Abstract
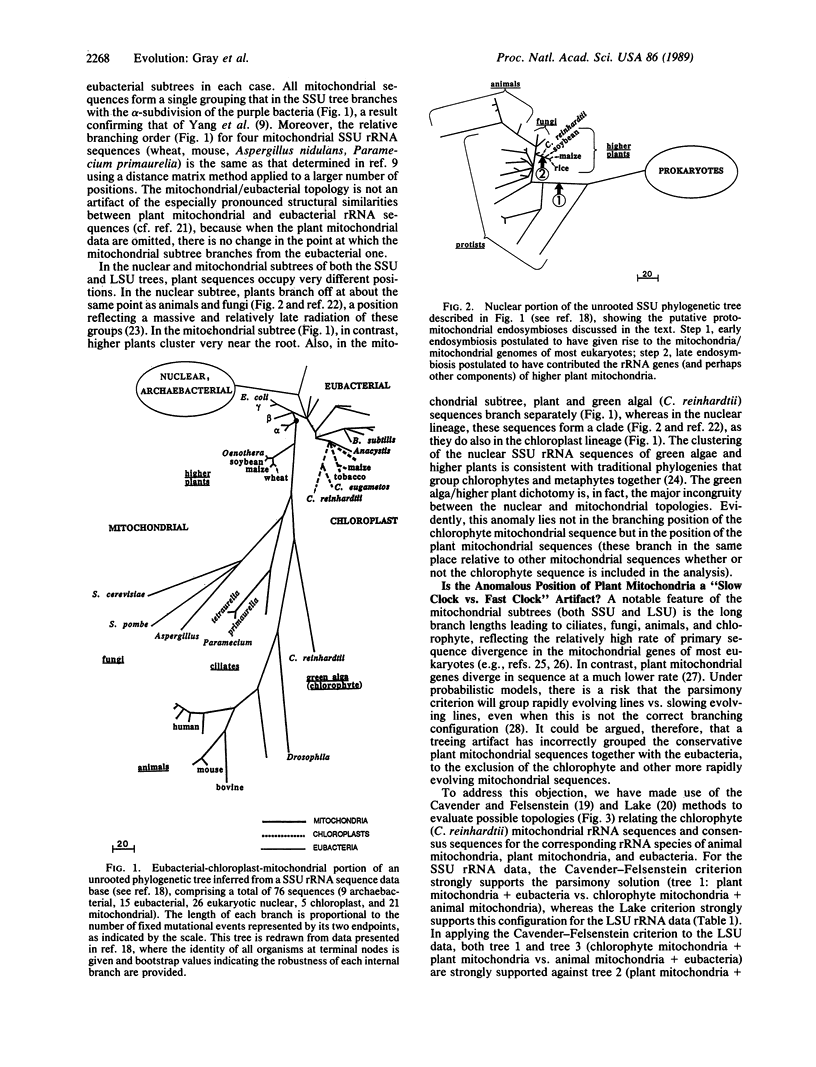
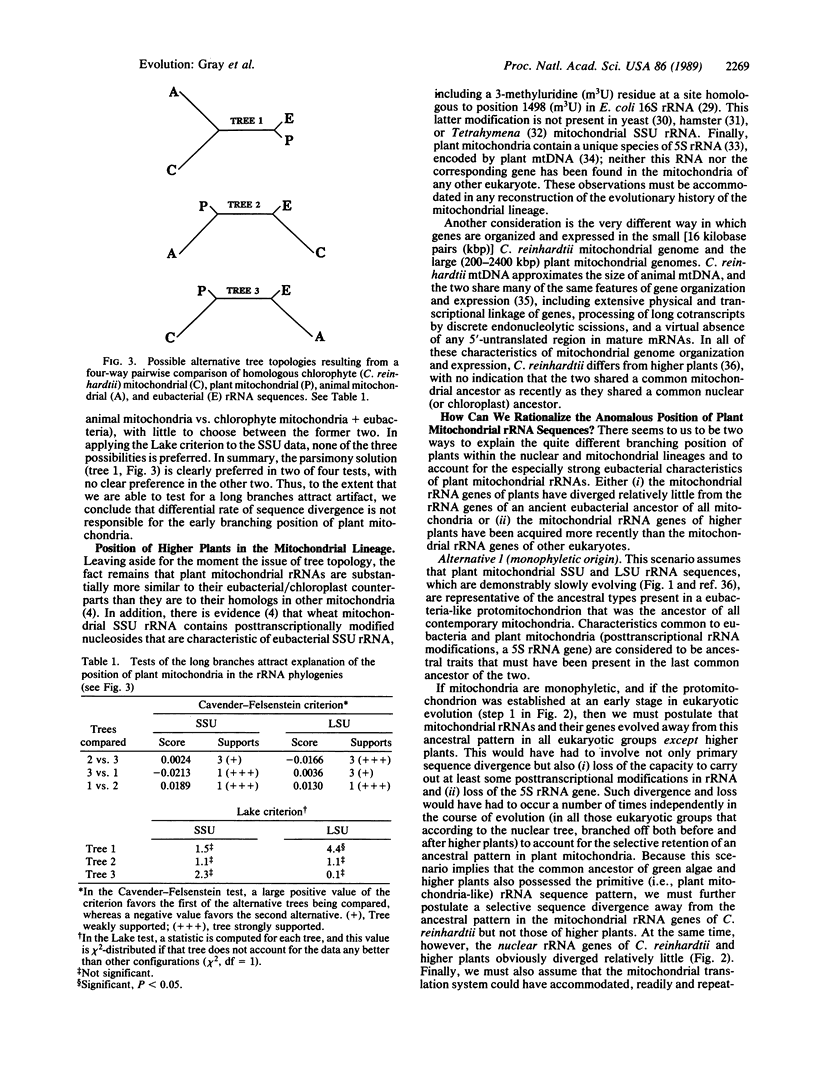
Higher plants occupy very different positions in the mitochondrial and nuclear lineages of global phylogenetic trees based on conserved regions of small subunit (SSU) and large subunit (LSU) rRNA sequences. In the nuclear subtree, plants branch off late, at a position reflecting a massive radiation of the major multicellular (and some unicellular) groups; in the mitochondrial subtree, in contrast, plants branch off early, near the point of connection between the mitochondrial and eubacterial lineages. Moreover, in the nuclear lineage, plants branch together with the unicellular green alga Chlamydomonas reinhardtii, whereas in the mitochondrial lineage (in both SSU and LSU trees), metaphytes and chlorophyte branch separately. Statistical evaluation indicates that the anomalous branching position of higher plants in the mitochondrial lineage is not a treeing artifact attributable to the relatively rapid rate of sequence divergence of non-plant mitochondrial rRNA sequences. In considering alternative biological explanations for these results, we are led to propose that the rRNA genes in plant mitochondria may be of more recent evolutionary origin than the rRNA genes in other mitochondria. This proposal has implications for monophyletic vs. polyphyletic scenarios of mitochondrial origin and is consistent with other evidence indicating that plant mtDNA is an evolutionary mosaic.
Keywords: ribosomal RNA genes, chlorophytes, metaphytes, phylogeny
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bonen L., Gray M. W. Organization and expression of the mitochondrial genome of plants I. The genes for wheat mitochondrial ribosomal and transfer RNA: evidence for an unusual arrangement. Nucleic Acids Res. 1980 Jan 25;8(2):319–335. doi: 10.1093/nar/8.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann N Y Acad Sci. 1987;503:55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- Cedergren R., Gray M. W., Abel Y., Sankoff D. The evolutionary relationships among known life forms. J Mol Evol. 1988 Dec;28(1-2):98–112. doi: 10.1007/BF02143501. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Taylor R. H., Davenport L. W. Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 1978 Nov;5(11):4385–4397. doi: 10.1093/nar/5.11.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Gillham N. W., Boynton J. E., Harris E. H. Specific elimination of mitochondrial DNA from Chlamydomonas by intercalating dyes. Curr Genet. 1987;12(1):41–47. doi: 10.1007/BF00420726. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Boer P. H. Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial DNA. Philos Trans R Soc Lond B Biol Sci. 1988 May 31;319(1193):135–147. doi: 10.1098/rstb.1988.0038. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Doolittle W. F. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982 Mar;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. Mitochondrial genome diversity and the evolution of mitochondrial DNA. Can J Biochem. 1982 Mar;60(3):157–171. doi: 10.1139/o82-022. [DOI] [PubMed] [Google Scholar]
- Gray M. W. Organelle origins and ribosomal RNA. Biochem Cell Biol. 1988 May;66(5):325–348. doi: 10.1139/o88-042. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Sankoff D., Cedergren R. J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984 Jul 25;12(14):5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Gunderson J. H., Elwood H., Ingold A., Kindle K., Sogin M. L. Phylogenetic relationships between chlorophytes, chrysophytes, and oomycetes. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5823–5827. doi: 10.1073/pnas.84.16.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon K. W. Change of cellular "pathogens" into required cell components. Ann N Y Acad Sci. 1987;503:359–371. doi: 10.1111/j.1749-6632.1987.tb40622.x. [DOI] [PubMed] [Google Scholar]
- Klootwijk J., Klein I., Grivell L. A. Minimal post-transcriptional modification of yeast mitochondrial ribosomal RNA. J Mol Biol. 1975 Sep 25;97(3):337–350. doi: 10.1016/s0022-2836(75)80044-1. [DOI] [PubMed] [Google Scholar]
- Köchel H. G., Küntzel H. Mitochondrial L-rRNA from Aspergillus nidulans: potential secondary structure and evolution. Nucleic Acids Res. 1982 Aug 11;10(15):4795–4801. doi: 10.1093/nar/10.15.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küntzel H., Köchel H. G. Evolution of rRNA and origin of mitochondria. Nature. 1981 Oct 29;293(5835):751–755. doi: 10.1038/293751a0. [DOI] [PubMed] [Google Scholar]
- Lake J. A. A rate-independent technique for analysis of nucleic acid sequences: evolutionary parsimony. Mol Biol Evol. 1987 Mar;4(2):167–191. doi: 10.1093/oxfordjournals.molbev.a040433. [DOI] [PubMed] [Google Scholar]
- Marechal L., Runeberg-Roos P., Grienenberger J. M., Colin J., Weil J. H., Lejeune B., Quetier F., Lonsdale D. M. Homology in the region containing a tRNA(Trp) gene and a (complete or partial) tRNA(Pro) gene in wheat mitochondrial and chloroplast genomes. Curr Genet. 1987;12(2):91–98. doi: 10.1007/BF00434662. [DOI] [PubMed] [Google Scholar]
- Morin G. B., Cech T. R. Phylogenetic relationships and altered genome structures among Tetrahymena mitochondrial DNAs. Nucleic Acids Res. 1988 Jan 11;16(1):327–346. doi: 10.1093/nar/16.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven P. H. A multiple origin for plastids and mitochondria. Science. 1970 Aug 14;169(3946):641–646. doi: 10.1126/science.169.3946.641. [DOI] [PubMed] [Google Scholar]
- Schnare M. N., Gray M. W. 3'-Terminal sequence of wheat mitochondrial 18S ribosomal RNA: further evidence of a eubacterial evolutionary origin. Nucleic Acids Res. 1982 Jul 10;10(13):3921–3932. doi: 10.1093/nar/10.13.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M. N., Heinonen T. Y., Young P. G., Gray M. W. A discontinuous small subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J Biol Chem. 1986 Apr 15;261(11):5187–5193. [PubMed] [Google Scholar]
- Schuster W., Brennicke A. Plastid, nuclear and reverse transcriptase sequences in the mitochondrial genome of Oenothera: is genetic information transferred between organelles via RNA? EMBO J. 1987 Oct;6(10):2857–2863. doi: 10.1002/j.1460-2075.1987.tb02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L., Elwood H. J., Gunderson J. H. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer D. F., Bonen L., Gray M. W. Primary sequence of wheat mitochondrial 5S ribosomal ribonucleic acid: functional and evolutionary implications. Biochemistry. 1981 Jul 7;20(14):4022–4029. doi: 10.1021/bi00517a011. [DOI] [PubMed] [Google Scholar]
- Spencer D. F., Schnare M. N., Gray M. W. Pronounced structural similarities between the small subunit ribosomal RNA genes of wheat mitochondria and Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(2):493–497. doi: 10.1073/pnas.81.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1946–1950. doi: 10.1073/pnas.81.7.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart K. D., Mattox K. R. The case for a polyphyletic origin of mitochondria: morphological and molecular comparisons. J Mol Evol. 1984;21(1):54–57. doi: 10.1007/BF02100627. [DOI] [PubMed] [Google Scholar]
- Wiseman A., Gillham N. W., Boynton J. E. Nuclear mutations affecting mitochondrial structure and function in Chlamydomonas. J Cell Biol. 1977 Apr;73(1):56–77. doi: 10.1083/jcb.73.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Li W. H., Sharp P. M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Oyaizu Y., Oyaizu H., Olsen G. J., Woese C. R. Mitochondrial origins. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]


