Abstract
Background/Aims: Recent studies have implicated a region on chromosome 1q21-23, including the NOS1AP gene, in susceptibility to schizophrenia. However, replication studies have been inconsistent, a fact that could partly relate to the marked psychopathological heterogeneity of schizophrenia. The aim of this study is to evaluate association of polymorphisms in the NOS1AP gene region to schizophrenia, in patients from a South American population isolate, and to assess if these variants are associated with specific clinical dimensions of the disorder. Methods: We genotyped 24 densely spaced SNPs in the NOS1AP gene region in a schizophrenia trio sample. The transmission disequilibrium test (TDT) was applied to single marker and haplotype data. Association to clinical dimensions (identified by factor analysis) was evaluated using a quantitative transmission disequilibrium test (QTDT). Results: We found significant association between eight SNPs in the NOS1AP gene region to schizophrenia (minimum p value = 0.004). The QTDT analysis of clinical dimensions revealed an association to a dimension consisting mainly of negative symptoms (minimum p value 0.001). Conclusions: Our findings are consistent with a role for NOS1AP in susceptibility to schizophrenia, especially for the ‘negative syndrome’ of the disorder.
Key Words: NOS1AP, Schizophrenia, Clinical heterogeneity, Genetic association, Psychiatric genetics
Introduction
Schizophrenia is a severe and debilitating psychiatric disorder which is among the leading causes of morbidity and long-term disability world-wide [1]. Family, twin, and adoption studies indicate that schizophrenia has an important genetic component [2], and linkage studies have implicated a number of genomic regions potentially involved in disease susceptibility [3, 4, 5, 6, 7, 8, 9]. A region of 12 cM on chromosome 1q21-22 produced a parametric multipoint LOD score of 6.5 in a sample of 22 Canadian families [5]. This region harbours several candidate genes for schizophrenia, including the Regulator of G Protein Signalling 4 (RGS4), the Nitric Oxide Synthase 1 (neuronal) Adaptor Protein (NOS1AP; also called CAPON), and the U2AF Homology Motif Kinase 1 (UHMK1). The NOS1AP gene is of particular interest due to its involvement in the glutamatergic neurotransmission system, a pathway thought to play a role in the aetiology of schizophrenia [10]. Several studies have shown significant association between markers in the NOS1AP gene and schizophrenia, in samples from Canada (p = 0.0016), and China (p = 0.000017) [11, 12]. Also, we recently analysed marker D1S1679, located 23.5 kb downstream of the NOS1AP gene, in a schizophrenia trio sample from a South American population isolate (Antioquia) [13, 14, 15] and found a significant association (p = 0.019) [16]. However, other studies have failed to detect such an association [17]. The discordance of the NOS1AP-schizophrenia association results could relate to the marked psychopathological heterogeneity of schizophrenia. Patients can differ widely with respect to age of onset, inter-episode recovery and prominence of both positive and negative symptoms [18]. Since this clinical variability could reflect an underlying genetic heterogeneity, it seems interesting to explore the analysis of clinical dimensions – quantifiable symptom complexes varying across affected individuals – in studies aimed at the identification of genetic loci predisposing to schizophrenia [19, 20].
To follow-up our previous report implicating marker D1S1679, we obtained data for SNPs in the NOS1AP gene typed in an expanded collection of Antioquian schizophrenia trios. We performed single-marker as well as haplotype-based TDT analyses. In view of the psychopathological heterogeneity of schizophrenia, we also used a dimensional approach to evaluate association between clinical features and the NOS1AP gene. These analyses confirmed association of NOS1AP polymorphisms both to disease and to a clinical dimension comprising negative symptoms of schizophrenia.
Methods and Materials
Study Sample and Clinical Procedures
The study sample consisted of 102 patients diagnosed with schizophrenia (79 males, 23 females; mean age 34.3 ± 10.2 years; mean age of onset 21 ± 7.2 years; mean duration of the illness 13.4 ± 8.7 years) and their nuclear families. For 38 of the patients, both parents were available (complete trios); for 45, one parent and one full sibling were available (parent-sibling-trios), for 3 patients, one parent and two full sibs were available, and for 13 patients, only one parent could be obtained (duos). Three families were of a different structure, two of them comprising the index case, a sibling and a paternal uncle resp. aunt, one consisting of the index case and two full siblings.
Patients with a diagnosis of schizophrenia were recruited in the municipalities of Medellín and Envigado (Antioquia, Colombia) at Hospital Mental de Antioquia, Hospital San Vicente de Paúl, Clínica Sameín, Clínica Insam, and the Mental Health Centre of Envigado.
Patients were assessed by a psychiatrist (J.G., C.L., M.R.) using the Spanish version of the Diagnostic Interview for Genetic Studies (DIGS) previously validated in Colombia [21, 22]. All patients were in remission during the entire assessment procedure. A final diagnosis using DSM-IV-TR criteria [23] was reached through a best estimate procedure which required two experienced psychiatrists (other than the one conducting the interview; C.P., G.R.) to independently reach the same diagnosis of schizophrenia based on the DIGS and clinical records. If no consensus diagnosis could be obtained, a third psychiatrist (J.O.) was consulted. If still no consensus could be reached, the subject was excluded from the study. Additionally, inclusion criteria required at least six out of the eight great-grandparents to be of Antioquian origin. Patients with mental retardation and/or neurological lesions were excluded from the study.
The clinical dimensions were obtained from the Scales of Assessment of Positive and Negative Symptoms (SAPS and SANS) [24, 25, 26] by factor analysis. The SANS and SAPS were applied to all schizophrenic patients by an experienced psychiatrist. To comply with the minimum sample size recommended for factor analysis [27, 28], the scales were also applied to an additional sample of 150 schizophrenic patients diagnosed according to the same diagnostic criteria and with similar clinical parameters, making for a total sample of 252. However, these additional individuals were not available for genotyping.
This study was approved by the Bioethics Committee of the Universidad de Antioquia (Colombia) and by the Ethics Committees of all participating institutions.
Marker Selection and Genotyping
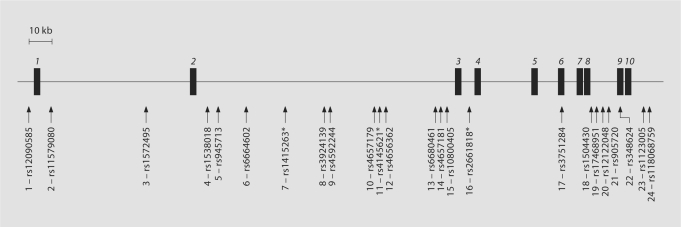
We selected 24 SNP markers that together cover 314 kb across the NOS1AP gene region. A schematic overview of the markers chosen and their location with respect to NOS1AP is given in figure 1. The 24 markers included 9 SNPs from the original association study of NOS1AP [11] (numbers in brackets refer to SNP numbering as in fig. 1): rs1572495 [#3], rs1538018 [#4], rs945713 [#5], rs1415263 [#7], rs3924139 [#8], rs4145621 [#11], rs2661818 [#16], rs3751284 [#17], and rs348624 [#22], and additional SNPs selected from evolutionary conserved regions within or close to NOS1AP [29]: rs12090585 [#1], rs11579080 [#2], rs6664602 [#6], rs4592244 [#9], rs4657179 [#10], rs4656362 [#12], rs6680461 [#13], rs4657181 [#14], rs10800405 [#15], rs1504430 [#18], rs17468951 [#19], rs12122048 [#20], rs905720 [#21], rs1123005 [#23], rs11806859 [#24).
Fig. 1.
Location of markers genotyped in the NOS1AP gene region (markers rs6664602 [#6] and rs4656362 [#12] were excluded from subsequent analyses; see text). The locations of NOS1AP exons are indicated by black boxes; exon numbers are in italics. Consecutive marker numbers from 1 to 24 are included for easier comparison with the LD plot in figure 2a. Markers significantly associated with schizophrenia in Brzustowicz et al. [11] are indicated with an asterisk.
Genomic DNA was extracted from blood samples using standard laboratory procedures.
A multiplex PCR assay was developed to genotype multiple SNPs. The multiplex PCR design was based on software described in [30] and genotyping was conducted via a ligation detection assay and scored on a Luminex 100 [31, 32]. Primer sequences and amplification conditions can be obtained from the authors.
Data Analysis
Hardy-Weinberg equilibrium was evaluated separately in founders and cases using the GENEPOP program [33] (available online at http://wbiomed.curtin.edu.au/genepop/). Allele frequencies of non-transmitted alleles were estimated using TDTPHASE v.2.4 [34].
The Transmission Disequilibrium Test (TDT) [35] for single markers was carried out as implemented in the TRANSMIT program, version 2.5.4 [36]. TRANSMIT carries out a generalised transmission/disequilibrium test that can be applied to situations of uncertain allele or haplotype transmission, allowing the analysis of data from patients with only one available parent, and to multilocus data even under uncertain phase, thereby permitting the use of all available genotypic data, including all nuclear family structures (data from unaffected siblings are used to infer parental genotypes and haplotypes). The statistic estimated by TRANSMIT follows a χ2 distribution and is calculated as (O – E)2 = Var(O – E), where O and E refer to the number of observed and expected transmissions of alleles/haplotypes. As an alternative method of determining the p value, the program employs a bootstrap method which provides more accurate results than the χ2 approximation for small samples.
Linkage disequilibrium across the NOS1AP gene was evaluated using Haploview v3.2 [37] based on parental genotypes. To compare the LD structure in our samples to the LD structure in the European population to which the Antioquian population is very close with 80% autosomal European ancestry [13], we downloaded the HapMap data for Europeans for the same region from the HapMap project website (http://www.hapmap.org) and displayed the LD plot using Haploview. Regions with the highest levels of linkage disequilibrium were used for haplotype-based TDT with WHAP v.2.09 [38]. For these analyses, the prevalence of the disease in the population was set to 0.5% [1], and the minimum frequency for a haplotype to be included in the analysis was fixed at 1%. As WHAP only accepts data from parent-offspring trios or duos and cannot accommodate other relatives, WHAP analyses included only 99 index cases: 38 complete trios and 61 duos. Two tests were performed: an omnibus haplotype test (testing the effects of each haplotype against a reference haplotype) and a haplotype-specific test (testing the effect of each haplotype against all others).
Clinical dimensions were obtained by principal component factor analysis performed on the individual items of the SANS and SAPS. Sampling adequacy was evaluated with the Kaiser-Meyer-Olkin measure (KMO) [39]. The resulting KMO of 0.89 indicated good variable factorability (i.e., the variables under study have a low partial correlation coefficient). Factor analysis was then performed using SPSS 13.0, using the Scree criterion for factor selection [40]. In order to assign items to factors, only the items with a loading of 0.40 or greater were taken into account. The factor solution was then rotated (using the VARIMAX procedure)and factor scores calculated using regression. All factors have eigenvalues >1.0, indicating that they account for more variance than any single SANS or SAPS item. Four of the index cases had no quantitative data available, thus reducing the sample size for these analyses to 98.
The factor scores were normalised before QTDT analysis. First, all 98 individuals were ranked based on their dimensional score. Ranks were converted to percentiles [rank/(N + 1), where N is the number of individuals] and z-scores obtained using the inverse standard normal cumulative distribution. The z-scores were then used as input for the quantitative TDT analysis with the QTDT program v.2.5.1 [41].
Results
All markers tested were found to be in Hardy-Weinberg equilibrium, with exception of marker rs4656362 [#12] (p = 0.0197). Marker rs6664602 [#6] was found not to be polymorphic in the Antioquian population. These two markers were therefore excluded from all further analyses.
LD Analysis
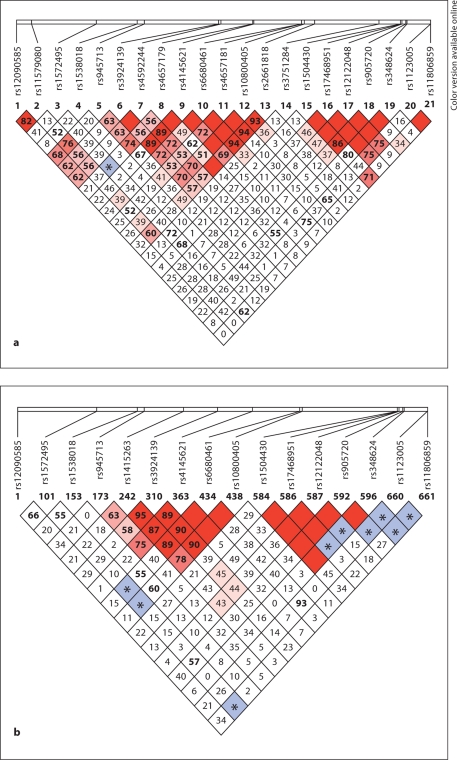
The pattern of LD between the SNPs genotyped in the Antioquian sample, as determined by Haploview, is represented in figure 2a. Two regions of strong LD are apparent, the first one covering markers rs945713 [#5] to rs2661818 [#16], spanning 129.1 kb and comprising introns 2 and 3 of NOS1AP (region 1), the second one covering markers rs1504430 [#18] to rs348624 [#22] and spanning 5.1 kb mostly within intron 8 (region 2).
Fig. 2.
a LD structure between 22 genotyped SNPs across the NOS1AP gene in the Antioquian sample. Marker numbers (1 to 24) are as in figure 1. Values in the boxes refer to % D′. Where no number is shown, D′ = 1.0. Red and pink (black and grey in printed version) boxes indicate a LOD score of ≥ 2.0 (D′ significant); blue (marked by asterisk) and white boxes indicate a LOD score of < 2.0 (D′ not significant). b LD structure across the NOS1AP gene based on data for the European HapMap population (CEU; http://www.hapmap.org). Only HapMap SNPs also genotyped in this study are shown. Since not all SNPs included here are HapMap SNPs, the set of SNPs displayed for the European population is smaller than the one typed in the Antioquian population.
For comparison, the LD structure of the European HapMap population (CEU) for a subset of the markers typed in this study is displayed in figure 2b. It shows great similarity to the pattern seen in the Antioquian sample, with two regions of high LD in the same locations as observed in Antioquia.
Single Marker Association Tests
The results of the single marker TDT are summarised in table 1. Of the 22 SNPs examined, eight showed significant association to schizophrenia (overtransmitted alleles are shown in parentheses): rs945713 [#5] (G), rs1415263 [#7] (T), rs3924139 [#8] (T), rs4592244 [#9] (A), rs4145621 [#11] (C), rs6680461 [#13] (G), rs4657181 [#14] (A), and rs3751284 [#17] (T). For all eight markers, both the p value based on the χ2 approximation and the bootstrap p value are nominally significant. Of these eight markers, seven are in high LD with each other and are located in LD region 1, while the remaining marker, rs3751284 [#17], is not in strong LD with any other marker (fig. 2a).
Table 1.
Single marker TDT (TRANSMIT) for 22 SNPs within the CAPON gene
| SNP | Numbera | allele | allele frequencyb | O | E | Var(O-E) | χ2 (1 d.f.) | p value | bootstrap p valuec |
|---|---|---|---|---|---|---|---|---|---|
| rs12090585 | 1 | G | 0.667 | 132 | 132.98 | 19.558 | 0.0487 | 0.83 | 0.82 |
| A | 0.333 | 68 | 67.024 | ||||||
| rs11579080 | 2 | G | 0.623 | 110 | 109.87 | 13.812 | 0.0012 | 0.97 | 0.97 |
| T | 0.377 | 64 | 64.129 | ||||||
| rs1572495 | 3 | G | 0.907 | 180 | 182.74 | 8.5861 | 0.8728 | 0.35 | 0.40 |
| A | 0.093 | 24 | 21.263 | ||||||
| rs1538018 | 4 | G | 0.650 | 146 | 142.31 | 18.396 | 0.7392 | 0.39 | 0.39 |
| C | 0.350 | 60 | 63.688 | ||||||
| rs945713 | 5 | G | 0.470 | 120 | 109.93 | 18.947 | 5.3500 | 0.021 | 0.011 |
| A | 0.530 | 82 | 92.068 | ||||||
| rs1415263 | 7 | C | 0.478 | 87 | 97.35 | 19.523 | 5.4866 | 0.019 | 0.015 |
| T | 0.522 | 117 | 106.65 | ||||||
| rs3924139 | 8 | C | 0.426 | 95 | 105.48 | 18.506 | 5.9341 | 0.015 | 0.007 |
| T | 0.574 | 107 | 96.521 | ||||||
| rs4592244 | 9 | G | 0.426 | 95 | 105.48 | 18.506 | 5.9341 | 0.015 | 0.007 |
| A | 0.574 | 107 | 96.521 | ||||||
| rs4657179 | 10 | G | 0.802 | 164 | 165.42 | 13.308 | 0.1518 | 0.70 | 0.70 |
| T | 0.198 | 40 | 38.579 | ||||||
| rs4145621 | 11 | C | 0.570 | 138 | 126.19 | 18.856 | 7.3991 | 0.007 | 0.004 |
| T | 0.430 | 64 | 75.812 | ||||||
| rs6680461 | 13 | T | 0.740 | 136 | 144.44 | 15.679 | 4.5391 | 0.033 | 0.028 |
| G | 0.260 | 68 | 59.564 | ||||||
| rs4657181 | 14 | A | 0.567 | 143 | 131.54 | 19.618 | 6.6972 | 0.010 | 0.006 |
| T | 0.433 | 63 | 74.462 | ||||||
| rs10800405 | 15 | C | 0.724 | 132 | 139.15 | 15.149 | 3.3715 | 0.07 | 0.05 |
| G | 0.276 | 66 | 58.853 | ||||||
| rs2661818 | 16 | C | 0.651 | 128 | 124.71 | 16.631 | 0.6498 | 0.42 | 0.39 |
| G | 0.349 | 62 | 65.287 | ||||||
| rs3751284 | 17 | C | 0.638 | 105 | 114.27 | 20.327 | 4.2276 | 0.040 | 0.019 |
| T | 0.362 | 95 | 85.73 | ||||||
| rs1504430 | 18 | C | 0.681 | 129 | 134.41 | 19.001 | 1.5430 | 0.21 | 0.25 |
| T | 0.319 | 75 | 69.585 | ||||||
| rs17468951 | 19 | A | 0.639 | 139 | 136.23 | 18.138 | 0.4228 | 0.52 | 0.54 |
| G | 0.361 | 65 | 67.769 | ||||||
| rs12122048 | 20 | G | 0.633 | 138 | 135.1 | 17.474 | 0.4817 | 0.49 | 0.47 |
| A | 0.367 | 64 | 66.901 | ||||||
| rs905720 | 21 | C | 0.696 | 143 | 139.31 | 16.922 | 0.8057 | 0.37 | 0.39 |
| T | 0.304 | 59 | 62.692 | ||||||
| rs348624 | 22 | C | 0.845 | 171 | 171.12 | 11.231 | 0.0013 | 0.97 | 0.98 |
| T | 0.155 | 33 | 32.877 | ||||||
| rs1123005 | 23 | A | 0.854 | 169 | 173.3 | 11.254 | 1.6394 | 0.20 | 0.28 |
| A | 0.718 | 55 | 56.138 | ||||||
| rs11806859 | 24 | G | 0.282 | 147 | 145.86 | 15.976 | 0.0810 | 0.78 | 0.80 |
| A | 0.718 | 55 | 56.138 | ||||||
Nominally significant χ2 and p values are italicized and bold. Markers belonging to LD regions 1 (top) and 2 (bottom) are shaded.
O = observed transmissions; E = expected transmissions; d.f. = degree of freedom.
SNP number as in figure 1
in untransmitted chromosomes
based on 1000 bootstrap samples.
Haplotype Association
Haplotype analyses were carried out based on the two LD regions shown in table 1 and figure 2a. The results of association tests for LD region 1 (markers rs945713 [#5] to rs2661818 [#16]) are shown in table 2. The omnibus haplotype test resulted in a p value of 0.35, indicating no significant overall association of this region with schizophrenia. In the haplotype-specific test, the p value for the most common haplotype was 0.04 with an odds ratio of 0.54, indicating a possible trend towards (negative) association between this particular haplotype and schizophrenia. For five of seven markers in LD region 1, the haplotype showing possible negative association consisted of alleles found to be undertransmitted in the single marker analyses (rs945713 [#5] – A; rs3924139 [#8] – C; rs4145621 [#11] – T; rs6680461 [#13] – T; and rs4657181 [#14] – T). No significant association was detected between LD region 2 (markers rs1504430 [#18] to rs348624 [#22]) and schizophrenia (data not shown).
Table 2.
Results of the transmission disequilibrium test of the haplotype containing the ten SNPs forming LD region 1
| Omnibus test (OT) overall p = 0.348 |
Haplotype-specific test (HS) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype # | rs945713 [#5] | rs1415263 [#7] | rs3924139 [#8] | rs4592244 [#9] | rs4657179 [#10] | rs4145621 [#11] | rs6680461 [#13] | rs4657181 [#14] | rs10800405 [#15] | rs2661818 [#16] | Frequency | OROT | lower limita | upper limita | ORHS | p value |
| 1 | A | C | T | A | G | T | T | T | C | G | 0.281 | reference haplotype | 0.54 | 0.04 | ||
| 2 | G | T | C | G | T | C | G | A | G | C | 0.131 | 1.79 | 0.73 | 4.37 | 1.13 | 0.75 |
| 3 | G | T | C | G | G | C | G | A | G | C | 0.096 | 2.29 | 0.85 | 6.18 | 1.48 | 0.38 |
| 4 | G | T | C | G | G | C | T | A | C | C | 0.084 | 3.37 | 1.21 | 9.40 | 2.24 | 0.09 |
| 5 | G | T | C | G | G | C | T | T | C | G | 0.059 | 3.40 | 1.07 | 10.82 | 2.22 | 0.14 |
| 6 | A | T | C | G | G | C | G | A | G | C | 0.056 | 1.23 | 0.36 | 4.24 | 0.77 | 0.65 |
| 7 | A | C | T | A | G | C | T | A | C | C | 0.053 | 1.67 | 0.43 | 6.42 | 0.99 | 0.99 |
| 8 | A | C | T | A | G | T | T | A | C | C | 0.048 | 0.63 | 0.16 | 2.40 | 0.39 | 0.15 |
| 9 | G | C | T | A | G | C | T | A | C | C | 0.03 | 1.33 | 0.25 | 6.98 | 0.87 | 0.86 |
| 10 | G | T | C | G | T | C | T | A | C | C | 0.03 | 1.12 | 0.21 | 5.93 | 0.72 | 0.68 |
| 11 | A | T | C | G | G | C | T | A | C | C | 0.028 | 4.86 | 1.06 | 22.24 | 3.03 | 0.14 |
| 12 | G | C | T | A | G | T | T | T | C | G | 0.027 | 1.55 | 0.25 | 9.78 | 0.88 | 0.88 |
| 13 | G | T | T | A | G | C | T | A | C | C | 0.025 | 4.19 | 0.84 | 20.80 | 2.63 | 0.23 |
| 14 | G | T | T | A | T | C | T | A | C | C | 0.015 | 3.30 | 0.40 | 27.33 | 2.00 | 0.51 |
| 15 | A | C | T | A | T | C | G | A | G | C | 0.014 | 0.95 | 0.06 | 14.07 | 0.56 | 0.66 |
| 16 | G | T | C | G | T | C | T | T | C | C | 0.013 | 0.92 | 0.08 | 10.21 | 0.59 | 0.65 |
| 17 | G | C | T | A | G | C | G | A | G | C | 0.011 | 0.28 | 0.03 | 2.77 | 0.19 | 0.15 |
OT = Omnibus test; HS = haplotype-specific test; OR = odds ratio.
Nominally significant p values are printed in bold italics.
Upper and lower limit of the 95% confidence interval for the OR of the omnibus test. WH AP does not calculate the 95% confidence intervals for the ORs of the haplotype-specific test; instead, a p value is given for each of the tested haplotypes.
QTDT Analysis on Clinical Dimensions
Principal component factor analysis identified six clinical dimensions accounting for 57.9% of the phenotypic variance: (1) affective flattening and alogia; (2) auditory, somatic and visual hallucinations, and first rank and paranoid delusions; (3) formal thought disorders; (4) avolition and social isolation; (5) bizarre behaviour, and (6) olfactory hallucinations and other delusions. Individual items and their factor loadings are listed in table 3.
Table 3.
Clinical dimensions obtained from SANS and SAPS. Item loadings after VARIMAX rotation. Bold type indicates the item loadings that contribute to each dimension (an item loading is counted into a specific dimension if its value >0.4).
| Dimension |
|||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| Affective flattening and alogia | Auditory, somatic and visual hallucinations, and first rank and paranoid delusions | Formal thought disorders | Avolition and social isolation | Bizarre behaviour | Olfactory hallucinations and other delusions | ||
| Eigenvalue | 12.5 | 7.3 | 3.7 | 2.04 | 1.9 | 1.5 | |
| % of variance | 25.1 | 14.7 | 7.3 | 4.1 | 3.7 | 3 | |
| Items | Facial expression | 0.80 | 0.05 | 0.003 | 0.37 | 0.03 | 0.004 |
| Spontaneous movements | 0.83 | 0.01 | −0.04 | 0.23 | 0.02 | −0.09 | |
| Expressive gestures | 0.84 | 0.04 | −0.05 | 0.34 | 0.01 | 0.01 | |
| Eye contact | 0.79 | 0.04 | 0.09 | 0.20 | 0.17 | −0.10 | |
| Affective non-responsiveness | 0.79 | 0.04 | 0.001 | 0.34 | 0.09 | 0.005 | |
| Vocal inflections | 0.84 | −0.009 | 0.02 | 0.27 | 0.07 | 0.11 | |
| Poverty of speech | 0.74 | −0.09 | 0.13 | 0.19 | 0.35 | −0.03 | |
| Poverty of content | 0.64 | −0.10 | 0.31 | 0.27 | 0.24 | 0.10 | |
| Blocking | 0.49 | 0.09 | 0.33 | −0.08 | 0.25 | 0.05 | |
| Latency of response | 0.71 | −0.02 | 0.14 | 0.07 | 0.27 | −0.13 | |
| Grooming and hygiene | 0.38 | 0.07 | 0.13 | 0.34 | 0.42 | 0.17 | |
| Impersistence at work | 0.23 | −0.01 | 0.04 | 0.60 | 0.20 | 0.08 | |
| Physical anergia | 0.37 | 0.08 | 0.07 | 0.64 | 0.01 | −0.04 | |
| Recreational interests | 0.36 | 0.16 | 0.07 | 0.71 | 0.073 | −0.04 | |
| Sexual interest | 0.38 | 0.05 | 0.02 | 0.66 | 0.01 | 0.05 | |
| Intimacy and closeness | 0.38 | 0.12 | 0.11 | 0.67 | 0.15 | 0.07 | |
| Relationship with friends | 0.37 | 0.11 | 0.08 | 0.72 | 0.03 | 0.03 | |
| Social inattentiveness | 0.49 | 0.001 | 0.34 | 0.33 | 0.42 | 0.02 | |
| Inattentiveness during testing | 0.30 | −0.05 | 0.26 | 0.29 | 0.46 | 0.03 | |
| Auditory hallucinations | −0.13 | 0.72 | 0.06 | 0.24 | 0.16 | 0.07 | |
| Voices commenting | −0.20 | 0.75 | 0.11 | 0.16 | 0.09 | −0.03 | |
| Voices conversing | −0.08 | 0.75 | 0.16 | 0.15 | 0.08 | 0.09 | |
| Somatic hallucinations | 0.03 | 0.51 | 0.12 | 0.18 | 0.13 | 0.41 | |
| Olfactory hallucinations | 0.01 | 0.29 | 0.05 | 0.26 | 0.06 | 0.48 | |
| Visual hallucinations | −0.02 | 0.50 | 0.06 | 0.12 | 0.19 | 0.39 | |
| Persecutory delusions | −0.07 | 0.75 | 0.04 | 0.16 | 0.17 | 0.12 | |
| Delusions of jealously | 0.08 | 0.05 | −0.03 | −0.12 | 0.16 | 0.55 | |
| Delusions of guilt | −0.12 | 0.08 | 0.02 | 0.0001 | 0.12 | 0.46 | |
| Grandiose delusions | −0.05 | 0.08 | 0.06 | 0.03 | 0.05 | 0.61 | |
| Religious delusions | 0.002 | 0.19 | 0.04 | −0.02 | −0.09 | 0.68 | |
| Somatic delusions | 0.002 | 0.37 | 0.17 | 0.15 | 0.07 | 0.52 | |
| Delusions of reference | −0.07 | 0.71 | −0.04 | 0.09 | 0.17 | 0.11 | |
| Delusions of being controlled | 0.06 | 0.77 | 0.19 | 0.01 | 0.06 | 0.16 | |
| Delusions of mind reading | 0.14 | 0.78 | 0.06 | −0.08 | −0.004 | 0.12 | |
| Thought broadcasting | 0.14 | 0.75 | 0.07 | −0.13 | −0.01 | 0.07 | |
| Thought insertion | 0.13 | 0.78 | 0.04 | −0.03 | 0.004 | 0.13 | |
| Thought withdrawal | 0.15 | 0.77 | 0.06 | −0.14 | −0.06 | 0.10 | |
| Clothing and appearance | 0.24 | 0.21 | 0.23 | −0.12 | 0.57 | 0.07 | |
| Social and sexual behaviour | 0.22 | 0.25 | 0.17 | 0.08 | 0.59 | 0.15 | |
| Aggressive behaviour | 0.09 | 0.26 | 0.02 | 0.13 | 0.49 | 0.20 | |
| Stereotyped behaviour | 0.19 | 0.11 | 0.05 | 0.21 | 0.54 | 0.29 | |
| Derailment | 0.26 | 0.21 | 0.74 | 0.08 | 0.01 | 0.22 | |
| Tangentiality | 0.27 | 0.16 | 0.66 | 0.15 | 0.06 | 0.26 | |
| Incoherence | 0.27 | 0.04 | 0.76 | −0.01 | 0.08 | 0.11 | |
| Illogicality | 0.25 | 0.14 | 0.71 | 0.02 | −0.005 | 0.19 | |
| Circumstantiality | −0.19 | 0.14 | 0.68 | −0.04 | 0.05 | −0.07 | |
| Pressure of speech | −0.26 | 0.10 | 0.60 | 0.09 | 0.04 | 0.04 | |
| Distractible speech | 0.16 | −0.04 | 0.61 | 0.16 | 0.33 | −0.06 | |
| Clanging | −0.06 | 0.04 | 0.54 | 0.07 | 0.26 | −0.15 | |
| Inappropriate affect | 0.17 | 0.09 | 0.48 | −0.05 | 0.51 | −0.06 | |
The results of the QTDT analysis on these clinical dimensions are summarised in table 4. Since the QTDT program can only test markers for which 30 or more probands are informative, markers rs11579080 [#2], rs1572495 [#3], rs4657179 [#10], rs348624 [#22], and rs1123005 [#23] could not be included in the analysis. Nominally significant associations were obtained for the following marker-dimension combinations (the allele associated with increased trait values is given after the p value): dimension 1 (affective flattening and alogia) with marker rs3751284 [#17] (p = 0.016; C), dimension 2 (auditory, somatic and visual hallucinations, and first rank and paranoid delusions) with marker rs1415263 [#7] (p = 0.034; C), dimension 4 (avolition and social isolation) with markers rs6680461 [#13] (p = 0.004; G) and rs10800405 [#15] (p = 0.001; G), and dimension 6 (olfactory hallucinations and other delusions) with marker rs6680461 [#13] (p = 0.048; C). Three of the markers showing nominally significant p-values in the analysis of clinical dimensions had also shown nominally significant p values in the categorical analyses (rs3751284 [#17], rs1415263 [#7], and rs6680461 [#13)). For markers rs3751284 [#17] and rs1415263 [#7], the alleles associated with increased trait values in the quantitative analysis are different from the ones shown to be overtransmitted to schizophrenic patients in the categorical analysis (tables 1 and 4), whereas for marker rs6680461 [#13], the allele shown to increase the trait value corresponds to the overtransmitted allele in the categorical analysis (allele G).
Table 4.
QTDT analysis of clinical dimensions for 17 SNPs in the NOS1AP gene region
| SNP | Numbera | Inf. trios | Dim. 1 | Dim. 2 | Dim. 3 | Dim. 4 | Dim. 5 | Dim. 6 |
|---|---|---|---|---|---|---|---|---|
| rs12090585 | 1 | 47 | * | * | * | * | * | * |
| rs1538018 | 4 | 40 | * | * | * | * | * | * |
| rs945713 | 5 | 44 | * | * | * | * | * | * |
| rs1415263 | 7 | 43 | * | 0.034 (C) | * | * | * | * |
| rs3924139 | 8 | 38 | * | * | * | * | * | * |
| rs4592244 | 9 | 38 | * | * | * | * | * | * |
| rs4145621 | 11 | 39 | * | * | * | * | * | * |
| rs6680461 | 13 | 34 | * | * | * | 0.004 (G) | * | * |
| rs4657181 | 14 | 45 | * | * | * | * | * | * |
| rs10800405 | 15 | 31 | * | * | * | 0.001 (G) | * | * |
| rs2661818 | 16 | 32 | * | * | * | * | * | * |
| rs3751284 | 17 | 50 | 0.016 (C) | * | * | * | * | * |
| rs1504430 | 18 | 42 | * | * | * | * | * | * |
| rs17468951 | 19 | 44 | * | * | * | * | * | * |
| rs12122048 | 20 | 39 | * | * | * | * | * | * |
| rs905720 | 21 | 43 | * | * | * | * | * | 0.048 (C) |
| rs11806859 | 24 | 31 | * | * | * | * | * | * |
Overall Bonferroni significance level: 0.10; overall empirical significance level: 0.08
SNP number as in figure 1. Inf. trios., number of informative trios for the marker; Dim., dimension. For the explanation of the dimensions, see text and table 3. p values are shown for each marker/dimension combination; the allele that increases the trait (dimension) value is shown in parentheses.
p value >0.05.
Discussion
NOS1AP is an interesting candidate gene for schizophrenia susceptibility because its product, the NOS1AP protein, seems to be involved in NMDA receptor mediated glutamatergic neurotransmission, which is thought to be implicated specifically in negative symptoms in schizophrenia [10, 42]. Intracellular NMDA receptor induced signal transmission relies on the interaction of the receptor molecule with the neuronal Nitric Oxide Synthase (nNOS) through a mediator protein, PSD95. NOS1AP competes with PSD95 for interaction with nNOS and is thought to be involved in the regulation of nNOS activity in the neuron [43]. Dysregulation of NOS1AP availability could therefore lead to a disruption of signalling processes following glutamatergic neurotransmission downstream of the NMDA receptor. In line with a possible role for abnormal NOS1AP expression in schizophrenia, Xu and colleagues have recently identified a short isoform of the NOS1AP protein and shown its increased expression in the schizophrenic brain in comparison to healthy subjects in a post-mortem brain study [44].
The observation of association of several SNPs in the NOS1AP gene to the schizophrenia phenotype and also to specific clinical dimensions of the disorder confirms and extends our previous results with microsatellite marker D1S1679 [16] and further strengthens the link between NOS1AP and schizophrenia.
Of the SNPs found to be associated with schizophrenia in our study, two were also found to be significant in the study by Brzustowicz and colleagues: rs1415263 [#7] and rs4145621 [#11] [11]. Moreover, the associated allele was the same in both studies: the T allele of rs1415263 [#7], and the C allele of rs4145621 [#11] (table 1). Interestingly, Xu and colleagues [44] found the expression levels of the short NOS1AP isoform overexpressed in the schizophrenic brain to be associated with the T allele of marker rs1415263 [#7).
The only statistically significant SNP outside LD region 1, rs3751284 [#17], is a synonymous change in exon 6 of the gene. The markers in LD region 1 showing association are located in intron 2 of NOS1AP, and it is possible that these markers are in LD with a further, unidentified susceptibility-conferring variant, such as a regulatory element upstream of the NOS1AP gene. This scenario could also explain the identification of a protective, rather than a risk-conferring, haplotype – the common protective haplotype 1 might be in LD with the protective allele at the susceptibility locus, whereas the risk allele at the same locus might be in LD with not one, but several of the remaining, rarer haplotypes, thereby diluting the susceptibility-conferring effect in the observed haplotypes. Another possible explanation for our findings is that intronic and synonymous exonic SNPs could affect posttranscriptional mRNA processes [45]. In both scenarios, genetic variation could lead to changes in the availability of functional NOS1AP in the neuron and thereby to alterations in glutamatergic neurotransmission.
The analysis of clinical dimensions showed an association between markers rs6680461 [#13] and rs10800405 [#15] and features of dimension 4 (avolition and social isolation), which captures negative symptoms of the disorder. For both markers, the allele associated with increased symptom severity is also overtransmitted to schizophrenic patients in the single marker categorical TDT analysis (although the results do not reach statistical significance for rs10800405 [#15); see tables 1 and 3). These findings are consistent with a role of NOS1AP in the NMDA receptor pathway and the NMDA receptor hypofunction theory of the negative syndrome of schizophrenia [10, 42].
For dimension 1 (affective flattening and alogia), the C allele of marker rs3751284 [#17] was found to increase trait values. However, in the categorical single marker analysis, this allele was found to be undertransmitted to schizophrenic patients. In a similar manner, the C allele of marker rs1415263 [#7] is associated with increased values for dimension 2 (auditory, somatic and visual hallucinations, and first rank and paranoid delusions) but was found to be undertransmitted to schizophrenic patients. The p values from the quantitative analyses are only marginallyally significant, particularly in view of the many tests carried out in this analysis. For marker rs905720 [#21], the direction of association is the same in both categorical and quantitative analyses. However, the categorical results are not significant, and the uncorrected p value in the quantitative analysis is only marginally significant. Further studies are needed to shed light on a possible role of NOS1AP in dimensions 1, 2 and 6.
While there is mounting evidence for a possible link between NOS1AP and schizophrenia, not all results are consistent. A large case-control study by Puri and colleagues failed to replicate the positive findings in a British sample [17]. Instead, these authors have found an association between schizophrenia and the UHMK1 gene, also located on chromosome 1q23, in the British sample and they suggested that the original linkage signal, as well as the subsequent association results for NOS1AP by Brzustowicz and colleagues might be due to UHMK1 [46]. However, the analysis of LD patterns in the European Hapmap population in the region encompassing NOS1AP and UHMK1 revealed no significant LD between these two genes (data not shown). Antioquian nuclear DNA has a highly predominant European ancestry, and LD patterns in Europeans are proxy for LD patterns in Antioquia, as seen in figure 2. We therefore conclude that our results are not due to LD between NOS1AP and UHMK1.
In conclusion, we have found significant association of markers within the NOS1AP gene to schizophrenia and to a clinical dimension capturing negative symptoms of the disorder (avolition and social isolation). Our results are consistent with previous studies conducted in different populations and provide further evidence for the implication of the NOS1AP gene in the aetiology of schizophrenia. Future work should aim to investigate whether this association, particularly the association of NOS1AP with negative symptoms of the disease. To shed further light on a possible role of NOS1AP in the deficit syndrome, it would be of special interest to focus follow-up studies on deficit patients.
URLs
GENEPOP homepage: http://wbiomed.curtin.edu.au/genepop/
The HapMap project: http://www.hapmap.org/
QTDT homepage: http://www.sph.umich.edu/csg/abecasis/QTDT/
WHAP homepage: http://pngu.mgh.harvard.edu/∼purcell/whap/index.shtml
Acknowledgements
This work was partly funded by Colciencias grant 1115-04-11944 (Colombia), CODI-Universidad de Antioquia grants E-00810 and CIM-E01242 (Colombia) and grant R01 MH62440 from the National Institutes of Mental Health (USA). We would like to thank Mari-Wyn Burley for technical assistance and Gonçalo Abecasis for advice on the use of his QTDT programme. We are particularly grateful to all the patients and family members who participated in this study.
References
- 1.Saha S, Chant D., Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLOS Medicine. 2005;2:413–433. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craddock N, O'Donovan MC, Owen MJ. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O'Neill FA, Walsh D, Kendler KS. Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry. 2002;7:542–559. doi: 10.1038/sj.mp.4001051. [DOI] [PubMed] [Google Scholar]
- 4.Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, Lamacz M, Thomas MG, Gehrig C, Radhakrishna U, Snyder SE, Balk KG, Neufeld K, Swartz KL, DeMarchi N, Papadimitriou GN, Dikeos DG, Stefanis CN, Chakravarti A, Childs B, Housman DE, Kazazian HH, Antonarakis S, Pulver AE. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 5.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, Bassett AS. Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet. 1999;65:1096–1103. doi: 10.1086/302579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Q, Martinez M, Zhang J, Sanders AR, Badner JA, Cravchik A, Markey CJ, Beshah E, Guroff JJ, Maxwell ME, Kazuba DM, Whiten R, Goldin LR, Gershon ES, Gejman PV. Suggestive evidence for a schizophrenia susceptibility locus on chromosome 6q and a confirmation in an independent series of pedigrees. Genomics. 1997;43:1–8. doi: 10.1006/geno.1997.4815. [DOI] [PubMed] [Google Scholar]
- 8.Lindholm E, Ekholm B, Shaw S, Jalonen P, Johansson G, Pettersson U, Sherrington R, Adolfsson R, Jazin E. A schizophrenia-susceptibility locus at 6q25, in one of the world's largest reported pedigrees. Am J Hum Genet. 2001;69:96–105. doi: 10.1086/321288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O'Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O'Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coyle JT. Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell Mol Neurobiol. 2006;26:365–384. doi: 10.1007/s10571-006-9062-8. [DOI] [PubMed] [Google Scholar]
- 11.Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet. 2004;74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y, Li H, Qin W, Chen W, Duan Y, Xiao Y, Li C, Zhang J, Li X, Feng G, He L. Association of the carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase gene with schizophrenia in the Chinese Han population. Biochem Biophys Res Commun. 2005;328:809–815. doi: 10.1016/j.bbrc.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 13.Bedoya G, Montoya P, Garcia J, Soto I, Bourgeois S, Carvajal L, Labuda D, Alvarez V, Ospina J, Hedrick PW, Ruiz-Linares A. Admixture dynamics in Hispanics: A shift in the nuclear genetic ancestry of a South American population isolate. Proc Natl Acad Sci USA. 2006;103:7234–7239. doi: 10.1073/pnas.0508716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvajal-Carmona LG, Ophoff R, Service S, Hartiala J, Molina J, Leon P, Ospina J, Bedoya G, Freimer N, Ruiz-Linares A. Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet. 2003;112:534–541. doi: 10.1007/s00439-002-0899-8. [DOI] [PubMed] [Google Scholar]
- 15.Carvajal-Carmona LG, Soto ID, Pineda N, Ortiz-Barrientos D, Duque C, Ospina-Duque J, McCarthy M, Montoya P, Alvarez VM, Bedoya G, Ruiz-Linares A. Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. Am J Hum Genet. 2000;67:1287–1295. doi: 10.1016/s0002-9297(07)62956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda A, Garcia J, Lopez C, Gordon D, Palacio C, Restrepo G, Ortiz J, Montoya G, Cardeno C, Calle J, Lopez M, Campo O, Bedoya G, Ruiz-Linares A, Ospina-Duque J. Putative association of the carboxy-terminal PDZ ligand of neuronal nitric oxide synthase gene (CAPON) with schizophrenia in a Colombian population. Schizophr Res. 2006;82:283–285. doi: 10.1016/j.schres.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Puri V, McQuillin A, Thirumalai S, Lawrence J, Krasucki R, Choudhury K, Datta S, Kerwin S, Quested D, Bass N, Pimm J, Lamb G, Moorey H, Kandasami G, Badacsonyi A, Kelly K, Morgan J, Punukollu B, Nadeem H, Curtis D, Gurling HMD. Failure to confirm allelic association between markers at the CAPON gene locus and schizophrenia in a British sample. Biol Psychiatry. 2006;59:195–197. doi: 10.1016/j.biopsych.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Tamminga CA, Holcomb HH. Phenotype of schizophrenia: a review and formulation. Mol Psychiatry. 2005;10:27–39. doi: 10.1038/sj.mp.4001563. [DOI] [PubMed] [Google Scholar]
- 19.Fanous AH, Kendler KS. Genetic heterogeneity, modifier genes, and quantitative phenotypes in psychiatric illness: searching for a framework. Mol Psychiatry. 2005;10:6–13. doi: 10.1038/sj.mp.4001571. [DOI] [PubMed] [Google Scholar]
- 20.Peralta V, Cuesta MJ. How many and which are the psychopathological dimensions in schizophrenia? Issues influencing their ascertainment. Schizophr Res. 2001;49:269–285. doi: 10.1016/s0920-9964(00)00071-2. [DOI] [PubMed] [Google Scholar]
- 21.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 22.Palacio CA, García J, Arbeláez MP, Sánchez R, Aguirre B, Garcés I, Montoya GJ, Gómez J, Agudelo A, López CA, Calle JJ, Cardeño CA, Cano JF, López MC, Montoya P, Herrera CP, González N, González A, Bedoya G, Ruiz A, Ospina J. Validación de la entrevista diagnóstica para estudios genéticos (DIGS) en Colombia. Biomédica. 2004;24:56–62. [PubMed] [Google Scholar]
- 23.American Psychiatric Association . 4. Washington, DC: American Psychiatric Association; 2000. Diagnostic and Statistical Manual of Mental Disorders, ed. [Google Scholar]
- 24.Andreasen NC. The Scale for the assessment of positive symptoms (SAPS) Iowa City: The University of Iowa; 1984. [Google Scholar]
- 25.Andreasen NC. The Scale for the Assessment of Negative Symtoms (SANS): conceptual and theorical foundations. Br J Psychiatry Suppl. 1989;7:49–58. [PubMed] [Google Scholar]
- 26.García J, Palacio CA, Garcés I, Arbeláez MP, Sánchez R, López CA, Montoya GJ, González N, González A, Ospina J. Inferencias fenomenológicas y nosoógicas en la esquizofrenia a partir de la validación de las escalas de síntomas positivos (SAPS) y de síntomas negativos (SANS) en Colombia. Revista de Neuro-Psiquiatría. 2003;66:195–214. [Google Scholar]
- 27.Dawson B, Trapp RG, Trapp R. Basic And Clinical Biostatistics, ed 4. Norwalk: Appleton & Lange; 2004. Statistical methods for multiple variables; pp. 245–279. [Google Scholar]
- 28.Kline P. The Handbook of Psychological Testing. London: Routledge; 1993. [Google Scholar]
- 29.Wratten N, Bruse SE, Azaro MA, Simone J, Hayter JE, Brzustowicz LM: Identifying schizophrenia-associated non-coding variants in the CAPON gene (abstract). 55th Annual Meeting of the American Society of Human Genetics, Salt Lake City, Utah 2005, Oct 25–29.
- 30.Wang HY, Luo M, Tereshchenko IV, Frikker DM, Cui X, Li JY, Hu G, Chu Y, Azaro MA, Lin Y, Shen L, Yang Q, Kambouris ME, Gao R, Shih W, Li H. A genotyping system capable of simultaneously analyzing >1000 single nucleotide polymorphisms in a haploid genome. Genome Res. 2005;15:276–283. doi: 10.1101/gr.2885205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor JD, Briley D, Nguyen Q, Long K, Iannone MA, Li MS, Ye F, Afshari A, Lai E, Wagner M, Chen J, Weiner MP. Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. Biotechniques. 2001;30:661–669. doi: 10.2144/01303dd04. [DOI] [PubMed] [Google Scholar]
- 32.Bruse SE, McGregor N, Azaro MA, Xu B, Brzustowicz LM: An inexpensive bead-based oligonucleotide ligation assay for SNP genotyping (abstract). 56th Meeting of the American Society of Human Genetics, New Orleans, La. 2006, Oct 9–13.
- 33.Raymond M, Rousset F. GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 34.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 35.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 36.Clayton D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 39.Kaiser HF. Index of Factorial Simplicity. Psychometrika. 1974;39:31–36. [Google Scholar]
- 40.Cattell RB. The scree test for the number of factors. Multivar Behav Res. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- 41.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coyle JT, Tsai G, Goff D. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann N Y Acad Sci. 2003 Nov;1003:318–327. doi: 10.1196/annals.1300.020. [DOI] [PubMed] [Google Scholar]
- 43.Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 44.Xu B, Wratten N, Charych EI, Buyske S, Firestein BL, Brzustowicz LM. Increased Expression in Dorsolateral Prefrontal Cortex of CAPON in Schizophrenia and Bipolar Disorder. PLoS Med. 2005;2:e263. doi: 10.1371/journal.pmed.0020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puri V, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Krasucki R, Lawrence J, Quested D, Bass N, Moorey H, Morgan J, Punukollu B, Kandasami G, Curtis D, Gurling H. Fine Mapping by Genetic Association Implicates the Chromosome 1q23.3 Gene UHMK1, Encoding a Serine/Threonine Protein Kinase, as a Novel Schizophrenia Susceptibility Gene. Biol Psychiatry. 2006;61:873–879. doi: 10.1016/j.biopsych.2006.06.014. [DOI] [PubMed] [Google Scholar]