Abstract
Human paraoxonase 1 (PON1) has broad substrate specificity and has been shown to protect against exposure to some organophosphorus (OP) insecticides due to its ability to hydrolyze toxic metabolites of some organophosphorothioate insecticides. PON1 status has been shown to be important in protecting against vascular disease, presumably due to the not-as-yet fully characterized role of the three PON proteins in modulating oxidative stress. More recently, all three PONs (1, 2, and 3) have been shown to inactivate the quorum sensing factor N-(3-oxododecanoyl)-L-homoserine lactone (3OC12-HSL) of Pseudomonas. Expression of human PON1 in Drosophila demonstrated the importance of PON1 in resistance to Pseudomonas infection. Many studies have examined only DNA single nucleotide polymorphisms as possible risk factors for disease or exposures. For all of the known functions of PON1, the level of PON1 enzyme is important and, in some cases, also the Q192R polymorphism. A simple high throughput two-substrate assay/analysis, plotting rates of diazoxon hydrolysis vs. paraoxon hydrolysis, provided both PON1 levels and functional Q192R phenotype/genotype. We have developed a new two-substrate assay/analysis protocol that provides PON1 status without use of toxic OP substrates. Factors were determined for inter-converting rates of hydrolysis of different substrates.
Keywords: PON1 status, Paraoxonase, Diazoxon, Diazinon, Chlorpyrifos, Chlorpyrifos oxon, Carotid artery disease, Quorum sensing factor, OP exposure
1 Introduction
Human paraoxonase 1 (PON1) has broad substrate specificity and has been shown to protect against exposure to some organophosphorus (OP) insecticides due to its ability to hydrolyze their toxic oxon metabolites at physiologically relevant rates (Costa et al. 1990; Li et al. 1993, 1995, 2000; Shih et al. 1998; Cole et al. 2005). The hydrolysis of other OPs, such as paraoxon (PO), while detectable with in vitro assays, is not physiologically relevant due to insufficient catalytic efficiency of hydrolysis (Li et al. 2000). The PON1 Q192R polymorphism affects the catalytic efficiency of hydrolysis of some PON1 substrates (Davies et al. 1996; Li et al. 2000). We introduced the term PON1 status to include both the functional PON1192 genotype as well as the plasma level of PON1, both of which can be important in determining risk of disease or exposure (Li et al. 1993). In all cases, rates of detoxication of both endogenous and xenobiotic substrates are determined by the plasma level of PON1, provided that the catalytic efficiency of hydrolysis is physiologically significant. For some cases, such as the detoxication of chlorpyrifos oxon (CPO), plasma PON1 level and the Q192R polymorphism are both important, with the PON1R192 alloform detoxifying CPO more efficiently than PON1Q192 (Li et al. 2000). The efficiency of detoxication of diazoxon (DZO) is nearly equivalent for both PON1192 alloforms (Li et al. 2000).
PON1 status has been shown to be important in protecting against vascular disease (Jarvik et al. 2000), presumably through the role of PON1 in modulating oxidative stress (reviewed in James 2006). More recently, all three PONs (1, 2, and 3) have been shown to inactivate the quorum sensing factor N-(3-oxododecanoyl)-L-homoserine lactone (3OC12-HSL) of Pseudomonas (Ozer et al. 2005). The definitive study by Stoltz et al. (2008; Chapter 17 in this book), where the expression of human PON1 in transgenic Drosophila resulted in increased resistance to infection by Pseudomonas aeruginosa, indicates that the PON family of proteins can also be considered as part of the innate immunity system (Chun et al. 2004).
After the genetic variability of PON1 was linked to cardiovascular disease, many studies have been carried out that examined only DNA single nucleotide polymorphisms (SNPs) as possible risk factors for disease or exposures. Some studies examined only the Q192R polymorphism, others both the Q192R and L55M polymorphisms and yet others have included the analysis of one or more promoter region polymorphisms, the most important of which appears to be the C-108T polymorphism that occurs in an Sp1 binding site (Deakin et al. 2003). Relatively few studies have examined the relationship between PON1 levels or activity and risk for disease. Mackness et al. (2001) found decreased levels of plasma PON1 (by ELISA) and paraoxonase (POase) activity among patients with coronary heart disease (CHD). They also carried out a meta-analysis of 18 previous studies, only three of which determined PON1 levels or activity. The assay of POase is not a good measure of risk for disease, since this activity is dramatically affected by the Q192R polymorphism with PON1R192 having much higher POase activity than PON1Q192. Since the gene frequencies for PON1Q192 and PON1R192 vary significantly among different ethnic groups, a mixture of individuals of different ethnic origin can significantly skew the data (Brophy et al. 2002). Mackness et al. (2001) recommended that “We, along with other authors, would strongly suggest that all further epidemiological studies into the role of PON1 and disease should include a measurement of the enzyme itself in addition to the genetic polymorphisms.” This same recommendation was echoed by two other experienced PON1 research teams (Deakin and James 2004; La Du 2003).
A second large study (Lawlor et al. 2004) examined association of the Q192R polymorphism with CHD in a large cohort (n = 3,266) combined with a meta-analysis of 38 other studies. These SNP analyses revealed no association with CHD; however, they suffered from the critical lack of data on plasma PON1 levels.
A third meta-analysis which examined four PON1 polymorphisms and one PON2 polymorphism included 43 genetic association studies (>11,000 cases and ~ 13,000 controls) and showed no significant association with CHD (Wheeler et al. 2000). This study also suffered from a lack of data on plasma PON1 levels or activity.
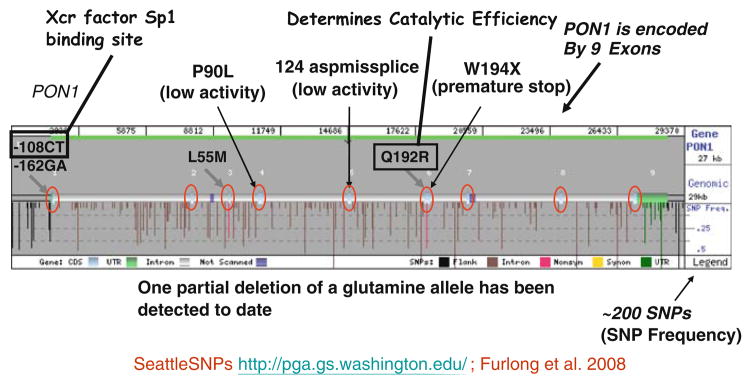
For all of the known functions of PON1, the level of PON1 is important and, in some cases, also the Q192R polymorphism. Figure 1 shows the PON1 polymorphisms and frequencies identified by the Seattle SNPs resequencing effort (Furlong et al. 2008). Characterizing all of the nearly 200 PON1 polymorphisms will not provide an accurate prediction of plasma PON1 levels. Measurement of the of the phenyl acetate hydrolysis activity of PON1 (AREase) is unaffected by the Q192R polymorphism and can serve as a surrogate measure of plasma PON1 protein levels (Furlong et al. 2006; Richter et al. 2008). Measurement of the AREase activity of plasma PON1 or determination of plasma PON1 protein levels by ELISA are the minimum measures that should be carried out in any epidemiological study. More useful measures are described below.
Fig. 1.
The human PON1 gene with known polymorphisms and their frequencies. The 5′ end of the gene is on the left. (Seattle SNPs, http://pga.gs.washington.edu/)
2 Two-Substrate Analyses of PON1 Status
Early studies by Eckerson et al. (1983) showed that a two-substrate assay/analysis plotting rates of PO hydrolysis vs. AREase would provide both plasma PON1 levels and a separation of low (PON1Q192 homozygotes) from the high metabolizers. This analysis, however, did not resolve PON1192 heterozygotes from PON1R192 homozygotes. When we made use of the two-substrate assay/analysis to examine rates of a number of different PON1 substrates, we found that plotting rates of diazoxon hydrolysis vs. paraoxon hydrolysis provided both relative PON1 levels for each PON1192 functional genotype/phenotype as well as a clear resolution of all three PON1192 phenotypes (Q/Q, Q/R, and R/R) (Davies et al. 1996; Richter and Furlong 1999). This analysis, however, uses two highly toxic OPs, DZO and PO.
3 Development of a PON1 Status Protocol with Non-OP Substrates
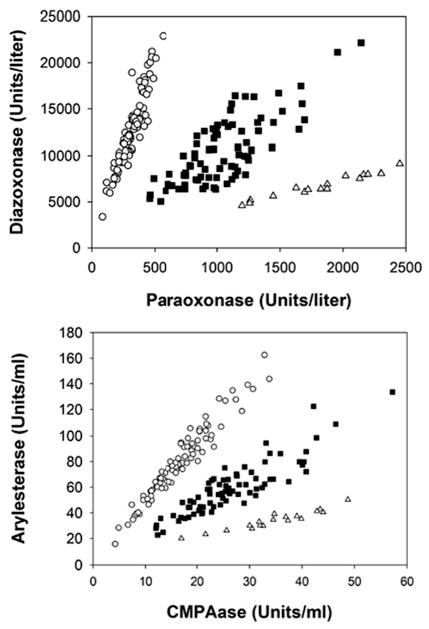
Since PON1 status appears to be an important risk factor for OP exposure as well as for a number of diseases, we examined rates of hydrolysis of more than 70 substrates under different conditions of salt concentration and pH to develop a high throughput PON1 status protocol that did not make use of the highly toxic OP substrates. Figure 2 shows a comparison of the DZOase vs. POase protocol for determining PON1 status and a new PON1 status protocol where rates of phenyl acetate hydrolysis at high salt are plotted against rates of 4-(chloromethyl)phenyl acetate (CMPA) in buffer alone (Richter et al. 2008, 2009). Both of these protocols resolve all three PON1192 phenotypes; however the new assay with non-OP substrates is more suitable for laboratories that are not equipped for using highly toxic compounds.
Fig. 2.
Comparison of the two protocols for determining PON1 status. (a) Assays using the highly toxic OP substrates DZO and PO; and (b) assays using the non-OP substrates phenyl acetate and CMPA. The 183 plasma samples included 86 PON1Q192 homozygotes, 79 heterozygotes and 18 PON1R192 homozygotes with genotypes verified by PCR. Reproduced from Richter et al. (2009) with permission
Since many assay protocols have been used by different laboratories over the years, we determined conversion factors that will allow inter-conversion of rates of hydrolysis of one substrate to another for each of the three PON1192 phenotypes (Richter et al. 2009) (Table 1). Also, we described how to determine physiologically relevant rates of in vivo hydrolysis of CPO and DZO. This new assay should encourage epidemiologists to measure the parameters that are important in relating genetic variability of PON1 to risk of disease or exposure.
Table 1.
Conversion factors for rates of substrate hydrolysis
| Phenotype | Conversion factors | r2 a |
|---|---|---|
| AREaseHSb (U/ml) × 172 = DZOasephys (U/L)c | 0.93 | |
| QR | AREaseHS (U/ml) × 204 = DZOasephys (U/L) | 0.82 |
| RR | AREaseHS (U/ml) × 286 = DZOasephys (U/L) | 0.87 |
| AREaseHS (U/ml) × 69 = CPOasephysd (U/L) | 0.87 | |
| QR | AREaseHS (U/ml) × 103 = CPOasephys (U/L) | 0.88 |
| RR | AREaseHS (U/ml) × 189 = CPOasephys (U/L) | 0.89 |
| AREaseLSe (U/ml) × 110 = DZOasephys (U/L) | 0.84 | |
| QR | AREaseLS (U/ml) × 100 = DZOasephys (U/L) | 0.72 |
| RR | AREaseLS (U/ml) × 83 = DZOasephys (U/L) | 0.93 |
| AREaseLS (U/ml) × 45 = CPOasephys (U/L) | 0.73 | |
| QR | AREaseLS (U/ml) × 50 = CPOasephys (U/L) | 0.84 |
| RR | AREaseLS (U/ml) × 55 = CPOasephys (U/L) | 0.92 |
| AREaseHS (U/ml) × 3.8 = POase (U/L) | 0.75 | |
| QR | AREaseHS (U/ml) × 15.9 = POase (U/L) | 0.50 |
| RR | AREaseHS (U/ml) × 47.6 = POase (U/L) | 0.90 |
| QQf | AREaseHS (U/ml) × 1.6 = AREaseLS (U/ml) | 0.85 |
| QRf | AREaseHS (U/ml) × 2.0 = AREaseLS (U/ml) | 0.66 |
| RRf | AREaseHS (U/ml) × 3.5 = AREaseLS (U/ml) | 0.83 |
| DZOasephys (U/L) × 1.08 = DZOaseHSg (U/L) | 0.90 | |
| QR | DZOasephys (U/L) × 1.01 = DZOaseHS (U/L) | 0.91 |
| RR | DZOasephys (U/L) × 0.84 = DZOaseHS (U/L) | 0.87 |
Correlation coefficient squared
AREaseHS = Arylesterase activity measured in buffer and 2 M NaCl
DZOasephys = Diazoxonase activity measured under physiological conditions
CPOasephys = Chlorpyrifos oxonase activity measured under physiological conditions
AREaseLS = Arylesterase activity measured in buffer
DZOaseHS = Diazoxonase activity measured at 2 M NaCl, pH 8.5 (reproduced from Richter et al. 2009, with permission)
We have shown previously that discrepancies between PON1192 SNP analysis and the functional PON1 status analysis can reveal mutations in the PON1 gene that can be characterized by sequencing the entire PON1 gene (Jarvik et al. 2003). The effects of polymorphisms in the 3′-untranslated region of the PON1 gene have yet to be characterized. While there is still much to learn about the effects of PON1 SNPs on expression, it will be important to couple such studies with the functional PON1 status analysis. Although the effects of some environmental influences on plasma PON1 levels are known (reviewed in Costa et al. 2005), the effects of epigenetic modifications on PON1 expression are yet to be explored.
Acknowledgments
This work was supported by grants from the National Institute of Environmental Health Sciences ES09883, ES04696, ES07033, ES09601 – EPA: RD-83170901-OE and the National Heart, Lung, and Blood Institute, HL67406 and HL074366.
References
- Brophy VH, Jarvik GP, Furlong CE. PON1 Polymorphisms. In: Costa LG, Furlong CE, editors. Paraoxonase (PON1) in Health and Disease: Basic and Clinical Aspects. Boston: Kluwer Academic Press; 2002. pp. 53–77. [Google Scholar]
- Costa LG, Vitalone A, Cole TB, Furlong CE. Modulation of paraoxonase (PON1) activity. Biochem Pharmacol. 2005;69:541–550. doi: 10.1016/j.bcp.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Chun CK, Ozer EA, Welsh MJ, Zabner J, Greenberg EP. Inactivation of a Pseudomonas aeruginosa quorum sensing signal by human airway epithelia. Proc Natl Acad Sci USA. 2004;101:3587–3590. doi: 10.1073/pnas.0308750101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TB, Walter BJ, Shih DM, Tward AD, Lusis AJ, Timchalk C, Richter RJ, Costa LG, Furlong CE. Toxicity of chlorpyrifos and chlorpyrifos oxon in a transgenic mouse model of the human paraoxonase (PON1) Q192R polymorphism. Pharmacogenet Genomics. 2005;15:589–598. doi: 10.1097/01.fpc.0000167327.08034.d2. [DOI] [PubMed] [Google Scholar]
- Costa LG, McDonald BE, Murphy SD, Omenn GS, Richter RJ, Motulsky AG, Furlong CE. Serum paraoxonase and its influence on paraoxon and chlorpyrifos-oxon toxicity in rats. Toxicol Appl Pharmacol. 1990;103:66–76. doi: 10.1016/0041-008x(90)90263-t. [DOI] [PubMed] [Google Scholar]
- Davies H, Richter RJ, Keifer M, Broomfield C, Sowalla J, Furlong CE. The human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nature Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- Deakin S, Leviev I, Brulhart-Meynet M-C, James RW. Paraoxonase-1 promoter haplotypes and serum paraoxonase: a predominant role for polymorphic position −107, implicating the Sp1 transcription factor. Biochem J. 2003;372:643–649. doi: 10.1042/BJ20021670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin SP, James RW. Genetic and environmental factors modulating serum concentrations and activities of the antioxidant enzyme paraoxonase-1. Clin Sci (Lond) 2004;107:435–447. doi: 10.1042/CS20040187. [DOI] [PubMed] [Google Scholar]
- Eckerson HW, Wyte CM, La Du BN. The human serum paraoxonase/arylesterase polymorphism. Am J Hum Genet. 1983;35:1126–1138. [PMC free article] [PubMed] [Google Scholar]
- Furlong CE, Richter RJ, Li W-F, Brophy VH, Carlson C, Meider M, Nickerson D, Costa LG, Ranchalis J, Lusis AJ, Shih DM, Tward A, Jarvik GP. The functional consequences of polymorphisms in the human PON1 gene. In: Mackness B, Mackness M, Aviram M, Paragh G, editors. The Paraoxonases: Their Role in Disease, Development and Xenobiotic Metabolism. Dordrecht, The Netherlands: Springer; 2008. pp. 267–281. [Google Scholar]
- Furlong C, Holland N, Richter R, Bradman A, Ho A, Eskenazi B. PON1 status of farm-worker mothers and children as a predictor of organophosphate sensitivity. Pharmacogenet Genomics. 2006;16:183–190. doi: 10.1097/01.fpc.0000189796.21770.d3. [DOI] [PubMed] [Google Scholar]
- James RW. A long and winding road: defining the biological role and clinical importance of paraoxonases. Clin Chem Lab Med. 2006;44:1052–1059. doi: 10.1515/CCLM.2006.207. [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Rozek LS, Brophy VH, Hatsukami TS, Richter RJ, Schellenberg GD, Furlong CE. Paraoxonase phenotype is a better predictor of vascular disease than PON1192 or PON155 genotype. Atheroscler Thromb Vasc Biol. 2000;20:2442–2447. doi: 10.1161/01.atv.20.11.2441. [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Jampsa R, Richter RJ, Carlson C, Rieder M, Nickerson D, Furlong CE. Novel paraoxonase (PON1) nonsense and missense mutations predicted by functional genomic assay of PON1 status. Pharmacogenetics. 2003;13:291–295. doi: 10.1097/00008571-200305000-00009. [DOI] [PubMed] [Google Scholar]
- La Du BN. Future studies of low-activity PON1 phenotype subjects may reveal how PON1 protects against cardiovascular disease. Arterioscler Thromb Vasc Biol. 2003;23:1317–1318. doi: 10.1161/01.ATV.0000082600.42562.7F. [DOI] [PubMed] [Google Scholar]
- Li W-F, Costa LG, Furlong CE. Serum paraoxonase status: a major factor in determining resistance to organophosphates. J Toxicol Environ Health. 1993;40:337–346. doi: 10.1080/15287399309531798. [DOI] [PubMed] [Google Scholar]
- Li W-F, Furlong CE, Costa LG. Paraoxonase protects against chlorpyrifos toxicity in mice. Toxicol Lett. 1995;76:219–226. doi: 10.1016/0378-4274(95)80006-y. [DOI] [PubMed] [Google Scholar]
- Li W-F, Costa LG, Richter RJ, Hagen T, Shih DM, Tward A, Lusis AJ, Furlong CE. Catalytic efficiency determines the in vivo efficacy of PON1 for detoxifying organophosphates. Pharmacogenetics. 2000;10:767–780. doi: 10.1097/00008571-200012000-00002. [DOI] [PubMed] [Google Scholar]
- Mackness B, Davies GK, Turkie W, Lee E, Roberts DH, Hill E, Roberts C, Durrington PN, Mackness MI. Paraoxonase status in coronary heart disease: are activity and concentration more important than genotype? Arterioscler Thromb Vasc Biol. 2001;21:1451–1457. doi: 10.1161/hq0901.094247. [DOI] [PubMed] [Google Scholar]
- Ozer EA, Pezzulo A, Shih DM, Chun C, Furlong C, Lusis AJ, Greenberg EP, Zabner J. Human and murine Paraoxonase 1 are host modulators of P. aeruginosa quorum-sensing. FEMS Microbiol Lett. 2005;253:29–37. doi: 10.1016/j.femsle.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Richter RJ, Furlong CE. Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics. 1999;9:745–753. [PubMed] [Google Scholar]
- Richter RJ, Jarvik GP, Furlong CE. Determination of paraoxonase 1 status without the use of toxic organophosphate substrates. Circ Cardiovasc Genet. 2008;1:147–152. doi: 10.1161/CIRCGENETICS.108.811638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter RJ, Jarvik GP, Furlong CE. Paraoxonase 1 (PON1) status and substrate hydrolysis. Toxicol Appl Pharmacol. 2009 doi: 10.1016/j.taap.2008.11.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DM, Gu L, Xia Y-R, Navab M, Li W-F, Hama S, Castellani LW, Furlong CE, Costa LG, Fogelman AM, Lusis AJ. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394:284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- Stoltz DA, Ozer EA, Taft PJ, Barry M, Liu L, Kiss PJ, Moninger TO, Parsek MR, Zabner J. Drosophila are protected from Pseudomonas aeruginosa lethality by transgenic expression of paraoxonase-1. J Clin Invest. 2008;118:3123–3131. doi: 10.1172/JCI35147. [DOI] [PMC free article] [PubMed] [Google Scholar]