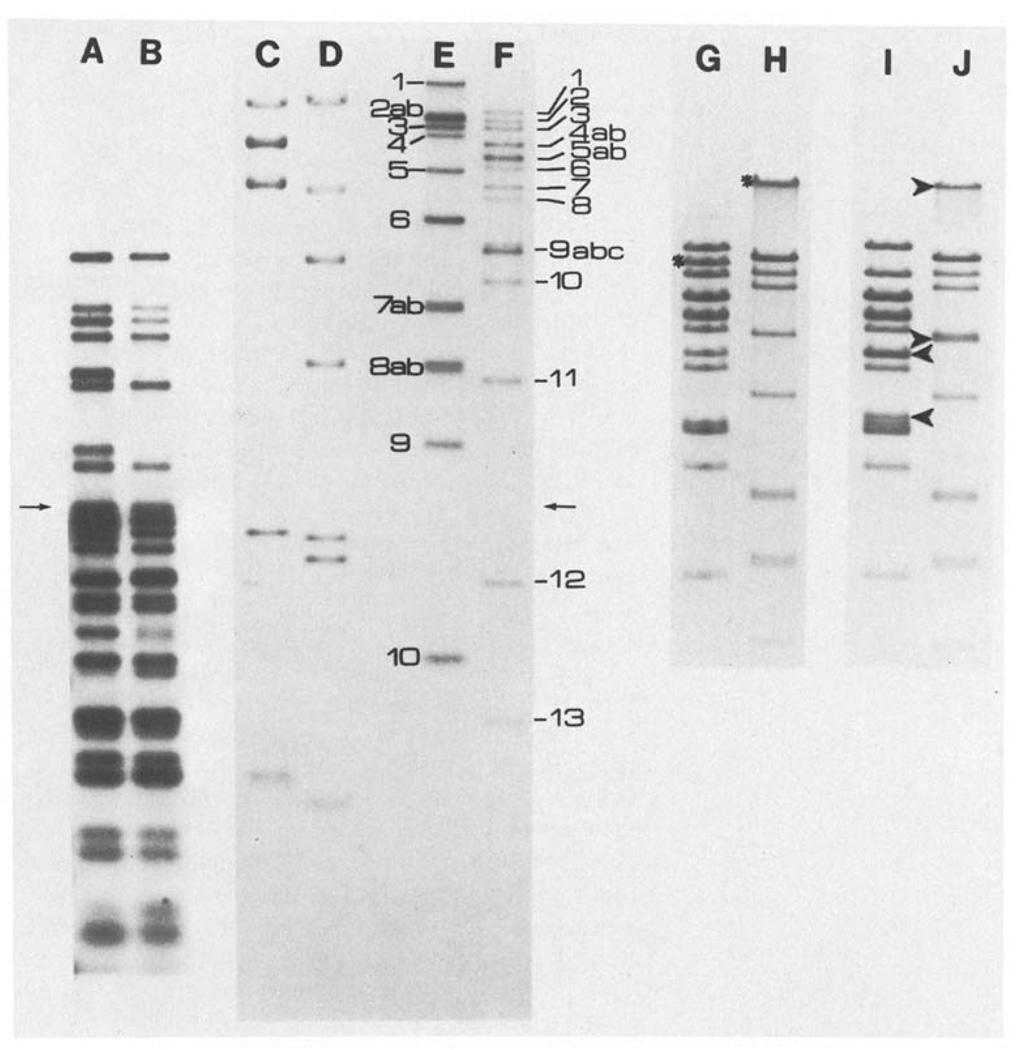
The number and size of fragments produced by various restriction enzyme digestions of T4 DNA were first determined by one-dimensional agarose electrophoresis using digests of the bacteriophage DNA’s of lambda and ØX174 as molecular-weight standards. Generally, the fragments were end-labeled to improve detection of small fragments and simplify analysis of multiple bands. This particularly aided in analysis of multiple digests when only the ends created by cleavage with one of the enzymes were labeled (see Materials and Methods). Figure 3 shows samples of these separations. As indicated on the figure, bands are numbered consecutively from high to low molecular weight. When two or more restriction fragments are found to comigrate they are distinguished by lower-case-letter subscripts. For those restriction enzymes making relatively few cuts, the fragments have also been given a second name – a letter designation. Here, fragments are ordered alphabetically according to their map position. For example, XhoI 8-C is the eighth band on a gel and the third on the map. Fragments can still be uniquely identified by either of the abbreviated names, (ie. XhoI 8 or XhoI C). This naming system greatly facilitates reference to both gel data and the map. Our band-numbering system coincides with that used by Rüger et al. (1977), but not with that used by Carlson et al. (1979) or Takahashi et al. (1978).
Fig. 3.
A–J. Representative one dimensional separations. The samples shown in lanes A–F were electrophoresed through a composite gel, the first half being 0.5% and the second half being 1.5% agarose (the arrows indicate the junction of the two gel concentrations). Lanes A–F are from an autoradiogram of this gel. Lane A shows fragments which were produced by XhoI cleavage of T4 DNA, labeled and then further cut with EcoRI (XhoI(32P)/EcoRI). Lane B shows the same sample as A but further digested with BglII (XhoI(32P)/EcoRI, BglII). The sizes determined for the fragments in lane A give the distance between XhoI sites and the adjacent EcoRI sites. These sizes were also used for internal calibration of two dimensiional separations where XhoI cut fragments are separated in the first dimension, recut with EcoRI and separated in the second dimension (XhoI-EcoRI). For molecular weight markers, λ CI857 S7 DNA obtained from Rae Lynn Burke was cut with BglII (lane C) or HindIII (lane D) and labeled. Lanes E and F show respectively, T4 DNA digested with BglII or XhoI and labeled. The band assigments are indicated. Lanes G–J show stained patterns obtained from T4 DNA digested with XhoI (Lane G), BglII (lane H), XhoI and BamHI (lane I) or BglII and BamHI (lane J) and electrophoresed on a 0.5% agarose gel. The stars indicate the bands formed by fragments containing the BamHI site; in the double digests (lines I and J) the arrowheads indicate the positions of the bands generated by BamHI cleavage. The cleavage of BglII 1 is not obvious because fragments in this molecular weight range do not separate well
The one additional complication in terms of nomenclature results from the fact that most C-T4 strains carry a deletion of some sort near the origin (Fig. 1). This generally will not affect our letter designations. However, these deletions affect the sizes of fragments, and thus their number designation. Fragments have been numbered here according to our own analyses. The expected sizes of the fragments in the absence of any deletions are also indicated, using the data of Takahashi et al. (1978 and personal communication) to determine the approximate location of additional restriction enzyme sites.
For enzymes making many cuts, such as EcoRI and HindIII, we have simply designated the fragments according to the map position (in kb) of their distal end and their approximate size. It should be noted that the method of determining the location of these sites, discussed below, does not involve the identification of specific fragments with specific bands in the total EcoRI or HindIII digests. Where such identification is known from cloning data (cf. Wilson et al. 1977), the band number is also indicated in Table 3, using the assigments of Wilson et al. (1977).
Table 3.
Positions of HindIII sites and correlation with data from analysis of cloned T4 DNA fragments
| Distal terminus |
Size 1 (kb) |
Known gene content |
Cloned fragment size 2 |
Ref. a | Distal terminus (kb) |
Size (kb) |
Known gene content |
Cloned fragment size 2 |
Ref. a |
|---|---|---|---|---|---|---|---|---|---|
| 0.4 | rIIA | 0.8 | [3] | 80.3 | 11.05 | ||||
| 0.75 | 0.35 | rIIA | 0.4 | [3] | 91.8 | 11.5 | |||
| 3.95 | 3.2 | 60 | [3] | 93.3 | 1.5 | ||||
| 5.6 | 1.65 | 39 | 1.7 | [11] | 94.3 | 1.0 | |||
| 6.3 | 0.7 | 95.2 | 0.9 | ||||||
| 10.65 | 4.35 | 96.5 | 1.3 | ||||||
| 11.8 | 1.15 | 97.8 | 1.3 | ||||||
| 12.95 | 1.15 | 99.5 | 1.7 | ||||||
| 14.00 | 1.05 | 102.65 | 3.15 | 18–22 | [3, 7] | ||||
| 17.1 | 3.1 | 103.7 | 1.05 | 22–23 | 1.1 | [3, 7] | |||
| 18.7 | 1.6 | 105.1 | 1.4 | 23 | [3, 7] | ||||
| 21.1 | 2.4 | 106.4 | 1.3 | ||||||
| 22.1 | 1.0 | 106.5 | 0.1 | ||||||
| 23.0 | 0.9 | 111.25 | 4.75 | ||||||
| 23.4 | 0.4 | 112.2 | 0.95 | ||||||
| 23.5 | 0.1 | 112.55 | 0.35 | ||||||
| 25.8 | 2.3 | 114.2 | 1.65 | 26 | |||||
| 26.8 | 1.0 | 117.5 | 3.3 | 26–29 | |||||
| 29.05 | 2.25 | 43 | 2.1 | [4, 5] | 120.4 | 2.9 | |||
| 31.25 | 2.2 | 43–44 | 2.0 | [4, 5] | 121.15 | 0.75 | |||
| 32.8 | 1.55 | 44–45 | 1.5 | [4, 5] | 121.6 | 0.45 | |||
| 34.2 | 1.4 | 46 | 1.3 | [5] | 122.4 | 0.8 | |||
| 36.3 | 1.9 | 123.5 | 1.1 | ||||||
| 38.8 | 2.5 | 125.4 | 1.9 | 30 | 1.9 | [11] | |||
| 39.35 | 0.55 | 137.3 | 11.9 | [8] | |||||
| 39.9 | 0.55 | 142.5 | 5.2 | nrdB-td | 5.3 | [8, 9] | |||
| 43.8 | 3.9 | 143.6 | 1.1 | frd | 1.05 | [8, 9] | |||
| 45.65 | 1.85 | 144.3 | 0.7 | 0.8 | [8, 9] | ||||
| [unmapped] | 146.85 | 2.55 | 32–33 | 2.65 | [10] | ||||
| 58.6 | 147.8 | 0.95 | 33 | [10] | |||||
| 60.1 | 1.5 | 149.2 | 1.4 | 1.5 | [10] | ||||
| 62.95 | 2.85 | 149.9 | 0.7 | 34 | 0.6 | [10] | |||
| 63.7 | 0.75 | 150.4 | 0.5 | 34 | 0.5 | [10] | |||
| 64.5 | 0.8 | 152.95 | 2.55 | 34–36 | 2.4 | [3, 10] | |||
| 65.15 | 0.65 | 156.5 | 3.55 | 36–37 | 3.5 | [3, 10] | |||
| 65.65 | 0.5 | 0.4 | [6] | 157.4 | 0.9 | 38 | 0.8 | ||
| 68.75 | 3.1 | e | 3.1 | [6] | 158.95 | 1.55 | |||
| 69.25 | 0.5 | 160.75 | 1.8 |
The sizes listed are those assigned to the map
The measured sizes of some cloned fragments of T4 DNA are listed for comparison. In addition, where it is known we have indicated the assignment of particular restriction fragments to an individual band observed after electrophoretic separation of EcoRI digested whole C-T4 DNA (cf. Wilson et al. 1977)
[3] Young and Mattson, pers. commun.
[4] Karam, pers. commun.
[5] Tanyashin, pers. commun.
[6] Owen, manuscript in preparation.
[7] Albright and Geiduschek, pers. commun.
[8] Mileham, Revel and Murray, man. in prep.
[10] Revel, manuscript in preparation
[12] Mattson et al. 1977
[13] This paper: these cloned fragments were analyzed by hybridization experiments (see text)
[14] Wilson, Tanyashin and Murray (1977)
[15] Fukada (1979)
The Use of Two-Dimensional Gel Analysis for Mapping DNA Fragments
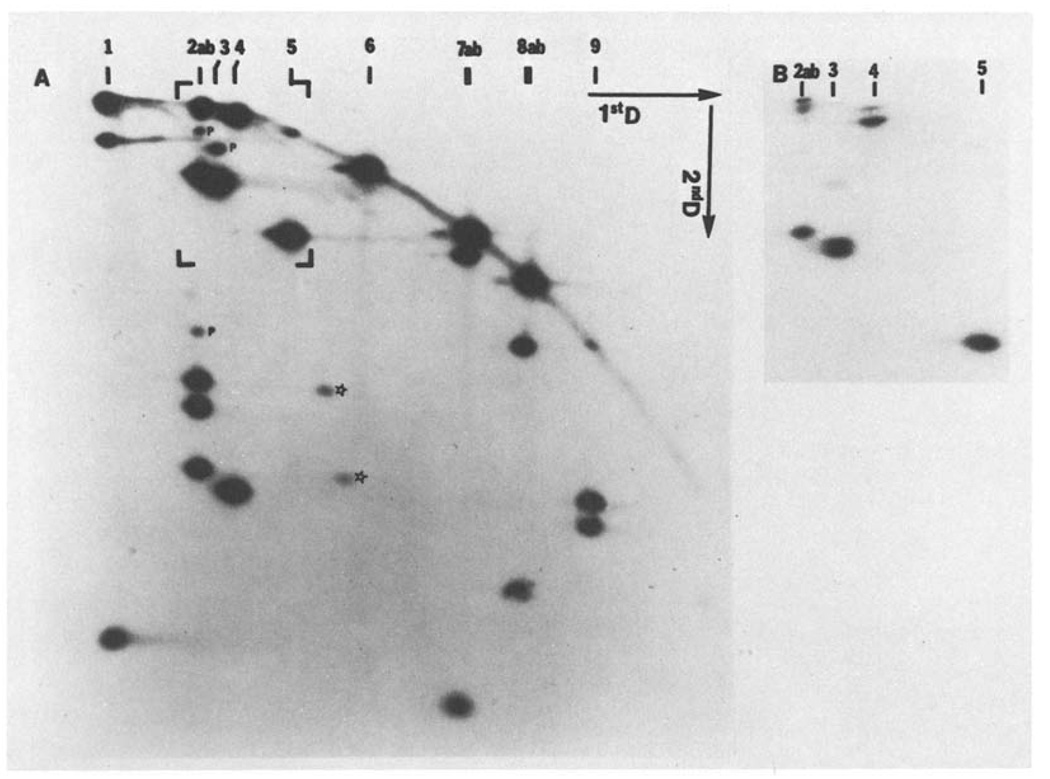
After digestion of the T4 DNA to completion with one restriction enzyme, the resulting fragments were transferred to a slab gel for in situ digestion with the second enzyme and separation in a second dimension, as described in “Materials and Methods”. Different enzyme combinations were used in this way; thus, the BglII(32p)- XhoI separation shown in Fig. 4 represents a separation of end-labeled BglII fragments in the first dimension followed by XhoI cleavage and separation in the second dimension. In an identical fashion, we have carried out XhoI(32P)-BglII, BglII(32P)- EcoRI, EcoRI(32P)-BglII, XhoI(32P)-EcoRI, and EcoRI(32P)-XhoI separations to derive the initial map of the BglII and XhoI sites.
Fig. 4.
A and B. A representative two dimensional separation: T4 BglII fragemnts recut with XhoI for separation in the second dimension. BglII cut T4 DNA was end-labeled and separated in a 0.5% agarose gel. The separated fragments were digested in situ with XhoI and electrophoresed in the second dimension (1% agarose). Panel A shows an autoradiogram of the entire gel and B shows a small region of the same gel (the area indicated by brackets in A) at lower autoradiographic exposure. The positions of BglII bands after separation in the first dimension are indicated along the top of the figure. A variety of different behaviors typical of such separations can be seen: BglII 1 was only partially digested by XhoI and in the second dimension some of the intact fragment is seen in addition to the two end fragments. Both fragments of the doublet BglII 2ab are partially digested by XhoI to produce 4 end-labeled fragments, some faint partials (p), and some undigested material. BglII 3 is digested to give two end fragments of very similar size, which can be seen as resolved on the lower exposure (B). BglII 3 also generates a faint partial. The lower molecular weight BglII fragments are digested to near completion. BglII 4 is cut once to generate one easily detected end fragment and one end fragment almost as big as the intact fragment (B). The two end fragments of BglII 5 have not been resolved. Fragments BglII 6, 7a and 8b are not cut by XhoI. Fragments 7b, 8a and 9 are cut to give clearly resolved end fragments. BglII 10, which was electrophoresed off the first dimension gel, contains no XhoI sites. The stars indicate two anomalous spots which appear to have been produced by a low level of BglII cleavage at an additional site within BglII 2b, to produce two fragments which each share one end fragment with BglII 2b. The additional site is located at 145.2 and behaves like a BglII star site
In principle, the deduction of map order from such a set of gels is straightforward. For example, the sizes of the terminal fragments produced by XhoI cleavage of each BglII fragment can be determined (Fig. 4). Likewise the size of each terminal fragment produced by BglII cleavage of XhoI fragments can be determined from the reciprocal gel (XhoI(32P)-BglII). In theory, it should then be possible to identify a unique set of overlaps of the XhoI and BglII fragments and determine a map order. In practice, the analysis is more complex due to several factors: (1) Even with only 13 BglII fragments and 17 XhoI fragments, certain fragments comigrate in the first dimension (Fig. 3); as a result, some of the end fragments detected in the second dimension could not be unambiguously assigned to individual fragments. For example (Fig. 4), the second band in the BglII digest contains two fragments, which generate four end-labeled fragments in the second dimension (1.6 kb, 2.2 kb, 2.7 kb and 6.9 kb). (2) Not all end fragments are resolved in the second dimension (e.g., the two ends of BglII 3 in Fig. 4); this resolution problem was further complicated by difficulties in precisely comparing mobilities on opposite sides of the gel. (3) Because the method of end labeling actually labels a stretch of DNA near the ends of the fragments (see Materials and Methods), if the second digestion cleaves off a very small end fragment (< 200 base pairs), the fragment immediately internal to the end fragment may be labeled. Furthermore, such very small end fragments may be lost during electrophoresis in the second dimension. (4) Some of the fragments produced by the first enzyme digestion will not be cut by the second enzyme and will migrate in the second dimension with the same molecular weight as in the first dimension; if two or more consecutive fragments are not cut by the second enzyme, their order cannot be deduced from this analysis. (Note that if the same percentage of agarose is used for the first and second dimensions all such altered fragments should form a straight diagonal across the gel. However, a higher percentage gel is usually used in the second dimension, and the non-ideal electrophoretic behavior of high-molecular-weight DNA causes the unaltered DNA fragments – and any products of incomplete cleavage – to form an arc (see Fig. 4).)
We circumvented the above difficulties primarily by repeating the analysis with additional combinations of enzymes, in particular by performing two-dimensional analyses with XhoI and BglII combined with EcoRI. This gave an unambiguous order to the fragments produced by XhoI and BglII digestion, as well as determining the position of the EcoRI sites immediately adjacent to these sites. The accuracy of the positioning of these sites should be quite high (probably less than 5% error) since calibrated one-dimensional gels were used to determine the sizes of all the XhoI, BglII and XhoI/BglII double-digest fragments, as well as most of the XhoI(32P)/EcoR1 and BglII(32P)/EcoRI fragments. The calibration of the two-dimensional gels was generally accurate enough to allow assignment of particular spots to bands in the one-dimensional double digest for size determination.
At this stage in the analysis, we received a preprint of Carlson’s (1979) map ordering the T4 KpnI and SalI sites. These were aligned and oriented with our map using one-dimensional analysis of digests with combinations of BglII, XhoI, KpnI and SalI. After accounting for the differences in the deletions, agreement was excellent. However, we saw one extra KpnI fragment, which Carlson has since confirmed (personal communication).
Information from several sources was combined to position the EcoRI sites. (1) Internally labeled DNA was analyzed on two-dimensional separations using XhoI-EcoRI ang BglII-EcoRI combinations; appropriate end-labeled fragments, at a higher concentration, were run to aid in analysis of the rather complicated pattern. (2) Fragments end-labeled at sites of KpnI cleavage or sites of SalI cleavage were further cut with BglII and then analyzed on two-dimensional gels with EcoRI cleavage prior the second dimension. These gels (KpnI(32P)/BglII-EcoRI and SalI(32P)/BglII-EcoRI) allowed us to position the EcoRI sites that are immediately adjacent to SalI and KpnI sites. (3) The alignment with the genetic map allowed us to take advantage of partial restriction maps obtained from cloned regions of the genome, the references for which are in Table 4. (4) Procudcts obtained by partial EcoRI digestion of SalI(32P) fragments were analyzed using methods which will now be described.
Table 4.
Positions of EcoRI sites and correlation with data from cloned T4 DNA fragments
| Distal terminus (kb) |
Size (kb) |
Cloned fragment No. (size) 2a |
Known gene content |
Ref.a | Distal terminus (kb) |
Size (kb) |
Cloned fragment No. (size) 2a |
Known gene content |
Ref.a |
|---|---|---|---|---|---|---|---|---|---|
| 3.1 | 4.0 | rIIA, 60 | [3] | 95.1 | 5.4 | 5a (5.5) | 12–16 | [3, 13, 14] | |
| 3.4 | 0.3 | 39 | [3, 12, 13] | 95.9 | 0.8 | 35 (0.8) | [14] | ||
| 6.25 | 2.85 | (2.8) | 39 | [3, 12, 13] | 97.45 | 1.55 | 29 (1.5) | 17–18 | [14] |
| 7.8 | 1.55 | 98.1 | 0.65 | 39 (0.6) | 18 | [3, 14] | |||
| 8.4 | 0.6 | 101.35 | 3.25 | 13 (3.25) | 18–20 | [3, 14] | |||
| 9.65 | 1.25 | 104.8 | 3.45 | (3.6) | 20–23 | [3] | |||
| 11.3 | 1.65 | 106.9 | 2.1 | (2.3) | 24 | [3] | |||
| 12.8 | 1.5 | 107.3 | 0.4 | ||||||
| 15.6 | 2.8 | 110.8 | 3.5 | ||||||
| 16.45 | 0.85 | 111.2 | 0.4 | ||||||
| 16.95 | 0.5 | 117.25 | 6.05 | 3 (6.1) | W-29 | [3, 13, 14] | |||
| 21.65 | 4.7 | 121.9 | 4.65 | ||||||
| 24.6 | 2.95 | (3.0) | 40, 41 | [3, 12] | 124.1 | 2.2 | 22 (2.15) | 30 | [12, 16] |
| 26.25 | 1.65 | 26 (1.77) | βgt, 42 | [3, 13, 14] | 124.6 | 0.5 | 30 | [12, 16] | |
| 27.5 | 1.25 | 43 | 125.05 | 0.45 | 30 | [12, 16] | |||
| 29.5 | 1.55 | 30 (1.44) | 43 | [4, 5, 14] | 126.45 | 1.4 | (1.5) | ||
| 34.65 | 5.6 | 4 (5.6) | 43–46 | [4, 5, 14] | 129.4 | 2.95 | (3.0) | 31 | [3] |
| 35.5 | 0.65 | 130.6 | 1.2 | (1.2) | |||||
| 41.6 | 6.1 | 2 (6.3) | 130.75 | 0.15 | (0.1) | ||||
| 42.2 | 0.6 | 133.05 | 2.3 | (2.15) | Pset | [8] | |||
| 42.9 | 0.7 | 134.5 | 1.45 | (1.35) | 63 | [8] | |||
| 54.45 | 11.55 | 1 (11.5) | 136.1 | 1.6 | (1.6) | 63 | [8] | ||
| 55.1 | 0.65 | 139.2 | 3.1 | (3.1) | nrdB | [8, 9] | |||
| 55.4 | 0.3 | 139.9 | 0.7 | (0.7) | nrdA | [8, 9] | |||
| 59.55 | 4.15 | 142.7 | 2.8 | (2.7) | nrdA, td | [8, 9] | |||
| 61.4 | 1.85 | 146.5 | 3.8 | frd, 32 | [8, 9] | ||||
| 62.05 | 0.65 | 147.6 | 1.1 | 33 | [10] | ||||
| 64.1 | 2.05 | 150.4 | 2.8 | (3.1) | 34 | [10] | |||
| 65.7 | 1.6 | e | [6] | 151.4 | 1.0 | 34 | [10] | ||
| 69.45 | 3.75 | e | [6] | 151.6 | 0.2 | (0.2) | 34 | [10] | |
| 70.05 | 0.6 | 152.3 | 0.7 | (0.8) | 35 | [10] | |||
| 70.6 | 0.55 | (0.5) | tRNA | [15] | 154.4 | 2.1 | (2.0) | 35–37 | [10] |
| 76.1 | 5.5 | 5b (5.3) | tRNA-64 | [11, 15] | 157.5 | 3.1 | (3.0) | 37–38 | [3, 10] |
| 80.5 | 4.4 | 7 (4.7) | 50-6 | [3, 13, 14] | 158.3 | 0.8 | (0.8) | t | [10] |
| 82.4 | 1.9 | 25 (1.8) | 6–7 | [14] | 159.4 | 1.1 | |||
| 86.1 | 3.7 | 11 (3.7) | 7–9 | [3, 14] | 160.5 | 1.1 | |||
| 89.1 | 3.0 | 16 (3.0) | 9–11 | [3, 14] | 161.3 | 0.8 | 52 | ||
| 89.7 | 0.6 | 41 (0.5) | 11–12 | [14] | 161.85 | 0.55 | (0.5) | 52 | [3, 13] |
See footnotes to Table 3
Two-Dimensional Analysis of Partial Digestion Products
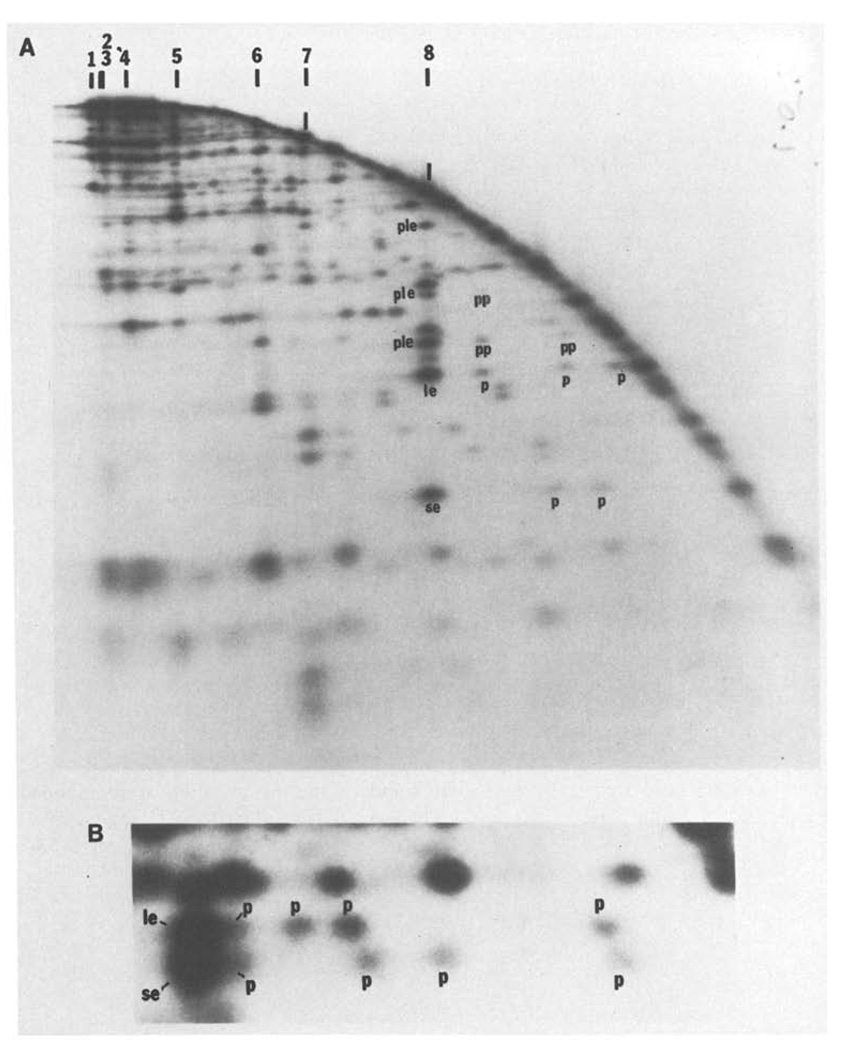
As described by Smith and Birnstiel (1976), a DNA fragment labeled at one end and partially digested with a restriction enzyme can be separated into a series of labeled bands whose sizes correspond to the distances between the labeled end and each restriction site. While this has proved to be a powerful tool for restriction mapping, its usefulness has been limited by the considerable effort required to purify starting restriction fragments specifically labeled at only one end. If a mixture of terminally labeled restriction fragments were partially digested with a second restriction enzyme and separated in one dimension, it would not be possible to determine which of the labeled ends generated a particular band. If, however, these separated partials are digested to completion with the second enzyme and electrophoresed in a second dimension, each labeled end generates a distinctive row of fragments in the resulting pattern. Thus, a labeled end fragment of particular size in the second dimension can be used to identify the positions of all partials extending from that end. (Fig. 5). This approach allows the simultaneous mapping of many sites on a number of restriction fragments. Kovacic and Wang (1979) independently described a similar use of two-dimensional analysis of partial digests.
Fig. 5.
A and B. An example of two-dimensional analysis of partial digests. T4 DNA was digested with SalI, end-labeled and then redigested with a low level of HindIII to generate a partial digest. This partial digest was separated on a 0.5% agarose first dimensional gel. The separated fragments were then digested further with HindIII and electrophoresed through a second dimensional 1.5% agarose gel. Panel A shows an autoradiogram of the entire separation. The arc dominating the pattern is generated by those fragments which were not recut prior to electrophoresis in the second dimension. A fraction of each SalI fragment remains intact during the first partial HindIII digestion and in the first dimension migrates to a position characteristic of the intact SalI fragment. These positions are indicated along the top of the figure. When recut with HindIII and electrophoresed in the second dimension, two terminal labeled SalI HindIII fragments will be generated from each of these SalI fragments. For example, SalI 8 generates a small end fragment (se) and a large end fragment (le) whose positions are indicated in panel A. Furthermore, the first partial HindIII digestion will have generated some labeled partials extending from one end to internal HindIII sites. These partials will migrate faster in the first dimension than the parent SalI fragment. When redigested with HindIII and anlyzed in the second dimension, each of these partials will generate one of the same labeled end fragments produced from the intact SalI fragment (these are labeld p). Thus, SalI 8 generates three detectable partials extending from the end with the large end fragment, le, and two detectable partials extending from the other end, se (one of the se partials was not detected on this gel). The size of each partial gives the distance of a HindIII site from the terminus. Although not intended, the “in situ” digestion prior to running this particular second dimension was also a partial digestion. The resulting partials seen in the second dimension complicate the pattern but can be interpreted. For example, the intact SalI 8 fragment generates in addition to the end fragments a set of partials which, extend to the end defined as le (ple). The larger partials produced by the first partial digestion can also produce lower molecular weight partials in the second dimension. These partials of partials are labeled pp. Note that the molecular weight of fragments extending from, for example, the le end of SalI 8 can be determined from the positions of the partials, p, in the horizontal direction or from the positions of the partials produced in the second dimension, ple, or from the positions of partials of partials, pp. All of these determinations agree. Furthermore, the same map can be obtained from an analysis of fragments extended to the other end, se, of SalI 8. Again the data agrees. - Panel B shows an enlargement of a region of the separation in A, and includes both the large end fragment, le, and the small end fragment, se, generated from SalI 7. The area enlarged in panel B is from the low molecular weight region of the second dimension gel and the equivalent area cannot be readily seen in panel A because it depicts a lower level of autoradiographic exposure. For both le and se, three partials p are clearly resolved, a fourth is only partially resolved and a fifth which is nearly as large as SalI 7 cannot be detected. The size of the four resolved partials determines the positions of 4 HindIII sites and the size of the end fragment defines a fifth. Thus, the size of the small end fragment (se), 0.29 kb, shows that there must be a HindIII site at this distance from the end. The smallest partial, 1 kb, defines the position of the next site and so on. Similarly the positions of the HindIII sites can be read from the other direction by examining le. Independent maps are generated by reading from the two ends and these are consistent with each other. In this case the two maps overlapped for 4 HindIII sites and the map generated by se defines an additional site not seen in the map generated by the other end, le. Similarly, the map generated by le detected one site not seen in the map read from se. Thus, there are six HindIII sites. Although two partials went undetected, the redundancy of the data allows construction of a complete map, and as above this can be confirmed by measurement of partials in the second dimension. It should be noted that the two partials which were not detected would according to their predicted size not be resolvable from intact SalI 7. For large fragments, all the partials are not resolved, but a map of the sites near each end can be constructed. This particular gel could be used to determine all the HindIII sites in SalI 5, SalI 6, SalI 7 and SalI 8 and some of the sites in SalI 1 and SalI 4
In the map presented here, the HindIII and PstI sites, plus some of the EcoRI sites, were mapped by a series of such analyses of partials. In these experiments, C-T4 DNA was cut with an enzyme which makes relatively few cuts, such as SalI, and then end-labeled. The end-labeled SalI fragments were partially digested with a second enzyme (either PstI, EcoRI or HindIII) and the partials separated in the first dimension. Redigestion to completion with the second enzyme and separation in the second dimension then produced horizontal trails of fragments, each member of the trail having the same size in the second dimension. For example, Fig 5b shows the two end fragments generated from the SalI 7 fragment after HindIII digestion. The larger end fragment is denoted as le and the smaller as se. The HindIII partials of the SalI 7 fragment extending from end se or le to internal HindIII sites will migrate in the first dimension as fragments smaller than intact SalI 7. Each of these partials can be recognized by the fact that they generate fragment se or le after redigestion. Thus electrophoresis in the second dimension produces the observed trail of fragments (denoted P) of the same molecular weight. By calibrating the first dimension separation we can determine the molecular weights of all partials extending from the given end. In the case of SalI 7, each end generates three partials which are clearly resolved and a fourth partial which is partly resolved and a fifth which is nearly as large as intact SalI 7 and is not resolved from it. Reading the positions of the HindIII sites from both end le and end se produces redundant data confirming the existence of six HindIII sites within SalI 7; cleavage at one site produces the end fragment while cleavage at the remaining five sites produces the partials. Since HindIII should produce about 150 such partials, it is not surprising that they are not all resolved in a single analysis. We were able to map most of the HindIII sites by analyzing end-labeled BglII, XhoI and SalI fragments by this partial method. In each of these analyses, we were able to unambiguously identify only a fraction of the end-labeled fragments and their trails, but when combined these analyses gave an almost complete map of the HindIII sites. The realtive orientation of the HindIII maps of individual fragments was determined by overlaps of maps derived from the different enzyme combinations and by matching the sizes of fragments in the maps to cloned fragments of known map location and to HindIII fragments detected in our blot-hybridization experiments.
Certain features of the HindIII map should be noted. The HindIII sites closest to SalI, BglII and XhoI sites will be most accurately positioned because these positions are derived directly from the measured molecular weights of end labeled doubly cut fragments (SalI(32P)/HindIII, BglII(32P)/HindIII and XhoI(32P)/HindIII fragments). The positions of the internal HindIII sites are expected to be less precise because the calibration of the two-dimensional gels is less accurate. Additionally, the sizes derived from high-molecular-weight partials cannot be determined as accurately as those from lower molecular weight fragments, since we are often looking at relatively small differences between two fairly large numbers. A gap in the HindIII map between 49.3 and 57.9 kb remains because the appropriate end fragments and partials could not be resolved. In addition, two regions of the HindIII map (38.8–45.65 and 91.9–99.5 kb) are uncertain, because confirming data was not available, and errors could have been made in interpreting the complex patterns.
The PstI map was derived from a similar analysis using partial digests of end-labeled BglII and SalI fragments and from two-dimensional analyses where XhoI(32P) or KpnI(32P)-BglII fragments were separated in the first dimensions, cut with PstI and analyzed in the second dimension. In addition, a two-dimensional analysis of end-labeled SalI fragments partially digested with EcoRI allowed us to complete the EcoRI map.
Aligning the Physical and Genetic Maps
Initial alignment with the genetic map relied on six different clones of T4 DNA, obtained from Ted Young and Wai Mun Huang. The portion of the genetic map carried on each clone had been determined by marker-rescue experiments (cf. Mattson et al. 1977) and is indicated on the resultant map (Table 4). Plasmid DNA from each clone was labeled as described in Materials and Methods. Each of the labeled DNAs was then used as a hybridization probe to analyze one-dimensional separations of assorted restriction digests of T4 DNA which had been transferred to diazophenylthioether paper (“blots”) as described in Materials and Methods. The patterns of hybridization to fragments obtained by digestion with XhoI, BglII, EcoRI, HindII or PstI (or various combinations of these enzymes) allowed unambiguous assignment of each cloned fragment to a position on the restriction map we had derived. The major lack of precision here results from ambiguity as to the exact positions of genes within the cloned fragments.
Several methods were used to confirm and refine the alignment at various points arount the map:
Restriction of a C-T4 containing deletion rIIH23 instead of NB5060 altered the XhoI restriction pattern as expected.
T4 DNA contains a single BamH1 restriction site (Takahashi et al. 1979) between genes 7 and 8 (Wilson et al. 1980). Multiple digests (Fig. 3) showed that this site lies at position 84.9 kb on our map.
After the initial alignment, many of the EcoRI fragments could be matched with the characterized cloned EcoRI fragments on the basis of size correspondence and approximate position (cf. Wilson et al. 1977).
Since the DNA of T4 is circularly permuted (cf. MacHattie et al. 1967), the choice of origin for mapping purposes is arbitrary. For the genetic map, the boundary between the rIIA and rIIB genes was chosen because it has been well defined genetically and physically (cf. Wood and Revel 1976). We have been able to localize this point quite precisely on the physical map. A number of cloned fragments have been isolated in this region (see Mattson et al. 1977) and an 870-base-pair HindIII fragment spanning the rIIA-rIIB junction has been sequenced (D. Pribnow, personal communication). This sequenced fragment contains no BglII, XhoI, PstI, SalI, KpnI or EcoR1 sites. Alignment of adjacent cloned fragments has allowed us to deduce that the HindIII site defining one end of the sequenced fragment is absent in the rIIB deletion used here. On the basis of the sequence data, we can position the origin of the map at the start of the rIIB gene 429 base pairs to the left of the HindIII site defining the other end of the sequenced fragment.
The deletion used here (NB5060) has a right endpoint within the sequenced HindIII fragment spanning the map origin; genetic and sequencing arguments suggest that this endpoint will be about 300 base pairs to the left of the origin (Bautz and Bautz 1967 and Britta Singer, personal communication). The left endpoint is contained in a gene 52 clone obtained from Wai Mun Huang. Hybridization and restriction analysis showed that the deletion’s left end-point must be immediately to the right of the XhoI site at 162.35.
Joyce Emrich Owen and collaborators have sequenced the DNA corresponding to much of the lysozyme (e) gene. This gene, contained on a 3.2 kb HindIII fragment, has a single EcoRI site, located at the position corresponding to amino acids 77 and 78 at about 66 kb on the Wood and Revel (1976) map. By alignment of the restriction fragments, this site corresponds to the EcoRI site at 65.7 kb on our map.
The tRNA region has been extensively characterized by K. Fukada (1979). She finds that the main group of tRNA genes, 0.65 kb long, begins 0.5 kb from the left end of a 5.2 kb EcoRI fragment corresponding to that at 70.6– 75.9 kb on our map and further that the smaller tRNA cluster is on a 0.52 kb fragment immediately to its left. Wood and Revel indicate that the tRNA cluster maps at 69.9–72 kb.
Agreement with Other Available Data
Wilson et al. (1977) determined the sizes of the EcoRI fragments in the region between genes 50 and 20 (i.e., positions 76.1 and 101.35 on our map). Our values for the sizes of each of these fragments agree with theirs within 0.1 kb except for the fragment terminating at 80.5 (4.4 vs 4.7 kb, which also appears to be within the expected experimental error).
Revel et al. (1980a, b) have identified the EcoRI and HindIII fragments corresponding to many of the known genes between genes 30 and 38 (positions 124 and 158 kb on our map), and mapped many of them relative to each other. Their sizes and ours correspond within 0.15 kb for all fragments, except that they see an additional EcoRI cut a position corresponding to 130.3 kb on our map, producing an extra 0.1 kb fragment, which we have therefore included; fragments this small may often be missed by our techniques. In those cases where their fragments had not yet been ordered, our data provided this information.
All of the identified cloned fragments (c.f. Wilson et al. 1977) could be unequivocally matched with an appropriate-sized fragment in the right region of the map.
-
While this work was in progress, Kiko et al. (1979) published a T4 cleavage map, not yet correlated with the genetic map, for the enzymes SmaI, KpnI and BglII; their map differed in very significant ways from ours. However, they sent us a revised map (Rüger et al. 1979), including also XhoI, which agrees with our map within 0.3 kb for each neighboring intercut distance, except where the different deletions result in differences. This is particularly encouraging since their C-T4 strain, (amE51(dCTPase), nd28(denA), “rIIH-23” (denB), alc8), contains a different alc mutation and den' B deletion, derived from a totally independent parent strain. It should be noted that their 4th BglII fragment corresponds to our 5th, while our 4th correponds to their 5th plus the difference in lengths of the deletions.
R. Marsh (personal communication) has also determined the cleavage map for this same C-T4 strain, using the enzymes KpnI, SalI and BglII, as well as XbaI and PvuI. Their data for the first three enzymes also agrees well with ours, as does their general alignment with the genetic map. However, several pieces of evidence, including a careful comparison of our data to the maps of Rüger and of Marsh, have indicated that the alc8 C-T4 strain does not carry the deletion rIIH23, as they have believed, but rather a substantially longer deletion, extending as far as ac. This presumably is the deletion included in some of the dCTPase-endo IV strains initially constructed by Bruner and inadvertantly labeled “rIIH23”; the problem is discussed by Kutter et al. (1975).
Also while this work was in progress. Carlson and Nicolaisen (1979) reported a T4 cleavage map for the enzymes SalI and KpnI; we took advantage of their preliminary information in our analysis of the map for those enzymes, as mentioned above. There were some initial discrepancies between their results and ours, but these have been resolved in their more recent experiments (Carlson, person communication); in particular, they now also pick up the one additional KpnI fragment which we had observed between KpnI 1 and 2 on the map.
Takahashi et al. (1978) have constructed a C-T4 strain which carries a point endo IV mutation instead of a deletion. They have constructed cleavage maps for the enzymes BglII, XmaI, KpnI, SalI( = XhoI) and SalI for this mutant and for a similar T4 strain with deletion den B-rPT8 instead of the point mutation (Takahashi, personal communication). Although a detailed comparison has not yet been made, our order and approximate fragment sizes agree except that we believe that they have reversed BglII fragments 9 and 8 b. Of the enzymes they analyzed, only SalI cleaves in the region covered by deletion NB5060. The position of the SalI site within the deletion is approximate, and is based on our analysis of their data (Takahashi et al. 1979).
Correlation Between Physical and Genetic Map Distances
As discussed by Wood and Revel (1976), recombinational hot spots prevent a perfect correlation between the physical and genetic maps of T4. The map of Edgar and Wood (1966) is based on corrected recombination frequencies. Wood and Revel (1976) refined the relation between physical and genetic distances by adjusting the map distances based on physical measurements of the sizes and locations of various mapped deletions and the molecular weights of protein products. A map derived by Mosig (1976) gives physical distances based on marker-rescue using incomplete genomes, a technique indicating the approximate midpoints of genes. These previous attempts to correlate the physical and genetic maps allow a fairly accurate alignment of the genetic map with the restriction map once a few genetic markers have been localized on the physical map. Since the restriction map provides the true physical map, exact localization of additional genetic markers on this map should eventually allow a precise comparison of physical and genetic distances.
The alignment of our restriction map with the genetic map (see above) allowed us to check our data by comparing it to data obtained from cloned fragments of T4. Cloned fragments whose restriction map was known and whose genetic composition was known from marker-rescue experiments could be matched to both the restriction map and the genetic map. Table 5 summarizes our comparison between the positions of cloned fragments on the restriction map and their positions on the genetic map of Wood and Revel (1976). The correlation is very good, almost never differing by more than 2 kb. Only in three short stretches is the physical distance substantially less than the genetic distance: between genes 43 and 45 (1.5vs 4.5kb), between genes 45 and 47 (2.0 vs 3.9 kb) and between genes td and 32 (1.7 vs 3.2 kb). Our correction largely eliminates the previous gaps unassigned to genes in those three regions of the genome. (For the region around gene 32, Hänngi and Zachau (1980) and Mosig (personal communication) also have evidence for such an anomaly in the genetic map). It is possible that these three sites represent recombinational hot spots.
Table 2.
Look up table with restriction fragments listed in order of electrophoretic migration for enzymes KpnI, SalI, BglII, XhoI, PstI
| Fragment | Size | Fragment | Size | ||
|---|---|---|---|---|---|
| SalI | 1-F | 38.4 | XhoI | 1-B | 19.1 |
| 2-A | 33.7 | 2-J | 16.6 | ||
| 3-G | 31.55 | 3-G | 15.05 | ||
| 4-D | 23.7 | 4a-P | 13.2 | ||
| 5-E | 15.15 | 4b-O | 13.0 | ||
| 6-C | 9.2 | 5a-F | 11.6 | ||
| 7-H | 3.9 | 5b-M | 11.6 | ||
| 8-B | 3.9 | 6-Q | 10.8 | ||
| 7-I | 9.6 | ||||
| 8-C | 8.9 | ||||
| 9a-D | 7.0 | ||||
| 9b-K | 6.75 | ||||
| 9c-L | 6.75 | ||||
| KpnI | 1-F | 46.1 | 10-N | 5.9 | |
| 2-D | 39.25 | 11-H | 4.0 | ||
| 3-C | 36.6 | 12-A | 2.2 | ||
| 4-G | 24.25 | 13-E | 0.9 | ||
| 5-A | 12.4 | ||||
| 6-E | 3.05 | ||||
| 7-B | 1.4 | ||||
| PstI | 1-A | 23.25 | |||
| BglII | 1-D | 55.35 | 2a-DD | 11.8 | |
| 2a-C | 17.4 | 2b-H | 11.5 | ||
| 2b-L | 17.4 | 3-EE | 10.5 | ||
| 3-A | 15.9 | 4-G | 9.95 | ||
| 4-B | 13.95 | 5-GG | 9.65 | ||
| 5-G | 10.75 | 6-F | 8.2 | ||
| 6-K | 8.25 | 7a-E | 7.45 | ||
| 7a-H | 5.45 | 7b-CC | 7.15 | ||
| 7b-I | 5.3 | 8-D | 6.8 | ||
| 8a-J | 4.35 | 9-M | 5.8 | ||
| 8b-E | 4.2 | 10-Q | 5.3 | ||
| 9-F | 3.25 | 11-J | 4.3 | ||
| 10-M | 1.2 | 12-T | 4.0 | ||
| 13-N | 0.6 | ||||
| 14-P | 3.55 | ||||
| 15a-X | 2.8 | ||||
| 15b-L | 2.7 | ||||
| 15c-W | 2.6 | ||||
| 15d-V | 2.55 | ||||
| 15e-AA | 2.45 | ||||
| 15f-BB | 2.35 | ||||
| 16-Y | 2.1 | ||||
| (incomplete list)a | |||||
See the footnote to Table 1
Acknowledgments
We would like to thank Bruce Alberts in whose laboratory this work was initiated and gratefully acknowledge his advice and help with the manuscript. We also thank Ursula Hibner for her advice and generous donation of T4 DNA polymerase, Rae Lynn Burke for help with the manuscript, Ted Young and Wai Mun Huang for cloned T4 DNA fragments, G. Wilson for his kind donation of the strains, Burt Guttman for his help with the construction and illustration of the map, and the many individual who shared information with us prior to publication (L. Albright, K. Carlson, U. Hänggi, K. Jakobs, J. Karam, R. Marsh, N. Murray, D. Pribnow, H. Revel, W. Rüger, H. Takahashi, J. Velten, and G.S. Wilson).
The work was supported by NSF grants PCM 7825677 to P.H.O. and PCM 7905626 and an NIH grant GM 24020 to Bruce Alberts.
References
- Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci USA. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautz FA, Bautz EKF. Mapping of deletions in a non-essential region of the phage T4 genome. J Mol Biol. 1977;128:345–355. doi: 10.1016/s0022-2836(67)80014-7. [DOI] [PubMed] [Google Scholar]
- Carlson K, Nicolaisen B. Cleavage map of bacteriophage T4 cytosine-containing DNA by sequence-specific endonucleases SalI and KpnI. J Virol. 1979;31:112–123. doi: 10.1128/jvi.31.1.112-123.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew R, Snopek TJ, Cozzarelli NR. Characterization of a new class of deletions of the D region of the bacteriophage T4 genome. Virology. 1975;64:144–152. doi: 10.1016/0042-6822(75)90086-0. [DOI] [PubMed] [Google Scholar]
- Edgar RS, Wood WB. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci USA. 1966;55:498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund PT. The 3′-terminal nucleotide sequence of T7 DNA. J Mol Biol. 1972;66:209–224. doi: 10.1016/0022-2836(72)90474-3. [DOI] [PubMed] [Google Scholar]
- Fukada K. Ph D Dissertation. UC San Diego: 1979. Studies on the tRNA biosynthesis of bacteriophage T4. [Google Scholar]
- Hänggi UJ, Zachau HG. Isolation and characterization of DNA fragments containing the dihydrafolate reductase gene of coliphage T4. Gene. 1980;9:271–285. doi: 10.1016/0378-1119(90)90327-n. [DOI] [PubMed] [Google Scholar]
- Helling RB, Goodman HM, Boyer HW. Analysis of endonuclease R.EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. I974;14:1236–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiko H, Niggeman E, Rüger W. Physical mapping of the restriction fragments obtained from T4 dC-DNA with the restriction endonucleases SmaI, KpnI and BglII. Mol Gen Genet. 1979;172:303–312. doi: 10.1007/BF00271730. [DOI] [PubMed] [Google Scholar]
- Kovacic RT, Wang JC. Rapid mapping of restriction sites of a DNA: Restriction of DNA in agarose gel and two-dimensional analysis of end labeled DNA. Plasmid. 1979;2:394–402. doi: 10.1016/0147-619x(79)90023-4. [DOI] [PubMed] [Google Scholar]
- Kutter E, Beug A, Sluss R, Jensen L, Bradley D. The production of undegraded cytosine containing DNA by bacteriophage T4 in the absence of dCTPase and endonucleases II and IV, and its effects on T4-directed protein synthesis. J Mol Biol. 1975;99:591–607. doi: 10.1016/s0022-2836(75)80174-4. [DOI] [PubMed] [Google Scholar]
- Kutter EM, Wiberg JS. Degradation of cytosine-containing bacterial and bacteriophage DNA after infection of Escherichia coli B with bacteriophage T4 D wild type and with mutants defective in genes 46, 47 and 56. J Mol Biol. 1968;38:395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- MacHattie LA, Ritchie DA, Thomas CA. Terminal repetition in permuted T2 bacteriophage DNA molecules. J Mol Biol. 1967;23:355–363. doi: 10.1016/s0022-2836(67)80110-4. [DOI] [PubMed] [Google Scholar]
- Mattson T, VanHouwe G, Bolle A, Selzer G, Epstein R. Genetic identification of cloned fragments of bacteriophage T4 DNA and complementation by some clones containing early T4 genes. Mol Gen Genet. 1977;154:319–326. doi: 10.1007/BF00571289. [DOI] [PubMed] [Google Scholar]
- Morton D, Kutter EM, Guttman BS. Synthesis of T4 DNA and bacteriophage in the absence of dCMP hydroxymethylase. J Virol. 1978;28:262–269. doi: 10.1128/jvi.28.1.262-269.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CF, Hama-Inaba H, Mace D, Sinha NK, Alberts B. Purification of the gene 43, 44, 45, and 62 proteins of the bacteriophage T4 DNA replication apparatus. J Biol Chem. 1979;254:6787–6796. [PubMed] [Google Scholar]
- Mosig G. Linkage map and genes of bacteriophage T4. In: Fasman GD, editor. Handbook of biochemistry and molecular biology. 3rd ed. Cleveland, Ohio: CRC press; 1976. p. 664. [Google Scholar]
- Mosig G. Genetic map of bacteriophage T4. In: Brien JO, editor. Genetic Maps. Vol 1. Bethesda, Maryland: National Cancer Institute, National Institutes of Health; 1980. p. 20. [Google Scholar]
- Potter SS, Bott KF, Newbold JE. Two-dimensional restriction analysis of the Bacillus subtilis genome: Gene purification and ribosomal ribonucleic acid gene organization. J Bacteriol. 1977;129:492–500. doi: 10.1128/jb.129.1.492-500.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby PWJ, Dieckmann M, Rhodes C, Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts RJ. Restriction endonucleases. Crit Rev Biochem. 1976;4:123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Rosenvold EC, Honigman A. Mapping of AvaI and XmaI cleavage sites in bacteriophage DNA including a new technique of DNA digestion in agarose gels. Gene. 1977;2:273–288. [Google Scholar]
- Rüger W, Neumann W, Rohr U, Niggemann E. The complete maps of BglII, SalI and XhoI restriction sites on T4 dc-DNA. Mol Gen Genet. 1979;176:417–425. doi: 10.1007/BF00333106. [DOI] [PubMed] [Google Scholar]
- Sharp PA, Sugden B, Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose ethidium bromide electrophoresis. Biochemistry. 1973;12:3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Smith HO, Birnstiel ML. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976;3:2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L, Gold L, Kutter E. A gene of bacteriophage T4 whose product prevents true late transcription on cytosine-containing T4 DNA. Proc Natl Acad Sci USA. 1976;73:3098–3102. doi: 10.1073/pnas.73.9.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Shimizu M, Saito H, Ikeda Y. Studies of viable T4 bacteriophage containing cytosine-substituted DNA (T4dc phage) Mol Gen Genet. 1979;168:49–53. doi: 10.1007/BF00267932. [DOI] [PubMed] [Google Scholar]
- Velten J, Abelson J. The generation and analysis of clones containing bacteriophage T4 DNA fragments. J Mol Biol. 1980;137:235–248. doi: 10.1016/0022-2836(80)90327-7. [DOI] [PubMed] [Google Scholar]
- Wahl GM, Stern M, Stark GR. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization using dextran sulfate. Proc Natl Acad Sci. 1979;76:3683–3688. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GS, Murray NE. Molecular cloning of the DNA ligase gene from bacteriophage T4 I., Characterization of the recombinants. J Mol Biol. 1979;132:471–491. doi: 10.1016/0022-2836(79)90270-5. [DOI] [PubMed] [Google Scholar]
- Wilson GG, Neve RR, Edlin GJ, Konigsberg WH. The BamHI restriction site in the bacteriophage T4 chromosome is located in or near gene 8. Genetics. 1980;93:285–296. doi: 10.1093/genetics/93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GG, Tanyashin VI, Murry NE. Molecular cloning of fragments of bacteriophage T4 DNA. Mol Gen Genet. 1977;156:203–214. doi: 10.1007/BF00283493. [DOI] [PubMed] [Google Scholar]
- Wood WB, Revel HR. The genome of bacteriophage T4. Bacteriol Rev. 1976;40:847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]