1. Introduction
Molecular imaging techniques are now indispensable tools in modern diagnostics, because they are highly specific and can provide biological information at the molecular level in living systems.1,2 They have enabled visualization of some of the specific molecular events that play key roles in disease processes, and they have enabled earlier diagnosis, as well as monitoring of therapeutic responses. Various imaging modalities, including positron emission tomography (PET), single photon emission computed tomography (SPECT), optical fluorescence imaging, magnetic resonance imaging (MRI), computed tomography, and ultrasound imaging are all successfully employed in the field of molecular imaging. Specific imaging is generally created by contrast agents; however, most current clinical imaging remains at the anatomical and macro functional level, due to the low targeting efficiency of such agents. To support the unmet needs for in vivo clinical molecular imaging, there has been considerable interest in investigating the design of highly sensitive and specific molecularly-targeted imaging probes. To date, a large variety of sophisticated imaging probes have been developed by combining various imaging moieties (i.e., radioisotopes, fluorophores, and nanoparticles) and targeting ligands (i.e., small molecules, peptides, proteins, antibodies, as well as cells). These efforts have profoundly impacted the availability of imaging probes and significantly improved the performance of imaging modalities. Several review articles have discussed recent development and applications of molecular imaging probes2-7, particularly the utilization of peptide- and peptide hormone-based imaging probes.
An ideal imaging probe would have high affinity and specificity for the target of interest. However, requirements beyond targeting selectivity become determinants for the suitability of probes for in vivo applications, including in vivo metabolic stability, high target-to-background ratio, rapid clearance from non-target tissues, and safety. Furthermore, tolerance and flexibility towards bulky chemical modification are also needed, because imaging probes are often associated with labeling of radioisotopes, fluorophores, and materials such as linkers, polymers, and metals. From a practical standpoint, synthetic peptides have attracted much attention as molecular imaging probes for small molecules and macromolecules.8-10 Recent advances in phage display technology, combinatorial peptide chemistry, and biology have led to the development of robust strategies for the design of peptides as drugs and biological tools, resulting in identification of a rich variety library of bioactive peptide ligands and substrates.11-13 To date, peptides that target a number of disease-related receptors, biomarkers, and the processes of angiogenesis and apoptosis are in place. These peptides reveal high specificity for their target at nanomolar concentrations and have low toxicity. They can be easily synthesized, modified to optimize their binding affinity, and possibly further modified structurally to improve their stability against proteolytic degradation, to increase half-life in circulation, and to enhance capillary permeability. All of these attributes promote penetration into tissue and more effective targeting. Furthermore, established peptide synthesis processes are easy to scale up, and they yield reproducible products with well-defined structures.
With the combination of advanced imaging sciences, peptide chemistry, and the increasing availability of animal imaging instruments, various kinds of highly specific peptide-based imaging probes for different imaging modalities have been designed and validated in preclinical and clinical investigations. In the following review, an overview of molecular imaging probes associated with peptides and peptide hormones designed for in vivo applications, including those for nuclear imaging, optical imaging, and MRI, is provided. For the sake of focus, this article will not discuss imaging probes that have been tested only under in vitro cellular conditions, although many of these can be applied in vivo. Key peptides for selective targeting of biological receptors or biomarkers and modification strategies for these peptides will be summarized. Then, the unique concepts, characteristics, and applications of various peptide-based imaging probes will be discussed for each of several modalities.
2. Design of Peptide-based Imaging Probes
Biologically-active peptides are the products of genes, and their targets are proteins or protein-coupled receptors. These naturally-occurring peptides play a regulatory role in the body and generally mediate their function through specific binding to receptors, such as G-protein-coupled receptors (GPCrs).14 This peptide-receptor binding can activate or inhibit biological processes via different mechanisms, including activation of G-proteins, tyrosine kinases, or transcription processes.15 Importantly, many of these receptors have been shown to be significantly overexpressed in certain diseases, especially in particular tumor types.16 This is principally why these overexpressed receptors and their receptor-binding peptides have been chosen as potential targets for molecular imaging and therapy by probes based upon their specific receptor-binding peptides. Advances in molecular cancer biology have revealed and elucidated an increasing number of tumor receptors and their specific regulatory peptides. For example, somatostatin (SST), integrin, gastrin-releasing peptide (GRP), cholecystokinin (CCK), alpha-melanocyte stimulating hormone (α-MSH), and glucagon-like peptide-1 (GLP-1) receptors have been identified and characterized for tumor receptor imaging.16-20 Specific peptide molecules for these receptors represent a class of perfect ligands for cancer targeting and have accelerated interest in the development of peptide-based imaging probes.8,10 These peptide molecules can be identified and selected by modifying known native peptides, molecular modeling of peptide-receptor structures, or screening combinatorial peptide libraries.12 Once the imaging target has been identified, the development of targeting peptides can generally be sped up by rational design and the use of screening libraries. Established methods that have been developed to identify novel targeting peptides include the phage display peptide library method,13 the spatially-addressable parallel library method,21 synthetic library methods requiring deconvolution,22 the affinity selection method,23 and the one-bead one-compound (OBOC) combinatorial library method.24 The use of phage display libraries is a powerful technique for receptor-specific peptide discovery. This method enables the researcher to generate an array of 108 – 109 different phages that can be easily screened to isolate targeting peptides. A number of methods can be further utilized to improve physicochemical properties of selected peptides. All of these structural modification processes need to be validated with adequate chemical and biological evaluations. Detailed rational approaches for the discovery of targeting peptides have been summarized elsewhere.11,12,25-27
A peptide is any combination of amino acids linked by peptide bonds. Generally, peptides considered as imaging ligands have a low molecular weight, containing several to fewer than 50 amino acids. These small receptor-binding peptides have a number of distinct advantages over proteins and antibodies. Small peptides have favorable pharmacokinetic and tissue distribution patterns as characterized by rapid clearance from blood and non-target tissues. Some peptides have good permeability properties that can permit rapid access of the peptide to target tissues. Peptides have low toxicity and immunogenicity. An important factor in designing peptide-based imaging probes is in vivo stability. Typically, naturally-occurring peptides have a short biological half-life, due to rapid degradation by various peptidases and proteases found in plasma and in most tissues. So, once the key amino acid residues that are involved in the biological activity have been determined, most peptides are molecularly engineered to prolong their biological half-lives in vivo. The methods commonly used to improve metabolic stability of peptides are the introduction or substitution of D-amino acids, use of unusual amino acids or side-chains, incorporation of amino alcohol, and acetylation and/or amination of peptide N- and C-termini. Furthermore, the rate of excretion and permeability of peptides can be modified by altering hydrophilic and hydrophobic balances of peptide structures by the introduction of specific hydrophilic and/or lipophilic amino acids or other chemicals. Peptide cyclization can be applied to restrict mobility of the peptide and to improve receptor binding affinity and stability. It should be noted that the biological functions of peptides are characteristic of their amino-acid-determined three-dimensional conformation. Therefore, care needs to be taken that modification of a peptide does not cause a significant loss of its biological activity. In particular, labeling a peptide to permit a wide range of imaging moieties may potentially interfere with its binding region. To retain biological activity of peptides, a spacer can be incorporated between the active site of the particular peptide and the imaging moiety, and/or the imaging moiety can be site-specifically conjugated to a peptide sequence position not involved in binding to the receptor. A number of receptor-specific peptides and their analogs have been screened, synthesized, and labeled (mostly with radioisotopes), using a variety of techniques to modify the native peptides, and then their clinical potential has been determined.
Once the key amino acid structures have been determined and optimized, peptides can be directly or indirectly labeled with imaging moieties to provide or augment the imaging signal, depending on the modality. For instance, several radioisotopes (99mTc, 18F, 64Cu, 111In, 123I, 68Ga) for PET and SPECT, organic near-infrared (NIR) fluorophores, or quantum dots (QDs) for optical imaging, and magnetic nanoparticles (iron oxide nanoparticles) for MRI can be conjugated to peptides via organic linkers, macrocyclic or branched chelators, polymers, or nanoplatforms by well-established bioconjugation techniques.9,28-30
A general strategy is clear for development of a molecularly targeted imaging probe with a peptide. Schematically, a probe can be created through combination of a targeting peptide, linker, and imaging moiety. The new peptide-associated probe should have high in vivo uptake and retention in the target, with low background uptake in non-target tissues. Moreover, the probe should be safe and easy to prepare. Considering these factors, there has been a great deal of attention and acceleration in the development of molecularly targeted peptide-based probes. The next sections introduce selected targeting peptides and describe different design strategies and applications of peptide-based probes.
3. Peptide Probes for Nuclear Imaging
Despite the rapid progress of a number of imaging modalities, nuclear imaging remains the premier clinical method. Both PET and SPECT have been well-developed and are widely-used in daily practice. Two unparallelled advantages that PET and SPECT provide are their high sensitivity and need for the injection of only a minute quantity of tracer molecules. PET and SPECT imaging modalities are the most sensitive molecular imaging techniques, providing picomolar-range sensitivity.31,32 Such high sensitivity and the ability to provide diagnostically-valuable molecular and metabolic information unattainable by purely anatomical imaging have made PET and SPECT the methods of choice to elucidate pathological alternations at cellular or molecular levels. FDG-PET, in particular, is utilized to measure the activities of glucose metabolism, and has found wide applications in oncology, cardiology, neurology, and pharmacology.
A critical step in the development of peptide probes for nuclear imaging is the radio-labeling process. Due to characteristics of short-lived radioisotopes, labeling, purification, and characterization of radio-labeled peptides need be performed in a restricted time window, which limits the options of the candidate chemistry. A targeting peptide should be radio-labeled efficiently with high specific activity and be physiologically stable. Easy-handling and high throughput are essential to acceptable radio-labeling chemistry. Additionally, several other issues need to be taken into account. One is the in vivo stability of the radio-labeled peptide. The isotopes may detach from the host peptide during the circulation, resulting in read-outs that are not correlated with the peptide migration. Second, the impact of radio-labeling on the peptide-receptor interaction is important. The imparted radioisotopes may interfere with or even block the peptide-receptor interaction, leading to compromised binding affinities. Third, the overall conformational structure of the peptide of concern is important. Incorporation of a radioisotope, chelator and spacer to a relatively compact peptide may cause non-trivial alternations to its physical properties. Those changes can lead to a different pharmacokinetic profile, including increased renal and hepatic elimination that are unfavorable to imaging. Various techniques have been developed that allow efficient labeling of peptides with clinically useful radioisotopes via a chelating moiety or prosthetic group. In the following section, the common techniques that have been developed for peptide labeling of metal isotopes and 18F will be introduced and their applications will be reviewed.
3.1. Radio-labeling Strategies
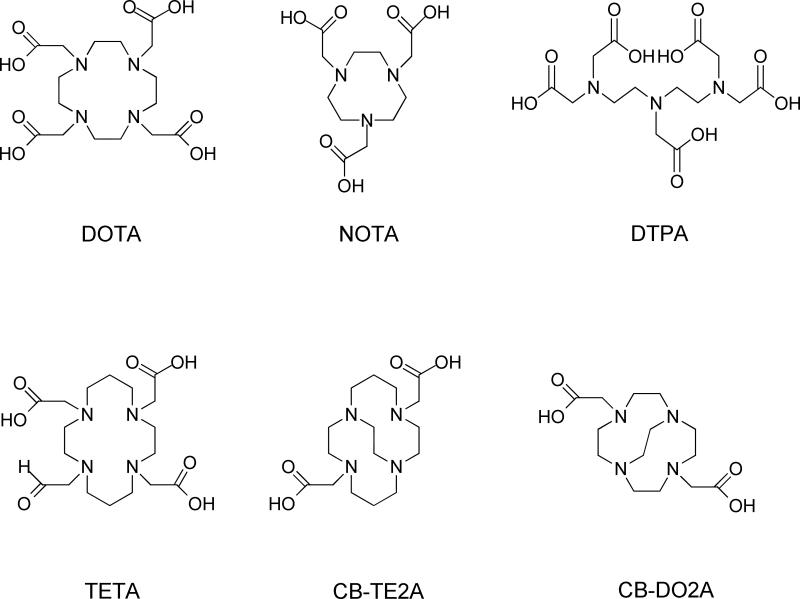
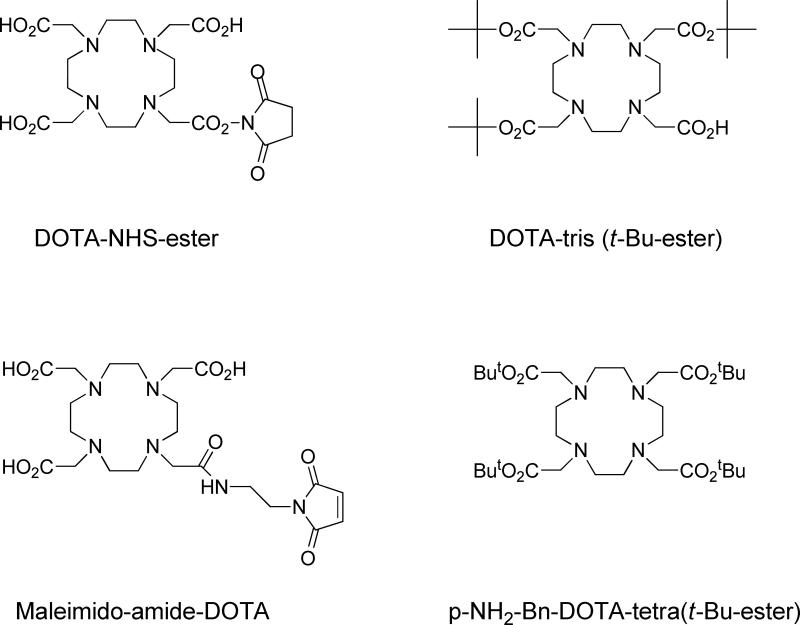
Metal isotopes including 64Cu, 68Ga, 177Lu, 111In, and 90Y are mainly introduced to the peptides with the aid of certain polyaminopolycarboxylic ligands, such as 1,4,7,10-tetraazacyclodecane-1,4,7,10-tetraacetic acid (DOTA), 2,2',2"-(1,4,7-triazacyclononane-1,4,7-triyl)triacetic acid (NOTA), diethylene triamine pentaacetic acid (DTPA), triethylenetetramine (TETA), and 4,11-bis(carboxymethyl)-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane (CB-TE2A) (Fig. 1). These chelators are efficient in coordinating the metals with all of the amino groups and some of the carboxyls. The multiple bonding sites lead to high binding strengths. As one example, in pure water, the equilibrium constant of the copper-TETA complex is 3×1015, which is comparable to the most stable non-covalent bonding known in biology (avidin-biotin, K≈1015).33 Furthermore, the complex typically forms within minutes upon mixing, independent of the peptide structure, which is highly favorable considering the fast decay of the isotopes. While several carboxyls might become involved in the complex formation, the remaining ones can be utilized to couple with the peptides, typically via formation of amide bonds with primary amines from either lysine or the N-terminus of the peptides. The carboxyl can be activated in situ with 1-ethyl-3-(3-dimethylaminopropyl (EDC) carbodiimide hydrochloride and N-hydroxysuccinimide (NHS, or its more water soluble derivative sulfo-NHS), yielding an intermediate that is reactive toward peptides.34 Even simpler, the chelators in carboxylic group modified form, such as DOTA-NHS ester and maleimido-amide-DOTA are now commercially available, cutting the coupling to one-step incubation. An alternative timing of introducing a DOTA-like chelator is during the solid-phase synthesis.35,36 For instance, tris-t-butyl-DOTA, which has one free carboxyl and three t-butyl protected ones, can be added at the N-terminal of the peptide subsequent to the assembly of the peptide sequence. Its protecting groups can be removed in parallel with amino acid side-chain protection groups at the end of the synthesis. Commonly, lysine lies in the center of the active site and coupling to it could seriously compromise binding affinity. In this context, solid-phase synthesis is advantageous over solution-phase synthesis for permitting N-terminus site specific coupling. The chemical structures of selected bifuctional DOTA analogs are shown in Fig. 2.
Figure 1.
Selected macrocyclic chelators.
Figure 2.
Selected bifunctional DOTA analogs.
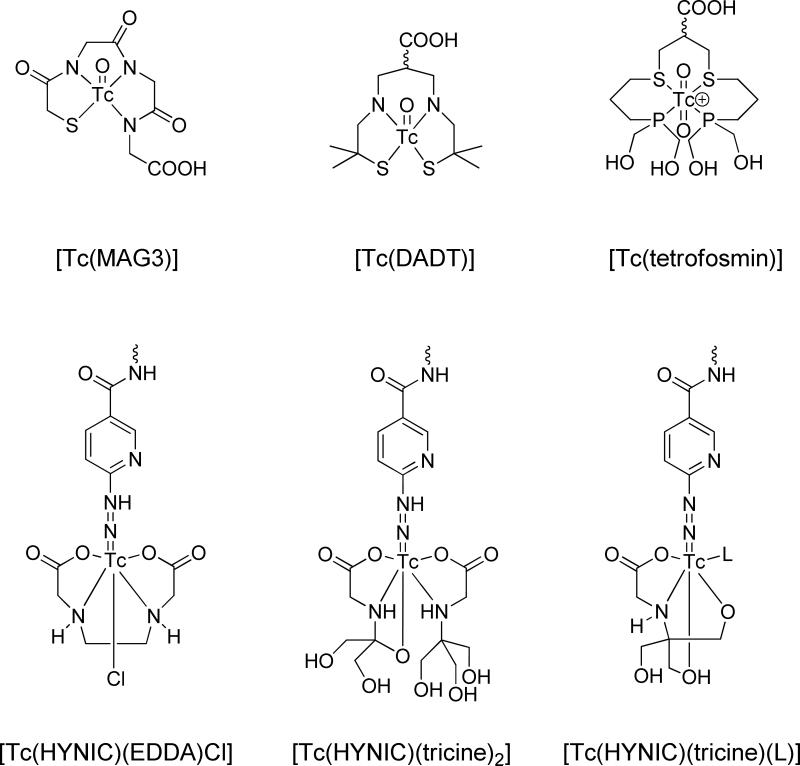
Metals like 99mTc and 186Re can be efficiently chelated by other chelating groups. 99mTc refers to the metastable nuclear isomer of 99Tc, which has a 6 h half-life.37 Although bare 99mTc can complex with polydentate chelators, such as boronic acid adducts or DTPA, the more favorable forms are [Tc=O]3+ or [Tc] 6-Hydrazinopyridine-3-carboxylic acid (HYNIC), which possess better stability. Various chelators with a combinational form of NxS4-x have proved effective in binding with [Tc=O]3+, resulting in a square pyramidal structure with the Tc in the center. For the sake of facile peptide coupling, other than imine/amine/amide and thiol-thioether groups, a fifth functional group, in most cases carboxyl, exists in the chelator. One of the most-adopted ligands is mercapto Ac-Gly-Gly-Gly (MAG3), which has three amide nitrogens and one sulfur from thiol coordinate with the [Tc=O]3+ core, while leaving a terminal carboxyl for peptide coupling.38-40 Likewise, 186Re can be complexed in the same manner. There are cases where [O=Tc=O]+ is utilized as the cores. In such a scenario, the complex constitutes an octahedral instead of a square pyramidal structure, with two O occupying the two apical sites, while the chelator occupies the four equatorial sites. One of the representative chelators is 1,2-bis[bis(2-ethoxyethyl)phosphino]ethane], in which the phosphine P serves as the donor atom.41,42 Similarly, HYNIC can take one or two binding sites of Tc, leaving four binding sites for the coligands such as tricine and ethylenediaminetetraacetic acid (EDTA). But, unlike the [Tc=O]3+, the chelator-peptide coupling was mostly achieved from the amine group of HYNIC instead of from the chelators. The chemical structures of selected chelating agents are shown in Fig. 3.
Figure 3.
Selected chelating agents for 99mTc.
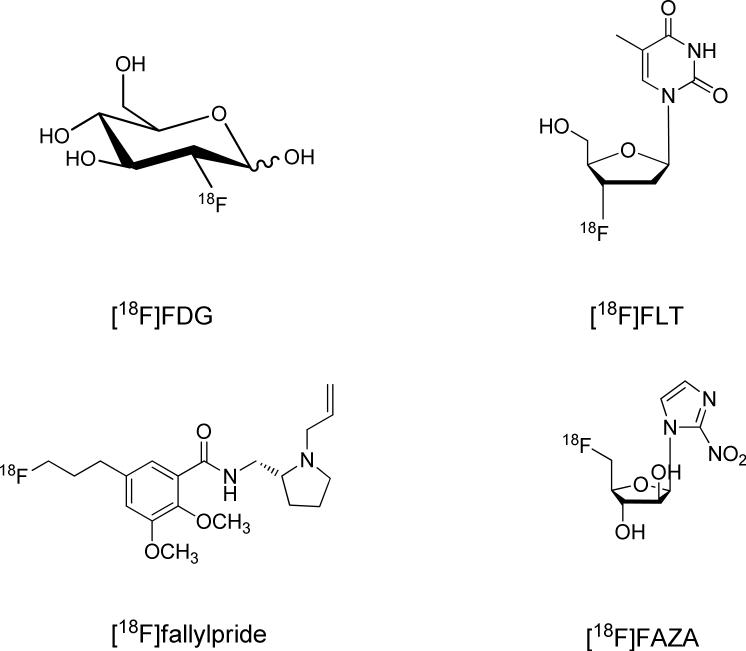
18F has so far been the mostly utilized radioisotope in PET imaging. Indeed, 2-[18F]fluoro-2-deoxy-D-glucose ([18F]FDG) is regarded as the “gold standard” in cancer staging and the diagnosis of a wide range of other diseases. Other 18F-labeled small molecular tracers that are of significance include 3’-deoxy-3’/[18F]fluorothymidine ([18F]FLT) for cell proliferation imaging, [18F]fallypride for dopamine receptor imaging, and [18F]fluoroazomycinarabinofuranoside ([18F]FAZA) for hypoxia imaging (Fig. 4).43 18F has a half-life of 109 minutes, which is short enough to cause negligible damage to the exposed living subjects. Meanwhile, 18F has a low β+-energy (0.64 MeV). Therefore, a short linear positron range in the tissue allows the acquisition of high-resolution images. Compared to the radiometal-labeling method, where the labeling process is no more than an instantaneous coordination, the 18F fluorination is far more complicated. There are generally two forms of 18F precursors, one is 18F− (such as K[18F] and Cs[18F] ) and the other is [18F]F2 or its derivative (such as acetyl hypofluorite (CH3COO[18F]F)). Although there are reports of using these two precursors for direct peptide labeling,44 generally these approaches are regarded as inefficient and lacking chemoselectivity, and are rarely employed. Practically, it is more common to convert the 18F precursors to certain forms of 18F-labeled prosthetic groups (called synthons), and to use those synthons for peptide labeling (Fig. 5).
Figure 4.
18F-labeled small molecule tracers.
Figure 5.
Radio-labeling of peptide with 18F.
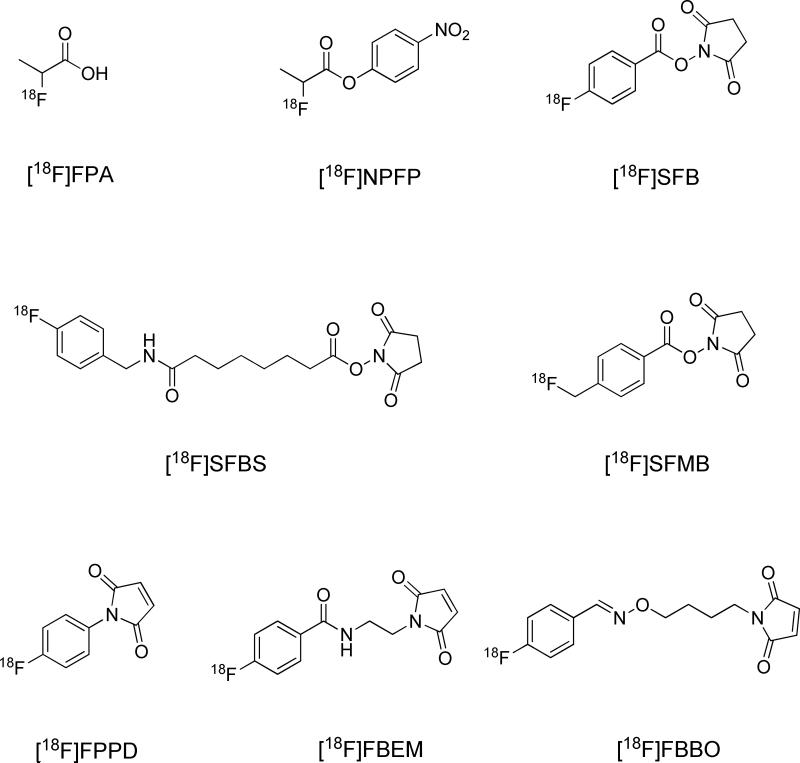
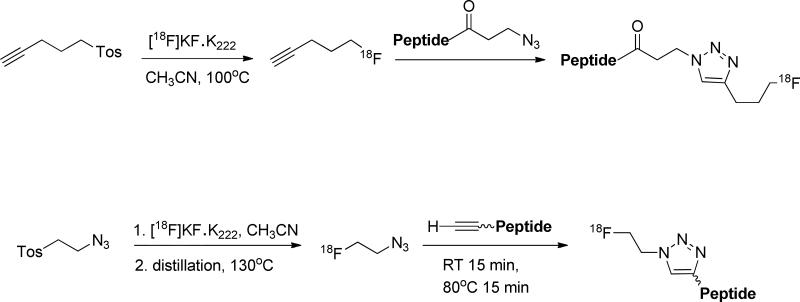
The primary amine from either the N-terminal amino acid or lysine side chain is widely utilized as the mounting site of 18F synthons (Fig. 6). Previously, several aliphatic prosthetic groups have been used. However, the leading role in 18F labeling has been gradually replaced by a group of [18F]fluoroaromatic compounds, with the most representative being 4-[18F]fluorobenzonic acid. Upon activation with N,N-disuccinimidyl carbonate, or, in solid phase synthesis, TSTU, 4-[18F]fluorobenzonic acid is converted to N-succinimidyl-4-[18F]fluorobenzoate ([18F]SFB), which can be efficiently coupled with peptides via acylation. Beside [18F]SFB, its analogs including 4-[18F]fluorobenzylamine succinimidyl ester ([18F]SFBS), N-succinimidyl-4-([18F]fluoromethyl)benzoate) ([18F]SFMB), 4-[18F]fluorophenacyl bromide ([18F]SPB), and 3-[18F]fluoro-5-nitrobenzimidate ([18F]SFMB) have also been exploited. Thiols are not as abundant as carboxyl and amine groups in peptides, so targeting thiols could afford better chemoselectivity. It is conceivable that cysteine is contained in the sequence, or, alternatively, that thiol molecules are artificially introduced. On the other hand, the efficient coupling between maleimide and thiol makes thiol the group of choice to put onto the synthons. In most cases, it is introduced with a maleimide-containing bifunctional molecule, which anchors onto the prosthetic group through the second functional group. For instance, [18F]SFB can be reacted with N-(2-aminoethyl)maleimide ([18F]FBEM),45 or, alternatively, [18F]FBA can be reacted with N-[4-(aminooxy)butyl]maleimide,46 with both yielding thiol reactive prosthetic groups. [18F]FBA is the precursor of [18F]SFB, which is derived from 18F− via a one-step fluoration reaction, and can be efficiently linked with aminooxy or hydrazine compounds in a subsequent one-step coupling. Since [18F]FBA-based peptide labeling takes fewer steps and leads to less activity loss during purification, there has been increasing interest in using it as a synthon. [18F]FDG itself can also be utilized as a synthon. [18F]FDG can be converted to [18F]FDG-maleimidehexyloxime ([18F]FDGMHO), which is highly reactive toward thiolated peptides.47 An elegant example of click chemistry is the Huisgen 1,3-dipolar cycloaddition that occurs between azides and alkynes under the catalysis of copper(I). Due to its simplicity, speed, selectivity, high yield, and mild reaction conditions, it has become an important bioconjugation technique and is also exploited for 18F radiolabeling. Clearly, there are two approaches to coupling 18F synthons and the peptides through click chemistry: [18F]fluoroalkynes reacting with peptide azides and the reverse, [18F]-azide with peptide alkyne (Fig. 7).48,49
Figure 6.
Selected 18F synthons.
Figure 7.
Radio-labeling of peptide with 18F via click chemistry.
Due to the small size of peptides, attaching a bulky radio-labeled chelating group or a prosthetic group may influence the biological activity of the peptides. Therefore, site-specific radiochemistry is needed and important for the preparation of biologically active peptide probes. In many cases, a spacer is generally present to separate the nuclide complexes from the peptide moiety. A wide variety of strategies have been developed in recent years for the convenient and efficient radiolabeling of peptides. In the next section, we discuss the significance of various peptide-receptors in molecular imaging, as well as recently-developed radiolabeled peptide-based probes for cancer targeting in vivo.
3.2. Radio-labeled Peptide Probes
3.2.1. Somatostatin Analogs
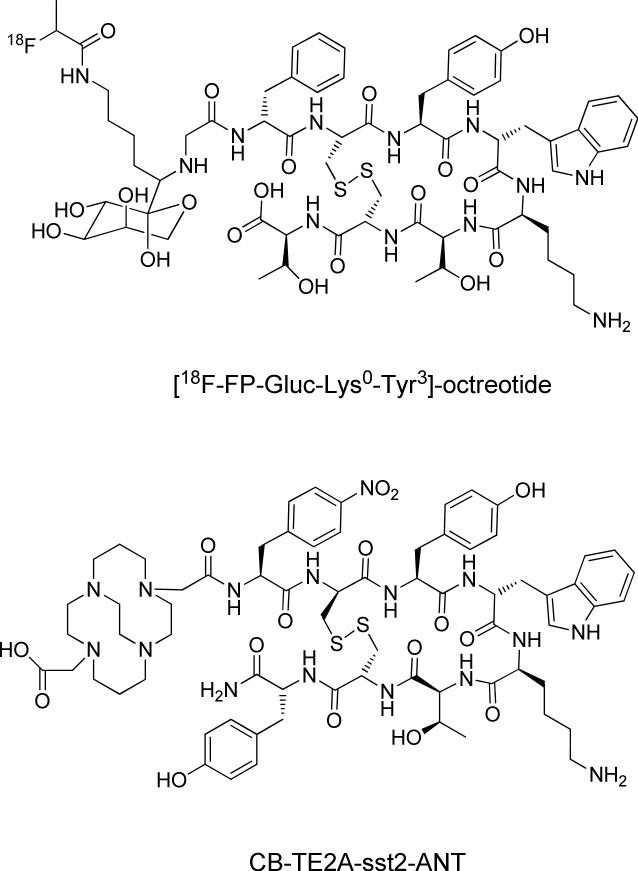
Somatostatins (SST) are a family of cyclopeptides that have inhibitory effects on the secretion of hormones such as growth hormone, insulin and glucogon.50 There are two naturally occurring active SSTs with SST-28 and SST-14 amino acid residues; their biological activities are mediated with G-protein coupled human somatostatin receptors (SSTrs). To date, five different subtypes of SSTrs (SSTr1–SSTr5) have been identified.51 SSTrs are overexpressed on a majority of tumors, such as the neuroendocrine tumors, gliomas, breast cancer, and small cell lung cancers (SCLC).52,53 SSTrs represent one of the best examples of targets for radio-labeled peptide probes. Because naturally occurring SSTs are vulnerable to rapid enzyme degradation in vivo, a large library of SST analogs with improved metabolic stability and biological activity has been developed. The most widely-investigated peptide is the cyclic octapeptide octreotide (D-Phe1-Cys2-Phe3-D-Trp4-Lys5-Thr6-Cys7-Thr8-ol) and its analogs. Octreotide lacks key enzyme cleavage sites and is more stable than native SSTs (half-life; 90 – 110 min vs. < 3 min). The first FDA-approved agent was [111In-DTPA]-octreotide (OctreoScan), which was originally designed for scintigraphy of neuroendocrine tumors. However, due to its moderate binding affinity to SSTr2 and because DTPA is not a suitable chelator for many other nuclides, the next generation of SST analogs has been introduced. Octreotide analogs with a higher affinity for SSTr2 have been discovered and labeled with the DOTA chelator, which forms thermodynamically and kinetically stable complexes with metals compared to DTPA. Representative DOTA-labeled SST analogs are [DOTA-Tyr3]-octreotide (DOTA-TOC). [DOTA-D-Phe1,Tyr3]-octreotide (DOTA-TATE), and [DOTA-Nal3]-octreotide (DOTA-NOC).54 99mTc-labeled SST analogs, such as [99mTc-EDDA/HYNIC]-TOC and [99mTc-EDDA/HYNIC]-TATE were designed for specific labeling of 99mTc.54,55 SST analogs have also been designed for PET imaging. Different DOTA-octreotide analogs including [68Ga-DOTA]-TOC, [68Ga-DOTA]-NOC, and [64Cu-DOTA]-TATE have been reported and their efficacy has been further confirmed in various preclinical and clinical settings.56 To improve probe pharmacokinetics and for clinical application in PET imaging, 18F-labeled carbohydrated conjugates of TOC such as [18F-FP-Gluc-Lys0-Tyr3]-octreotide have been developed. Such analogs provide better diagnostic performance, compared to OctreoScan, in patients with SSTr-positive tumors.57,58 Analogs with a high binding affinity to broader subtypes of SSTrs have been designed with the sequence of [DOTA-Nal3,Thr8]-octreotide and [DOTA-BzThi3,Thr8]-octreotide; they have shown 2-fold higher specific uptake in AR4-2J tumors in vivo compared with [111In-DOTA]-TOC.59 Recently, a series of DOTA-conjugated SSTr2 antagonists was prepared and their superior efficacy was demonstrated both in vitro and in an SSTr2-expressing tumor model (Fig. 8).60-62
Figure 8.
Selected somatostatin analogs.
3.2.2. Arg-Gly-Asp Analogs
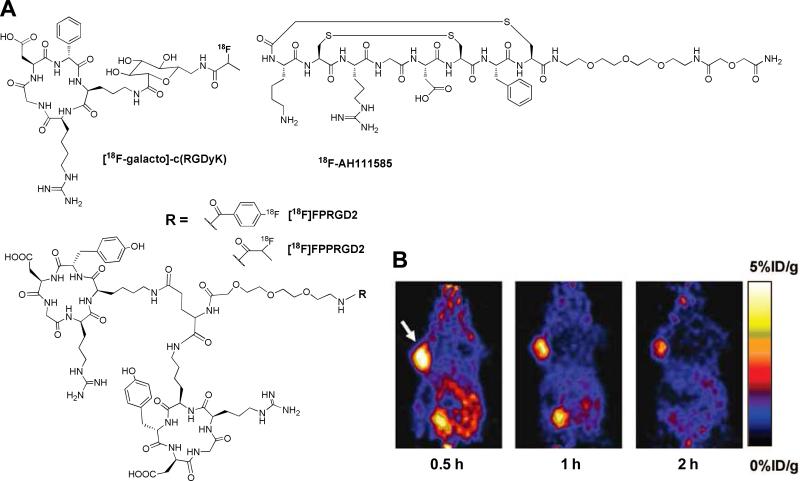
Integrins are a family of heterodimeric transmembrane glycoproteins consisting of two non-covalently associated receptor subunits, α and β.63,64 The αvβ3 integrin seems to be the most closely-related with tumor progression and plays important roles during tumor angiogenesis and formation of new blood vessels. The integrin αvβ3 receptor is normally expressed on endothelial cells and most normal organs. However, it is highly expressed on activated and proliferating endothelial cells during tumor angiogenesis and metastasis.17 The overexpression of integrin αvβ3 in several tumor forms, including melanomas, ovarian and lung carcinoma, neuroblastomas, glioblastomas, and breast cancer, has attracted much attention as an attractive target for diagnostic cancer imaging. The integrin receptor recognizes polypeptide domains containing the Arg-Gly-Asp (RGD) amino acid sequence, which is present in several extracellular matrix proteins such as vitronectin.65 Based on the knowledge that RGD sequences strongly bind with the αvβ3 receptor, a significant number of small linear and cyclic RGD peptide analogs have been designed.66-68 Of particular interest, a series of active and selective cyclic RGD analogs containing the sequence of cyclo(Arg-Gly-Asp-D-Tyr-Lys), referred to as “c(RGDyK)”, and cyclo(Arg-Gly-Asp-D-Phe-Lys), referred to as “c(RGDfK)”, were designed by backbone cyclization. To date, two different 18F-labeld RGD analogs, [18F-galacto]-c(RGDfK) and 18F-AH111585, are under clinical investigation.69-74 The glycopeptide [18F-galacto]-c(RGDfK) was synthesized by introducing SAA (7-amino-L-glycero-L-galacto-2,6-anhydro-7-deoxyheptanoic acid) to a lysine group of c(RGDfK) to improve the pharmacokinetics and reduce uptake by the liver. [18F-galacto]-c(RGDfK) specifically binds to the isolated αvβ3 receptor and has successfully identified αvβ3 receptor expression in patients with melanoma, sarcoma, breast cancer, and glioblastoma.70-72 18F-AH111585 is an 18F-labeled RGD4C peptide containing multiple disulphide bridges and a short poly(ethylene glycol) (PEG) spacer at the C-terminus to stabilize the peptide in vivo.75 These probes demonstrated comparable binding affinity with c(RGDfK) and their efficacy has been confirmed in patients with breast cancer.73,74 In order to provide enhanced binding affinity to integrin αvβ3 receptor and to improve pharmacokinetic profiles, several different forms of 18F-labeled RGD dimers have been developed by labeling Glu-[c(RGDyK)]2 ([18F]FRGD2) and NH2-PEG3-Glu-[c(RGDyK)]2 ([18F]FPRGD2) with [18F]SFB, and by labeling NH2-PEG3-Glu-[c(RGDyK)]2 ([18F]FPPRGD2) with 2-[18F]FPA.76-78 Both RGD dimers bind more specifically to αvβ3 receptor than to monomeric RGD peptide in vitro and in integrin αvβ3-positive U87MG tumor cells within mice. More recently, [18F]FPRGD2 and [18F]FPPRGD2 have been approved by FDA for first in human studies. Similarly, various multivalent dimers and tetramers of the cyclic RGD-based peptides, including Glu-[c(RGDyK)]2, Glu-[Gly-Gly-Gly-c(RGDfK)]2, and Glu-[Gly-[c(RGDfK)]2]2, were prepared and labeled with various radioisotopes, such as 64Cu, 68Ga, and 99mTc.79-83 Besides the conjugation of radioisotopes to the primary amine groups, labeling of RGD peptides through a thiol-reactive synthon, [18F]FBEM, (i.e., by click chemistry) have been reported.45,48 Of particular interest, a cyclic triazole-bearing RGD peptide, 18F-RGD-K5, was developed by using click chemistry and is now entering a clinical study.84,85 Most the reported RGD peptide probes have provided excellent tumor targeting efficiency in animal models (Fig. 9).
Figure 9.
(A) Selected RGD analogs for PET imaging. (B) Serial microPET images of U87MG tumor-bearing mice after intravenous injection of [18F]FPRGD2. Arrow indicates tumor. Modified with permission from ref. 77. Copyright 2007, Springer-Varlag.
3.2.3. Bombesin Analogs
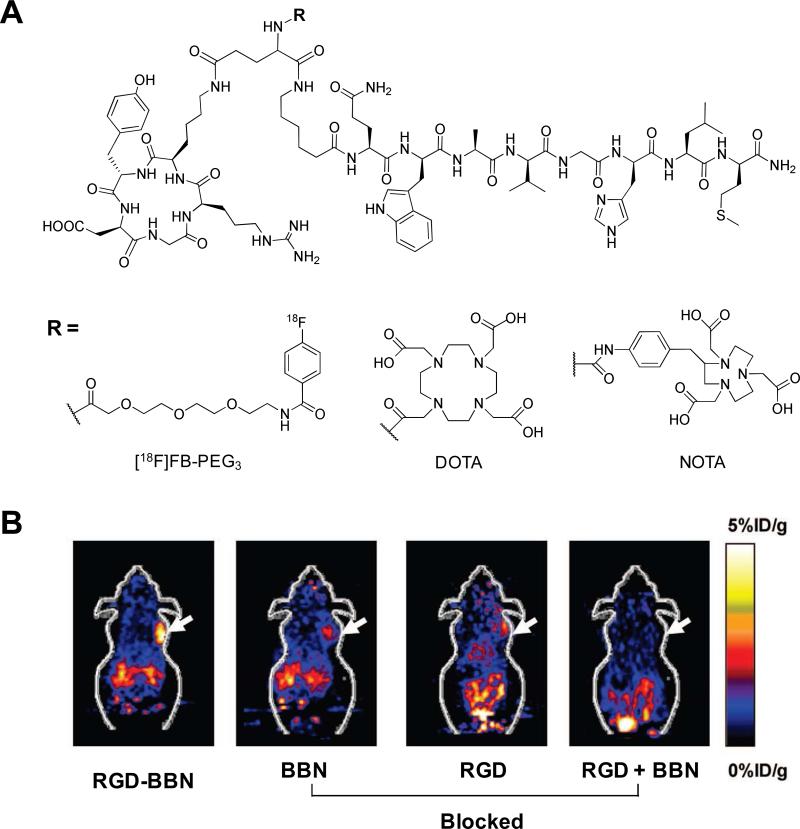
Bombesin (BBN) is an amphibian homologue of mammalian gastrin-releasing peptide (GRP; pGlu1-Gln2-Arg3-Leu4-Gly5-Asn6-Gln7-Trp8-Ala9-Val10-Gly11-His12-Leu13-Met14-NH2) that exhibits high affinity and specificity to the GRP receptor (GRPr).86 Four different GRPr subtypes have been characterized, and overexpression of GRPrs has been demonstrated in a variety of cancers including small cell lung cancer (SCLC), prostate, breast, pancreas, and gastrointestinal tumors.16 Like other naturally-occurring peptides, BBN has a very short circulation half-life (< 2 min). Various BBN analogs based on their key amino acid sequence, BBN7-14, were screened and developed as imaging probes, mostly by incorporating radioisotopes at the N-terminus of the peptide. For SPECT imaging of GRPr-expressing tumors, 99mTc-RP-527, a tripeptide N3S chelator coupled to BBN7-14 via Gly-5-aminovaleric acid linker, has been reported and evaluated in mice with PC-3 tumors and in breast cancer patients.87,88 The bifunctional chelator EDDA/HYNIC conjugated to a BBN analog for the preparation of [99mTc-EDDA/HYNIC-Lys3]-BBN was less lipophilic and had lower hepatobiliary and renal excretion in vivo.89 Macrocyclic chelator-labeled BBN analogs were developed to provide flexibility for both SPECT and PET applications. For instance, [111In-DOTA-11-Aun]-BBN7-14, [68Ga-DOTA-PEG4]-BBN7-14, [64Cu-DOTA-Pro1,Tyr4]-BBN, and [64Cu-NOTA-8-Aoc]-BBN7-14 were developed and their efficiency was successfully confirmed in vivo.90-93 [18F-Lys3]-BBN prepared by conjugating [18F]SFB to the lysine group of BBN analog demonstrated receptor specific uptake in PC-3 tumors in vivo.94 Potent BBN agonists and antagonists were designed and characterized both in vitro and in vivo. These included Demobesin 1 ([99mTc-N4-bzlg0,D-Phe6,Leu-NHEt13,des-Met14]-BBN6-14), Demobesin 4 ([99mTc-N4-Pro1,Tyr4,Nle14]-BBN, 68Ga-BZH3 ([68Ga-DOTA-PEG2-D-Tyr6,β-Ala11,Thi13,Nle14]-BBN6-14, and Z-070 ([111In-DOTA-PEG4-D-Tyr6,β-Ala11,Thu13,Nle14]-BBN6-14.95-98 As an alternative to the use of single or homogeneous multivalent peptides, peptide ligands recognizing both GRPr and integrin receptors have been designed and evaluated for their dual-receptor-targeting abilities. A BBN-RGD heterodimer was synthesized by coupling of Aca-BBN7-14 and c(RGDyK) peptides using glutamic acid as a linker.99 A Glu-BBN-RGD was prepared and labeled with various radioisotopes, such as 18F, 64Cu, and 68Ga, using combinations of PEG3 linker or chelating agents including DOTA and NOTA (Fig. 10).99-103 Dual-targeting of integrin and GRPr significantly improved tumor-targeting efficacy and pharmacokinetics compared with single RGD and BBN analogs in prostate and breast cancer models. The application of BBN peptide-based probes is considered to be more relevant clinically than SST peptides, due to the broad spectrum of BBN specificity for various tumors.
Figure 10.
(A) Selected RGD-BBN heterodimers. (B) Coronal microPET images of PC-3 tumor-bearing mice at 1 h after intravenous injection of [18F]FB-PEG3-Glu-RGD-BBN and a blocking dose of BBN peptide, c(RGDyK), or RGD + BBN peptides. Arrows indicate tumors. Modified with permission from ref. 100. Copyright 2009, American Chemical Society.
3.2.4 Cholecystokinin Analogs
Cholecystokinin (CCK) and gastrin are structurally and functionally related peptide hormones that function in the gastrointestinal tract and central nervous system.104 The biological action of these peptide hormones is mediated by CCK-gastrin receptors belonging to the super family of GPCrs. These receptors are distributed in a number of different tissues rather than as a selective expression of specific subtypes. In certain human tumors, overexpression of these receptor subtypes has been well demonstrated.18,105,106 CCK-2/gastrin receptors are overexpressd in a high percentage (> 90%) of medullary thyroid cancer (MTC) and in other tumors of SCLC, astrocytomas, stromal ovarian tumors, and gastroenteropancreatic cancers. Among the CCK analogs derived from a 115 amino acid precursor CCK protein, most CCK-related peptide probes were designed based on CCK-8.107 CCK-8 is a sulfated octapeptide C-terminus fragment of the biologically active CCK with the sequence of Asp-Tyr(SO3H)-Met-Gly-Trp-Met-Asp-Phe-NH2. Minigastrin (MG), a C-terminal truncated form of 13 amino acid peptide with the sequence of Leu1-Glu2-Glu3-Glu4-Glu5-Glu6-Ala7-Tyr8-Gly9-Trp10-Met11-Asp12-Phe13-NH2, is also frequently used to target CCK-2/gastrin receptors. CCK and gastrin share an identical sequence of five amino acids at their biologically active C-terminal region. Radiolabeling of CCK-8 peptides with 111In or 99mTc has been reported for CCK/2 receptor (CCK-2r) imaging in vivo. For instance, [111In and 99mTc-DTPA-Glu-Gly]-CCK-8 and [99mTc-tricarbonyl-Lys,His]-CCK-8 have been investigated and their high specificity for CCK-2rs demonstrated in A431 cells that overexpress CCK-2r and in an A431-CCK-2r xenograft model.108-110 MG analogs have also been combined with radioisotopes and shown to be suitable for CCK-2r/gastrin receptor targeting. [111In-DTPA-D-Glu1 or Leu1]-MG, [99mTc-EDDA/HYNIC-D-Glu1]-MG, and 111In-labeled small peptide libraries constructed based on the C-terminal sequences of CCK-8 or MG have been reported.111-113 Recently, DOTA-conjugated MGs with decreased numbers of glutamic acids demonstrated improved affinity in gastrin receptor-positive AR4-2J rat pancreatic tumor cells and showed tumor specificity in AR4-2J xenograft models.114 In another embodiment, cyclized MG analogs such as cyclo1,9[γ-D-Glu1,desGlu2-6,D-Lys9]-MG and cyclo1,9[γ-D-Glu1,desGlu2-6,D-Lys9,Nle11]-MG were synthesized and labeled with 99mTc-EDDA/HYNIC. The cyclized MG probes showed rapid internalization in cells expressing CCKr, and high tumor uptake with the low kidney retention in an animal model. However, cyclized MGs need improved metabolic stability and bioavailability in vivo to be useful clinically.
3.2.5. α-Melanocyte Stimulating Hormone Analogs
α-Melanocyte stimulating hormone (α-MSH) is a linear 13 amino acid peptide with the sequence Ac-Ser1-Tyr2-Ser3-Met4-Glu5-His6-Phe7-Arg8-Trp9-Gly10-Lys11-Pro12-Val13-NH2 produced in the pituitary gland, and is mainly responsible for the regulation of skin pigmentation.20 It has been reported that α-MSH receptors are distributed in more than 80% of human melanoma metastases.115 Therefore, radio-labeled peptides that target these receptors are attractive probes for diagnosis of melanoma. The rapid metabolization of α-MSH under physiological conditions led to the development of novel α-MSH analogs with improved stability and specificity in vivo. Most α-MSH analogs were designed and synthesized by amino acid substitution with D-and non-natural amino acids. For instance, replacement of Met4 with Nle4, and Phe7 with D-Phe7 in α-MSH results in a highly potent analog, NDP, with the sequence [Nle4,D-Phe7]-MSH.116 Several linear α/MSH analogs, such as [111In-DTPA]-NDP, [99mTc-Cys-Gly-Cys-Gly]-NDP, [99mTc-MAG2-Lys11]-NDP (MAG2: mercapto Ac-Gly-Gly), and [99mTc-Cys-Gly-Cys-Gly-D-Phe7]-MSH5-10, were reported some years ago.10,116 Despite their somewhat improved biological potency, many of these radio-labeled peptides were not developed further mainly due to poor tumor targeting characteristics in vivo. An alternative design for an α-MSH analog was developed using metal-cyclized peptides such as [Cys3,4,10,D-Phe7]-MSH3-13 (CCMSH).102 Subsequently, DOTA-conjugated and Re-mediated CCMSH analogs, including [DOTA]-ReCCMSH and [DOTA-Arg11]-ReCCMSH, were synthesized and labeled with various radioisotopes, such as 111In, 64Cu, and 68Ga.117-119 Compared with the linear α-MSH analogs, the metal-cyclized analogs displayed favorable pharmacokinetics and superior tumor targeting in vivo. Recently, a series of DOTA-conjugated lactam bridge-cyclized α-MSH analogs, including [DOTA-Glu-Glu]-cycloMSH and [Ac-Glu-Glu]-cycloMSH-DOTA, were designed and labeled with 111In, and were shown to possess superior tumor uptake and to prolong tumor retention in melanoma-bearing mice.120,121 For PET imaging of α-MSH expression, 18F-labeled peptide analog was also developed by labeling [18F]SFB to an α-MSH analog, Ac-Nle-Asp-His-D-Phe-Arg-Trp-Gly-Lys-NH2, and its tumor specificity was confirmed in mice bearing B16/F10 tumor. For application of dual-receptor-targeting, peptide ligands recognizing both integrin and α-MSH receptors have been designed.122 The RGD motif, c(RGDyD), was conjugated to [Arg11]-CCMSH through Lys to generate RGD-Lys-Arg11-CCMSH. The 99mTc-labeled peptide demonstrated enhanced cellular uptake in B16/F10 cells in vitro and rapid, high melanoma uptake in a B16/F10 xenograft model.
3.2.6. Other Peptide Analogs
Neurotensin (NT) is a 13 amino acid peptide with the sequence p-Glu1-Leu2-Tyr3-Glu4-Asn5-Lys6-Pro7-Arg8-Arg9-Pro10-Tyr11-Ile12-Leu13. NT is a neurotransmitter and a local hormone, and can be found in the central nervous system and in peripheral tissues.123 Three NT receptor subtypes have been cloned to date and overexpression of NT receptors has been found in many tumors. Examples include Ewing's sarcoma, meningiomas, SCLC, and MTC. In addition, > 70% of ductal pancreatic adenocarcinomas express NT receptors, and 90% of tumors are positive for NT receptors in patients with invasive ductal breast cancers.124,125 Since NT is rapidly degraded by peptidases in vivo, several approaches to stabilize the peptide were introduced mainly by replacement of the enzymatic cleavage sequence on the Arg8-Arg9 and Tyr11-Ile12 bond. Optimization of NT analogs has resulted in a number of NT8-13 analogs, in which the small C-terminal hexapeptide fragment contains various radioisotopes, including 111In, 99mTc, and 18F.126-129
Vasoactive intestinal peptide (VIP) is a 28 amino acid peptide initially isolated from porcine intestine. VIP is an important immunomodulator and stimulates the secretion of various hormones. VIP receptors have been detected on the cell membrane of normal intestinal and epithelial cells, and are overexpressed on various tumors cells, such as colonic adenocarcinomas, pancreatic carcinomas, and carcinoids.130 His1 plays an important role in receptor-binding affinity, therefore various VIP analogs were prepared by modifying the C-terminus of the peptide. VIP and its analogs labeled with various radioisotopes, including 99mTc, 64Cu, and 18F, have been evaluated in animal models and in humans.131-133
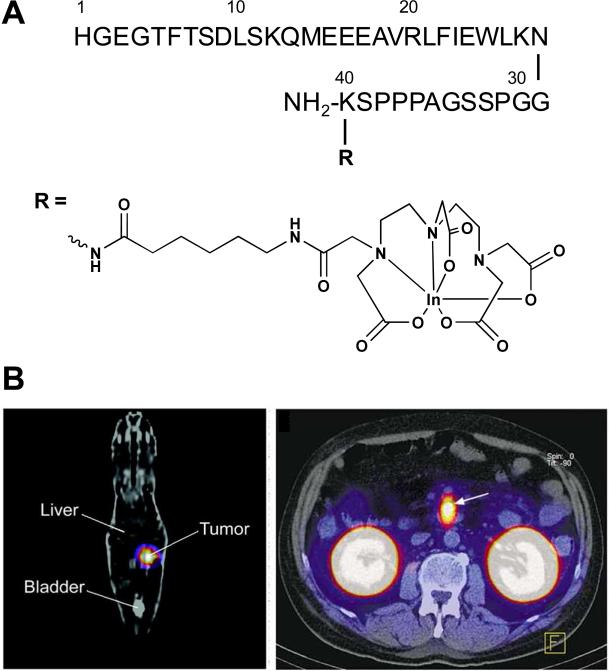
Glucagon-like peptide-1 (GLP-17-36) is a peptide hormone secreted from L-cells in the gastrointestinal tract that stimulates insulin secretion through GLP-1rs expressed on pancreatic beta cells of the islets.134,135 Recent studies have revealed that massive numbers of GLP-1rs are overexpressed in most human insulinomas and gastrinomas.19 Radio-labeled GLP-17-36 and its metabolically more stable agonist, [125I]-GLP-17-36, [111In-Ahx-DTPA-Lys40]-Exendin-4, [125I]-Exendin9-39 have been developed for GLP-1r-positive tissues in animal models.136-138 Among others, [111In-Ahx-DTPA-Lys40]-Exendin-4 is able to image targeted tumors in the Rip1-Tag2 mouse model of insulinomas and successfully detects tumors in patients with insulinomas that are not detected by conventional imaging methods (Fig. 11).137,139
Figure 11.
(A) Structure of [111In-Ahx-DTPA-Lys40]-Exendin-4. (B) GLP-1 receptor scanning of insulinoma; (left) coronal SPECT image of a tumor bearing Rip1Tag2 mouse and (right) transaxial SPECT image of an insulinoma in patient. Arrows indicate tumors. Modified with permission from ref. 137 and 139. Copyright 2006, the Society of Nuclear Medicine, Inc. Copyright 2008, the Massachusetts Medical Society.
Neuropeptide Y (NPY) is a 36 amino acid residue of the pancreatic polypeptide family, and NPYrs are widely produced in various neuroblastomas, breast cancers, and sarcomas.140,141 A number of radio-labeled NPY analogs have been designed and characterized, including NPYr-selective [111In-DOTA-Lys4, Phe7, Pro34]-NPY; however, more in vivo data are required to confirm their potential as tumor imaging agents.142,143 The chemokine receptor CXCR4 is a member of the GPCr family and has recently been identified as a target receptor for in vivo tumor imaging.144 The expression of CXCR4 is known to be up-regulated in a variety of cancer types, including breast, colon, lung, and prostate, and it plays a crucial role in tumor metastasis. Recently, a radio-labeled CXCR4 peptide inhibitor, [111In-DTPA]-Ac-TX14010, has been developed to image CXCR4 expression in metastatic tumors in vivo.145 Several peptide antagonists have been introduced as promising tools for receptor-targeted imaging. For many years, antagonists have not been considered for targeted receptor imaging, mainly due to the limited peptide internalization and accumulation in target cells, compared to that of agonists. However, recently developed SSTr and GRPr radio-labeled antagonists have shown preferable tumor uptake over agonists in vivo, as described in the previous section, and indicated the importance of developing antagonist peptides. Besides the development of numerous peptides for imaging receptors that are overexpressed in tumor cells, considerable attention has been directed towards the development of imaging agents for monitoring specific enzyme expression involved in tumor progression. For instance, matrix-metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases that play key roles in several biological processes and have been the focus of much interest as biomarkers in various diseases including cancers and inflammatory diseases.146 Since MMPs are significantly involved in cancer progression and certain inflammatory diseases, a variety of imaging modalities are utilized for the detection and imaging of MMPs in vivo.147 For example, new MMP cyclic peptide inhibitors containing a His-Trp-Gly-Phe sequence have been discovered and radio-labeled for imaging of MMP activity in vivo.148,149
Preclinical and clinical peptide-based probes for nuclear imaging have become essential tools for the detection and monitoring of various disease. A large number of novel peptide candidates can be designed, and any peptide or peptide hormone receptor that is overexpressed in key tumors can be targeted by well-established direct or indirect radio-labeling techniques on demand. Besides the use of single peptide ligands, flexible peptide chemistry can offer diverse peptide designs, such as multivalent dimers and tetramers, for improved binding affinity or heterodimers for dual-receptor-targeting. In addition, many of the diagnostic peptide probes described in this review can readily be redesigned as therapeutic radiopharmaceuticals by the replacement of diagnostic radioisotopes with therapeutic isotopes, for instance 90Y, 188Re, or 177Lu. Despite the significant progress in developing molecularly targeted peptides, their application in molecular imaging and therapy is still in the early stages. The clinical adaptation of molecular imaging will largely depend on the development of more sensitive and reliable radio-labeled peptide-based probes. Although significant efforts have been directed toward developing clinically applicable peptide-based probes, until now, there is only one FDA approved peptide probe on the market (OctreoScan). However, a variety of promising new candidates are under active development (Table 1), and other key players are likely to be available in the clinic in the near future. In the next section, we describe the different peptide-based probe design strategies for nonnuclear imaging modalities.
Table 1.
A selected list of radio-labeled peptides which are in clinical use or under investigation in clinical trials.
| Peptide Probes (receptor) | Peptide Sequence | Indication | Comments | Ref. |
|---|---|---|---|---|
| Somatostatin analogs (SSTr) | ||||
| Octreoscan | [111In-DTPA] -fCss(CFwKTC)T-ol | Neuroendocrine tumors | Approved | 52-54 |
| DOTA-TOC | [DOTA]-fCss(CYwKTC)T-ol | Neuroendocrine tumors | Clinical study | 52-54 |
| DOTA-TATE | [DOTA]-fCss(CYwKTC)T | Neuroendocrine tumors | Clinical study | 52-54 |
| DOTA-NOC | [DOTA]-fCss(C-Nal-wKTC)T-ol | Neuroendocrine tumors | Clinical study | 52-54 |
| 18F-FP-Gluc-TOCA | Nα-deoxyfructosyl-K([18F-FP])-fCss(CYwKTC)T | Neuroendocrine tumors | Clinical study | 58 |
| Arg-Gly-Asp analogs (ανβ3 integrin) | ||||
| [18F-galacto]-c(RGDfK) | c(RGDfK([18F-FP]-SAA)) | Head and neck cancers | Clinical study | 70-72 |
| 18F-AH111585 | RGD 4C analog containing a PEG spacer | Metastatic breast cancer | Clinical study | 73,74 |
| 18F-RGD-K5 | A cyclic triazole-bearing RGD peptide | Metastatic breast cancer | Clinical study | 85 |
| [18F]FPPRGD2 | [18F-FP]-PEG3-E-[c(RGDyK)]2 | n.d. | eIND | |
| Bombesin analogs (GRPr) | ||||
| 99mTc-RP-527 | [N3S] -G-Ava-QWAVGHLM-NH2 | Breast cancer | Clinical study | 88 |
| 68Ga-BZH3 | [DOTA]-PEG2-yQWAV-βA-H-Thi-Nle-NH2 | GIST | Clinical study | 98 |
| Neurotensin analog (NTr) | ||||
| 99mTc-NT-XI | (Nα-His)Ac-K(ψCH2-NH)RPY-Tle-L | PDAC | Clinical study | 129 |
| Vasoactive intestinal peptide analog (VIPr) | ||||
| 99mTc-TP3654 | HSDAVFTDNYTRLRKQMAVKKYLNSILN-Aba-GGaG | Various cancers | Clinical study | 131 |
| Glucagon-like peptide-1 analog (GLP-1r) | ||||
| [111In-Ahx-DTPA-Lys40]-Exendin-4 | HGEGTFTSDLSKQMEEEAVRLFIEWLKNGGPSSGAPPPSK(Ahx-DTPA)-NH2 | Insulinoma | Clinical study | 139 |
eIND: Exploratory Investigational New Drug; GIST: Gastointestinal stromal tumor; PDAC: Pancreatic ductal adenocarcinoma; n.d. not determined.
4. Peptide Probes for Optical Imaging
Real-time imaging of biological processes in live cells and in vivo is one of the basic goals of modern optical imaging techniques. Optical imaging methods provide many advantages over other imaging modalities that include high sensitivity, use of non-radioactive materials, and safe detection using readily available instruments at moderate cost. For many decades, microscopic optical imaging has been the gold standard for cellular imaging in molecular biology. Today, significant improvements in in vivo optical imaging techniques offer new paradigms and opportunities in preclinical and clinical molecular imaging applications. Recently developed optical imaging instruments and sophisticated optical imaging probes provide noninvasive, real-time imaging of small animals at the whole-body, tissue, and cellular levels. Certainly, the combination of various near-infrared (NIR) fluorophores and ligands, such as engineered peptides, has significantly expanded and improved the performance of optical imaging systems. Recent progress in the field of optical imaging has been discussed elsewhere.7,150-152 We will limit our discussions to design concepts for, and recent progress in the development of, peptide-based optical probes. After a brief introduction to fluorophores, we explain the basic concepts and applications of optical probes categorized as fluorophore-labeled and fluorophore-quenched molecular imaging probes.
4.1. Fluorophore-labeling Strategies
A fluorophore molecule has the capability to absorb photons of energy at one wavelength and subsequently emit the energy at a longer wavelength. This phenomenon has been widely utilized in preclinical research to track biological molecules or to study biochemical events. Fluorescent dye molecules typically contain an aromatic ring system as the generator of luminescence, and a larger aromatic ring system usually suggests a better quantum yield and a longer emission wavelength. Several important criteria for measuring the dye capacity are its efficiency of absorbing photons, emitting photons, and ability to undergo repeated excitation-emission cycles (i.e. photostability). Two critical indices are molar extinction coefficient (ε) and quantum yield (QY), which correlate with absorption and fluorescence, respectively, and which determine the overall fluorescence intensity of a fluorophore. For commonly utilized fluorophores, the two parameters are in the range of 5,000 - 250,000 cm−1M−1 for ε and 0.05 to 1.0 for QY.153 A wide selection of fluorophores with emission wavelengths spanning from visible to NIR spectrum has been developed. The choice of fluorophores is based on experimental needs, and the selection differs between in vitro and in vivo applications. Fluorophores with emission wavelengths between 400 nm and 600 nm, such as 7-amino-4-methylcoumarin (AMC), fluorescein isothiocyanate (FITC), and 5-carboxytetramethylrhodamine (TAMRA), are overwhelmingly used for in vitro cellular imaging. On the contrary, fluorophores with emissions in the near-infrared region (650 – 900 nm) are widely used for in vivo applications. This is because the latter group has better tissue penetration and interferes less with the background generated from water, hemoglobin, and deoxyhemoglobin (absorbance 560 nm).154
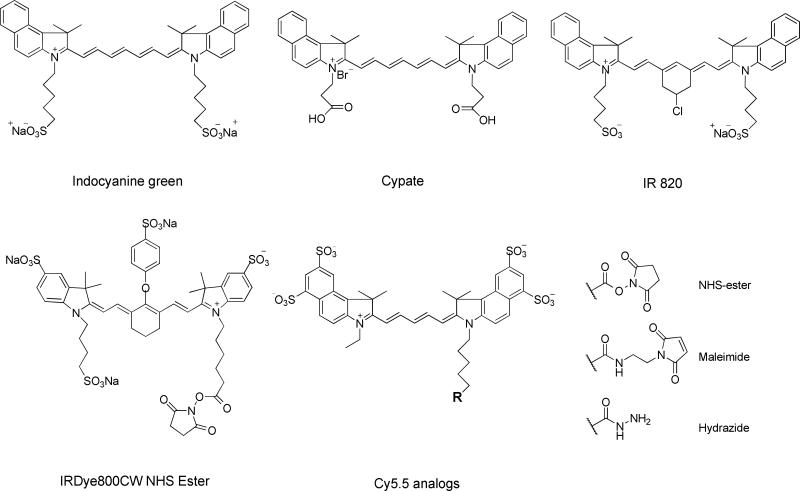
Most NIR fluorophores that are in practical use are essentially polymethines, and, in particular, heptamethine cyanines, which are comprised of benzoxazole, benzothiazole, indolyl, 2-quinoline, and 4-quinoline subclasses.29 These include indocyanine green (ICG, emission 830 nm), an FDA-approved tricarocyanine dye commonly used as an angiographic agent. Its analogs that have successively been developed include bispropylcarboxymethylinodocyanine (Cypate), Cy dyes from GE Healthcare, Alexa Fluor dyes from Invitrogen, and IRdye dyes from Li-COR Bioscience. Without charged groups, those heptamethine indocyanines are highly lipophilic and are toxic due to a high susceptibility of intracellular accumulation.155 To address that, the commercial formulas are di- or even tetra-sulfonated, which dramatically increases their solubility in physiological environments and reduces the risk of in vivo toxicity. At the same time, efforts to develop novel fluorophores have been continuous, aiming at achieving higher quantum yield, better photostability, and lower toxicity. Gremlich et al. synthesized a fluorescence oxazine dye AOI987, which has a maximum excitation at approximately 670 nm, and was demonstrated to have specific interaction with amyloid plagues in an APP23 transgenic mice model.156 Carreira et al. reported the preparation of conformationally restricted aza-dipyrromethene boron difluoride (aza-BODIPY) dyes, which demonstrate intense absorption, strong fluorescence, and high chemical- and photostability.157 In addition, Weissleder and his colleagues have developed a series of Nile Blue analogs, which may be 10-times brighter than Nile Blue and possess an excitation peak close to 680 nm.158 To date, a number of NIR fluorophores have been reported and their reactive intermediates for peptide bioconjugation are commercially available (Fig. 12). For instance, NIR fluorophore is provided in the forms of - NHS ester, /maleimide, and -hydrazide, with the targeting functional groups being amine, thiol, and carbonyl, respectively.
Figure 12.
Selected near-infrared (NIR) dyes.
The fluorophore molecules are sensitive to the environment, and their emission may vary with changes of pH, solvent, and buffer components.159 This can be beneficial, as when the fluorophore and quencher are arranged in close proximity (< 10 nm), strong fluorescence quenching can be induced by fluorescence resonance energy transfer (FRET)160 and dark quenching mechanisms.161 Fluorescence that has been quenched by FRET can be restored, and this manipulated fluorescence can report and image specific events. This principle has been employed in the design of molecular beacons or activatable imaging probes, which are effective in monitoring enzyme-based pathological alternations (Fig. 13).7 This probe is optically silent in its quenched state and becomes highly fluorescent following proteolysis of the peptide substrate linker by a target enzyme. Such a probe usually contains two (or more) identical or different fluorophores, which are joined together through a peptide linker that is the substrate of certain specific enzymes such as proteases. Proteases are enzymes that hydrolyze specific peptide substrates within proteins and are overexpressed in a number of pathologies.162 These probes can be activated by peptide cleavage induced by enzymes, which generates a strongly manipulated fluorescence signal at the target region (e.g., tumor). Compared with common targeting strategies, this activatable approach may provide a cleaner background and a better contrast/sensitivity, and hence has attracted increasing attention. To date, combination of NIR dyes and optical imaging instruments has expanded these techniques and allowed in vivo applications. The remainder of this section discusses recently developed optical imaging probes that combine various fluorophores with peptides or peptide hormones to detect and image target diseases in vivo. Optical probes associated with nanomaterials, including quantum dots and polymeric, inorganic nanoparticles, will be discussed in the next section.
Figure 13.
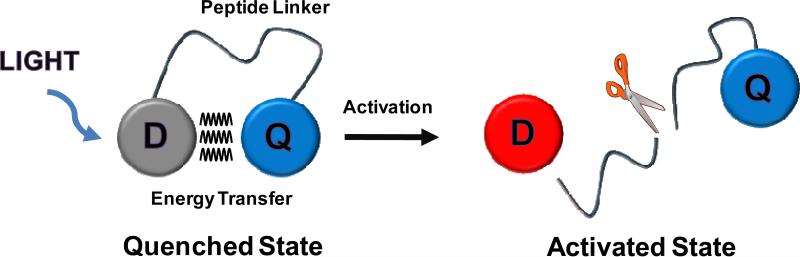
Simple schematic diagram of peptide-based activatable probe; D: Dye and Q: quencher.
4.2. Fluorophore-labeled peptide probes
Biologically-active peptides can be simply labeled with a fluorophore for optical imaging, which is similar to the probe design seen in radio-labeled peptide probes. Many active peptide analogs introduced in the previous section (dealing with nuclear imaging) have been developed as optical imaging probes by specific conjugation of NIR fluorophores. Receptor tyrosine kinases are important therapeutic targets for anti-neoplastic targeted therapies. Mesenchymal-epithelial transition factor (c-Met) is a receptor tyrosine kinase that is overexpressed in a number of malignancies, such as glioma.163 In order to investigate the possibility of c-Met receptor targeting by using optical imaging systems, Cy5.5-conjugated c-Met binding peptide (cMBP) was designed.164 cMBP with the sequence Lys-Ser-Leu-Ser-Arg-His-Asp-His-Ile-His-His-His containing Gly-Gly-Gly-Ser-Cys as a linker at the C-terminus was labeled with Cy5.5-maleimide through the thiol group of the cysteine residue. The binding affinity of the Cy5.5-cMBP was in the nanomolar range in U87MG cells and the probe displayed high tumor uptake at 24 h post-injection in a U87MG tumor model.164 In vivo use of a peptide-NIR dye conjugate consisting of an indotricarbocyanie (ITCC) dye and the octreotate has been reported for tumor imaging.165 The octreotate (D-Phe-c(Cys-D-Trp-Lys-Thr-Cys)-Thr) labeled at the N-terminus with ITCC and ITCC-octreotate showed strong receptor binding and cellular retention properties in RIN38 rat insulinoma cells overexpressing SSTr-2. When used in mice bearing RIN38-SSTr-2 tumors, the probe provided three-fold higher fluorescence in the tumor than in the muscle after intravenous injection. Libraries of SST peptide-dye conjugates differing in core peptide backbone cyclic structure, length of alkyl linker, and fluorophore moiety were prepared and tested in SSTr-positive H69 human SCLC tumors and a HT-29 colon cancer model.166,167 Optical probes associated with bombesin receptor have been developed by labeling Cypate and Alexa Fluor 680 to bombesin analogs, such as [Gly-Ser-Gly]-BBN7-14 and [Gly-Gly-Gly]-BBN7-14, respectively.168,169 Those probes have demonstrated receptor specific targeting ability in vitro and in pancreas, prostate, and breast tumor mice models. Various peptide probes with angiogenesis specific targets have been developed using NIR fluorophore-conjugated RGD peptide analogs. Cy5.5 dye conjugated to the ε-amino group of the lysine residue of c(RGDyK) or c(RGDfK) allows delineation of U87MG glioblastoma and KS167 Kaposi's sarcoma xenograft with high contrast (Fig. 14).170,171 In addition, conjugating a presumably inactive linear hexapeptide, Gly-Arg-Glu-Ser-Pro-Lys, with a Cypate yields a bioactive ligand, Cyp-GRD, that targets integrin-positive tumors.172 Besides imaging of tumors, Cy5.5-c(RGDyK) allows specific imaging of integrins expressed on macrophages recruited to vascular lesions and enables optical imaging of macrophage-rich atherosclerotic plaques in a mouse model of accelerated atherosclerosis.173 As described in the previous section, di-, and tetrameric RGD peptide analogs, such as Glu-[c(RGDyK)]2 and Glu-[Gly-[cyclo(RGDyK)]2]2, can also by labeled with NIR fluorophores, including Cy5.5 and Cy7, and demonstrate increased receptor binding affinity and imaging efficacy compared to the monomeric compound.174,175 Furthermore, a new type of Cy5 conjugated tetrameric molecule has been prepared by grafting four copies of c(RGDfK) molecules onto a cyclic decapeptide platform designated regioselectively addressable functionalized template (RAFT).176 Cy5-RAFT-c(RGDfK)4 has demonstrated higher uptake and prolonged retention in HEK293(β3) tumors in vitro and in vivo.177 Recently, Cy5.5-labeled integrin-binding knottin peptides--small (molecular weight approximately 3 kDa) and conformationally-constrained peptides that bind to integrins with high affinity (IC50, 10 – 30 nmolL−1)--have been developed and tested in U87MG glioblastoma xenograft models.178
Figure 14.
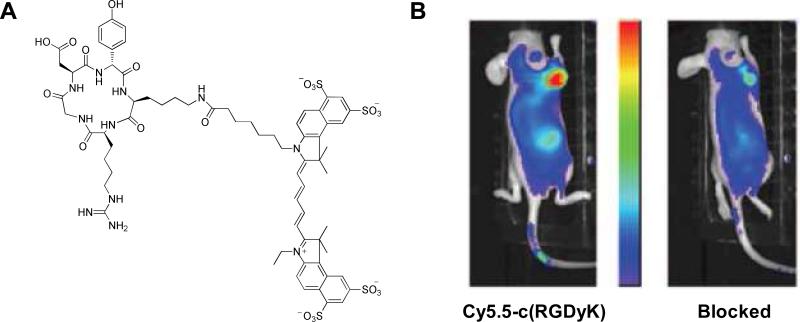
(A) Structure of c(RGDyK)-Cy5.5. (B) In vivo NIR fluorescence images of U87MG tumor-bearing mice at 4 h after intravenous injection of c(RGDyK)-Cy5.5 only (left) and a blocking dose of c(RGDyK) peptide (right). Modified with permission from ref. 170. Copyright 2004, the American Association for Cancer Research.
Fluorophore-labeled homing peptides selected from phage display have been successfully applied for in vivo optical imaging. Imaging of phosphatidylserine (PS) exposure is an extensively used molecular marker in noninvasive apoptosis imaging.179 A PS-recognizing peptide, Cys-Leu-Ser-Tyr-Tyr-Pro-Ser-Tyr-Cys, was identified by phage display and its binding of fluorescein-labeled peptide to apoptotic versus normal cells was assessed in vitro and in H460 cell-bearing tumor mice.180 Similarly, peptides that display sequences such as Cys-Leu-Trp-Thr-Val-Gly-Gly-Gly-Cys and Cys-Leu-Glu-Val-Ser-Arg-Lys-Asn-Cys have been successfully applied for imaging of atherosclerotic plaques in low-density lipoprotein receptor-deficient mice and apoptosis in a rat model of focal cerebral ischemia.181,182 Furthermore, a clinical trial of phage-selected peptide was reported using optical imaging techniques such as fluorescence endoscopy. A human colonic-adenoma-specific peptide, Val-Arg-Pro-Met-Pro-Leu-Gln, was screened, labeled with fluorescein, and successfully used for the detection and imaging of colonic dysplasia in humans by using a fluorescence confocal microendoscope.183 A phage-display-selected cyclic peptide containing the His-Try-Gly-Phe motif was used to identify stable and potent MMP inhibitor peptide for NIR imaging applications. Cy5.5-C6, Cy5.5-c(Lys-Ala-His-Trp-Gly-Phe-Thr-Leu-Asp)-NH2, was selectively taken up by MMP-2 expressing U87 cells in vitro and was visualized in mice with prostate PC-3 tumors.184 The use of phage display technique is a powerful method for identifying affinity peptides for various applications. However, in vivo validation of in vitro hits remains a challenge. A high-throughput method for identifying and optimizing peptide ligands to map and image biological targets of interest in vivo has been developed, using fluorophore-labeled phage clones as in vivo imaging probes.185 Novel phages with high affinity for osteonectin and vascular cell adhesion molecule/1 (VCAM-1), Ser-Pro-Pro-Thr-Gly-Ile-Asn, and Cys-Val-His-Ser-Pro-Asn-Lys-Lys-Cys have been identified and labeled with NIR fluorophores for a model target of invasive cancer and inflammatory endothelium, respectively.185 Each fluorophore-labeled phage clone demonstrates excellent in vivo targeting abilities in tumors and VCAM-1 expressing vessels, suggesting that they can be used as a platform for the development of imaging agents. Similarly, phage clones displaying the peptide sequence Ile-Ala-Gly-Leu-Ala-Thr-Pro-Gly-Trp-Ser-His-Trp-Leu-Ala-Leu have been identified and labeled with Alexa Fluor 680, and shown to bind and target PC-3 prostate carcinomas in a mouse model.186 Targeting peptide ligands selected by OBOC combinatory chemistry and high-throughput on-bead cell binding assays were labeled with Cy5.5 and applied to in vivo tumor imaging.187,188 Through screening OBOC peptide libraries against live MDA-MB-231 cells, a series of cyclic peptide ligands were identified, and their targeting efficiency to breast tumors in mouse was confirmed by in vivo NIR imaging. Because optical imaging is highly sensitive and able to detect picomolar to nanomolar concentrations of fluorophores, the approach of imaging by receptor-specific peptides could be successfully adopted by replacing the radionuclide with a fluorophore. It is well-suited for in vivo high-throughput screening studies and represents a potential alternative to nuclear imaging studies in the preclinical setting.
4.3. Fluorophore-quenched Activatable Peptide Probes
Until recently, the majority of peptide-based activatable probes targeted proteases. Various types of activatable probes targeting a wide range of proteases and protease-associated diseases, including cancer, inflammation, vascular disease, and infectious disease have been developed.7,189,190 Many activatable strategies utilize peptide chemistry to image protease activities in vivo. The simplest form of this peptide-based probe connects the NIR fluorophore and quencher with a protease-specific peptide spacer. A dual-labeled matrix metalloproteinase (MMP)-activatable probe was designed for potential use in imaging MMP-7 activities in tumor. The probe was synthesized by labeling Cy5.5 and NIRQ820 quencher to the MMP-7 substrate with a sequence of NIRQ820-Gly-Val-Pro-Leu-Ser-Leu-Thr-Met-Gly-Cys(Cy5.5)-Asp.191 Upon in vitro activation of this probe by MMP-7, a 7-fold increase in fluorescence was observed. Another example of a fluorophore-quencher pair involves the use of dark quencher.161,192 A dark-quenched, MMP-activatable peptide probe has been reported to image overexpressed MMP-13 in an osteoarthritis (OA) model.193 The fluorogenic peptide was prepared by a combination of Cy5.5 dye, MMP-13 specific peptide substrate, and the black hole quencher-3 (BHQ-3) (Fig. 15). Incubation of the probe, Cy5.5-Gly-Pro-Leu-Gly-Met-Arg-Gly-Leu-Gly-Lys(BHQ-3), with MMP-13 resulted in approximately a 30-fold increase in the NIR fluorescence signals, and the signal could be inhibited in the presence of MMP-13 inhibitor. Optical images in a rat model demonstrated that this probe can monitor and analyze the early and late stages of OA by imaging MMP-13 activities in vivo.193 Peptide probes have been reported that are selectively activated by enzymes and transported into the cells only after cell-specific activation based on the concept of activatable cell-penetrating peptide (ACPP) system.194,195 The ACPP is composed of a fluorophore-polycationic peptide (CPP, i.e. oligo(arginine)s) and a quencher-polyanionic peptide (polyanion, i.e. oligo(glutamic acid)s). These two strands are fused to a peptide sequence via various MMP-cleavable linkers, including Pro-Leu-Gly-Leu-Arg-Gly.195 This probe utilizes polycation-polyanion complexation to neutralize the charge of the CPP, thus inhibiting the cell-penetrating capabilities of the probe. Subsequent cleavage of the peptide linker by enzyme will dissociate the inhibitory polyanions, thereby allowing the fluoresce-activated CPP to enter the cells. These probes have been used successfully to identify MMP-2-9-positive tumors in vivo with a 3-fold increased fluorescence signal.
Figure 15.
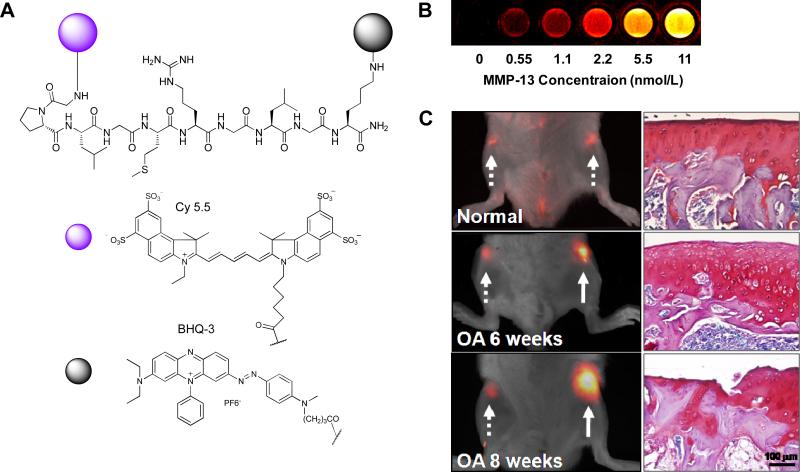
(A) Structure of MMP-13 activatable peptide-based probe, Cy5.5-GPLGGMRGLGK(BHQ-3)-NH2. (B) NIR fluorescence image of the probe in various concentrations of MMP-13 after a 40 min incubation at 37°C. In vivo imaging of upregulated MMP-13 in normal, six and eight week OA-induced cartilages 1 h after intracartilage-injection of the probe; (left) NIR fluorescence reflectance imaging of normal and OA cartilage after local injection of the probe, (right) histological evaluation of normal, six and eight week OA joints by Safranin-O staining, Arrows; dotted line (normal) and solid line (OA). Modified with permission from ref. 193. Copyright 2009, American Chemical Society.
Apoptosis is a programmed cell death process in multicellular organisms that plays key roles in the pathogenesis of many disorders, including autoimmune and neurodegenerative disorders, and cardiovascular disease, as well as in tumor responses to chemotherapy or radiotherapy.196 Because most anticancer therapies initiate apoptosis, imaging the progression of apoptosis could assist the clinical monitoring of apoptosis-related drug efficacy. A number of recent advances in optical imaging have enabled real-time imaging of apoptosis by monitoring specific apoptosis signaling molecules, such as caspases. Caspases are crucial mediators of apoptosis, and thereby represent obvious targets for apoptosis imaging.197 To image apoptosis in vitro and in vivo, a cell-permeable and caspase-activatable NIR probe, TcapQ, has been reported.198,199 To allow cell penetration, TcapQ pairs a membrane-penetration peptide, Lys-Lys-Lys-Arg-Lys-Val, with a caspase recognition peptide substrate, Asp-Glu-Val-Asp. Addition of the fluorophore-quencher pair, Alexa Fluor 647 and QSY 21, will strongly quench the cleavable caspase-specific substrate. An in vitro enzyme assay has demonstrated that TcapQ is efficiently cleaved by caspase-3 and caspase-7. Incorporation of the activatable strategy onto the cell-permeable caspase substrate has resulted in amplified fluorescence signals in apoptotic cells in vitro and in a rat model of glaucoma.198,199 Other caspase-3 activatable peptide probes have been designed using NIR dye-dye pairs, in which heptamethine carbocyanine dyes, cypate and Me2N-cypate are linked via the caspase-3 substrate Ac-Gly-Lys(Me2N-cypate)-Asp-Glu-Val-Asp-Ala-Pro-Lys(cybate).200 The probe has shown a nearly 2-fold increase in NIR fluorescence signal in paclitaxel-treated apoptotic A549 tumor cells and in caspase-3 rich tissues in a mouse model. As an alternative to using quenched peptide probes that become fluorescent after cleavage of the peptide by proteases, quenched activity-based probes (qABPs), which become fluorescent only after labeling by active proteases, have been developed.201 This probe contains a Cy5 and QSY21 quencher in close proximity via peptide acyloxymethylketones analogs that are quenched in their native state. When qABPs encounter the target enzyme, the probe binds to the enzyme and activity-dependent covalent modification releases the quencher, causing the probe to produce strong fluorescence. These cell-permeable qABP platforms have been successfully applied to labeling and imaging various proteases in living cells, and after intravenous injection have demonstrated their potential for real-time, whole-body imaging of capthepsin and caspase-3 activity in mice bearing grafted tumors.201-203
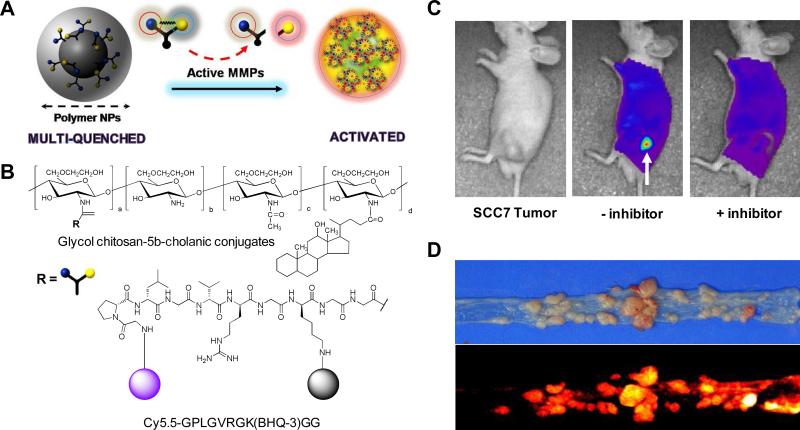
Recent polymer chemistry has generated a significant number of biocompatible polymers, including poly(amino acids), dendrimers, branched, graft, and block-co-polymers. Generally, polymer-based imaging probes have a number of enriched functional groups, which are ideal for efficient modification with imaging agents, prolonged plasma half-lives, improved stability, and targeting.204 A poly-L-lysine-associated peptide imaging probe has been developed for MMP imaging.205 This probe consists of multiple Cy5.5 fluorophores conjugated to a methoxy polyethylene glycol grafted poly-L-lysine backbone via a MMP-cleavable peptide linker Gly-Pro-Leu-Gly-Val-Arg-Gly. Cy5.5-peptide attaches to the polymer backbone closely enough to permit FRET induced by NIR dye-dye self-quenching. The probe can be activated by MMPs in vitro and in disease models of cancer and inflammatory disease.206-208 Another self-quenched peptide probe was designed based on a tetravalent, branched lysine core, where the dendritic arms that extend from the core incorporate a dipeptide, Lue-Arg, as the substrate for the targeted protease, cathepsin S.209 The four N-termini of the peptide are attached to the NIR fluorophore, CyTE-777 (emission 812 nm)210, via different lengths of PEG linkers (4, 8, and 12 ethylene oxide units, respectively). Upon proteolytic activation with cathepsin S, the probe displays in excess of a 70-fold increase in fluorescence emission. A dendrimer has also been used as the polymer backbone. A Cy5.5 fluorophore was conjugated to a PEGylated polyamidoamine (PAMAM)-Generation 4 dendrimer core via the MMP-7 cleavable peptide, Arg-Pro-Leu-Ala-Leu-Trp-Arg-Ser.211 When the probe was intravenously injected into MMP-7-positive tumor-bearing mouse, 2.2-fold higher fluorescence was observed than in a contralateral control tumor. Activatable peptide imaging probes with highly efficient quenching and low background noise can be used to overcome the limited optical resolution and low specificity of conventional optical imaging probes. The unique properties and applications of the various activatable imaging probes designed by combining fluorophores and materials, such as small molecules, polymers, and novel metals, have been summarized elsewhere.7,189,190.
Optical imaging techniques have become essential tools for small animal imaging and have already made a substantial impact on basic research, drug discovery and development, and preclinical studies. Furthermore, clinical adoption of optical imaging is currently underway in the fields of breast and endoscopic imaging, and its applications are expected to expand significantly. There is a clear need, however, for safe, specific, and reliable optical imaging probes. This will be facilitated by the recently developed of peptide-based optical imaging probes.
5. Nanoplatform-based Peptide Probes for Molecular Imaging
The combination of modern nanotechnology and molecular imaging science has yielded new strategies for designing nanoplatform-based imaging probes that efficiently detect and recognize disease-associated biomolecules.2 The most well-investigated nanomaterials include magnetic iron oxide nanoparticles (IONPs) for MRI, quantum dots (QDs) for optical imaging, polymeric nanoparticles, and carbon nanotubes. The attractiveness of such marriages arises from the unique size scale and physical properties provided by the nanomaterials, which afford unprecedented probe-biomolecule interactions, and allow visualization of biological events at subcellular or molecular levels.2 Besides the physical properties, nanoparticles also afford a large surface area that allows docking of one, or even multiple types of functional groups. Such multiple-peptide docking could lead to a much improved binding profile via a so-called polyvalent effect.212,213 Also, successful demonstrations have been made of integrating different imaging modalities into a single nanoplatform to achieve comprehensive lesion characterization.212,214 Prospectively, the nanoplatforms may provide an all-in-one solution that combines versatile targeting ligands, imaging probes, and therapeutic drugs, and allows simultaneous diagnosis, drug delivery, and monitoring of therapeutic response. In the next section, we will introduce various emerging approaches enveloped by the term “nanoparticle-labeled peptide probes” for in vivo imaging. There are many other nanomaterials that are under intensive studies for in vitro or cellular imaging that are beyond the scope of this review article.
5.1. Nanoparticle-labeling Strategies
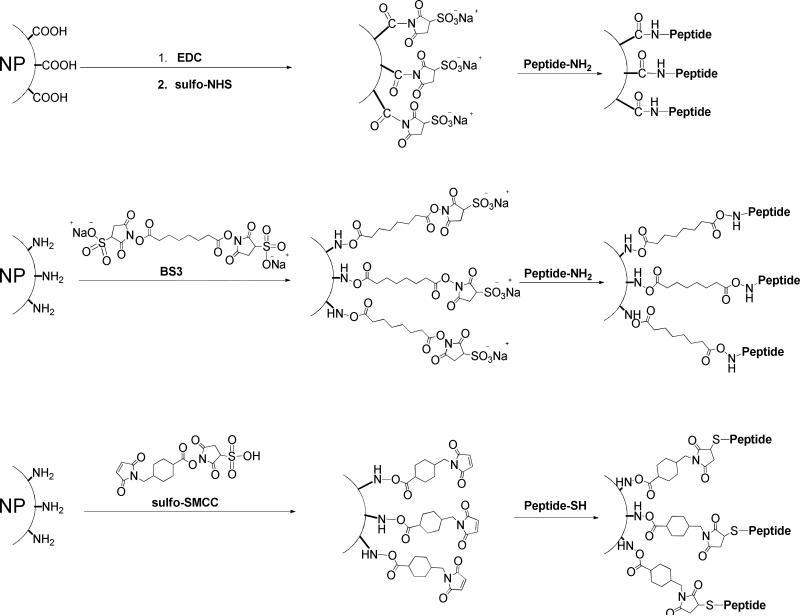
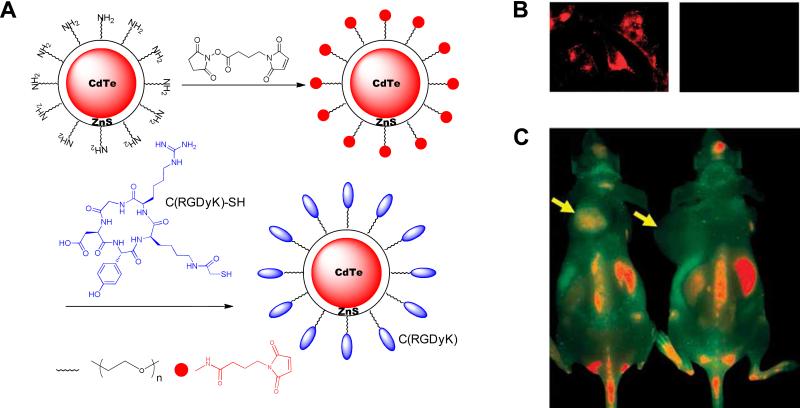
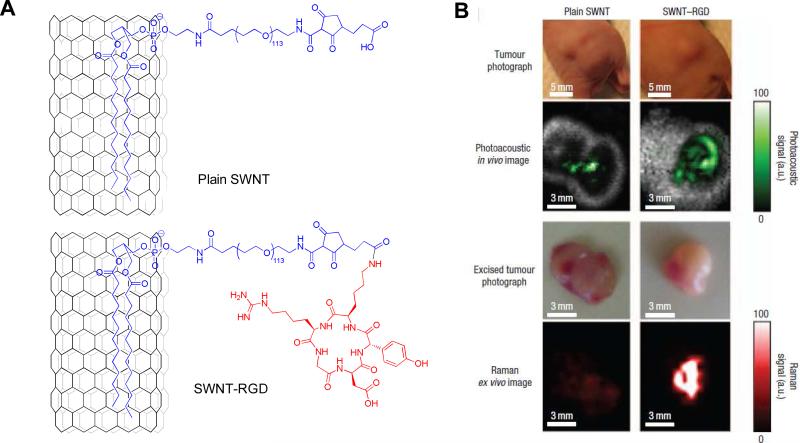
Due to the differences among the nature of particle surfaces, nanoparticle coating strategies vary accordingly. A broad range of materials, ranging from synthetic polymers, such as poly(vinylpyrrolidone) (PVP), poly(lactic-co-glycolic acid) (PLGA), poly(ethyleneglycol) (PEG), to natural macromolecules, such as oligonucleotide, polysaccharides, peptides, and proteins, have been investigated as particle coating materials.215-220 However, despite the complexity of these strategies for coating nanoparticles, the purpose of particle surface decoration is always to yield nanoplatforms with high stability and facile docking sites for functional molecules. In most cases, this refers to the presence of multiple carboxyls/amines/thiols on the particle surface, which facilitates the linkage between particle and peptide, independent of the cores (Fig. 16). For example, regardless of the core materials, the coupling between carboxylated nanoparticles and peptides is predominantly achieved by forming amide bonds between particle carboxyls and amines from either lysine side chain or N-termini amino acid. Most such reactions are conducted in buffer solution at low temperature (4°C to room temperature) using EDC-NHS as catalysts. The linkage can be achieved in a single step, where all the reactants and catalysts are added at the same time. A more common strategy, however, is a two-step process where the activation step is separated from the coupling step. Instead of direct coupling, the aminated-thiolated particle conjugation with peptides is in most cases accomplished with a bifunctional linker, which forms bonds with complementary moieties. For example, 2,3-dimercaptosuccinic acid (DMSA)-coated IONPs, which have a thiolated surface, can be coupled with peptides-antibodies in a two-step procedure via the mediation of sulfosuccinimidyl 4-[N-maleimidomethyl]cyclohexane-1-carboxylate (sulfo-SMCC).221-223 Also, bis[sulfosuccinimidyl] suberate (BS3) or its analogs can be utilized to link aminated quantum dots and peptides, by forming amide bonds with both sides. However, in the latter case, there is a high risk of crosslinking that may occur among the peptides/particles. To avoid this, one moiety is pre-thiolated, after which the coupling can be treated as a thiol-amine coupling. For instance, c(RGDyK) treated with S-acetylthioglycolic acid N-hydroxysuccinimide ester (SATA), followed by thiol deprotection with hydroxylamine under neutral conditions yields the thiolated RGD peptide c(RGDy(ε-acetylthiol)K). Subsequently, these peptides are coupled onto amine-QDs with a heterobifunctional linker, 4-maleimidobutyric acid N-hydroxysuccinimide ester, which contains a maleimide group and an ester group that are reactive toward thiol and amine, respectively.224
Figure 16.
Nanoparticle-labeling of peptides.
Currently utilized nanoparticles are in the range of 10 to 100 nm. Compared with peptide probes that are less than a few nanomaters, these particles are more difficult to extravasate and diffuse in the tumor interstitial space. This may explain the overwhelming number of studies on nanoprobes targeting markers such as integrin ανβ3 on vasculature. Also, conjugation with nanoparticles may dramatically change the biodistribution of the parent peptides. While peptides are mainly cleared through renal secretion, most nanoparticles end up being trapped in the liver, spleen and even lung by reticuloendothelial system (RES) uptake. A common solution is to couple antifouling agents, such as PEG, onto nanoparticles to extend their circulation half-life and to facilitate target organ-tissue uptake. A number of targeting and imaging moieties conjugated on a single nanomaterial will demonstrate improved specificity and amplified imaging signals at the target region. Peptides and peptide hormones can be engineered as nanoplatforms for targeted delivery of imaging agents to enhance targeting efficacy, plasma half-lives, and stability, and to reduce non-specific binding. There has been significant advancement in the field of nanoplatform-based probes, and several comprehensive review articles have summarized and discussed their application.2,28,30,151 In the next section, we discuss nanoplatform-based probes for in vivo imaging that have been generated by peptide-based approaches.
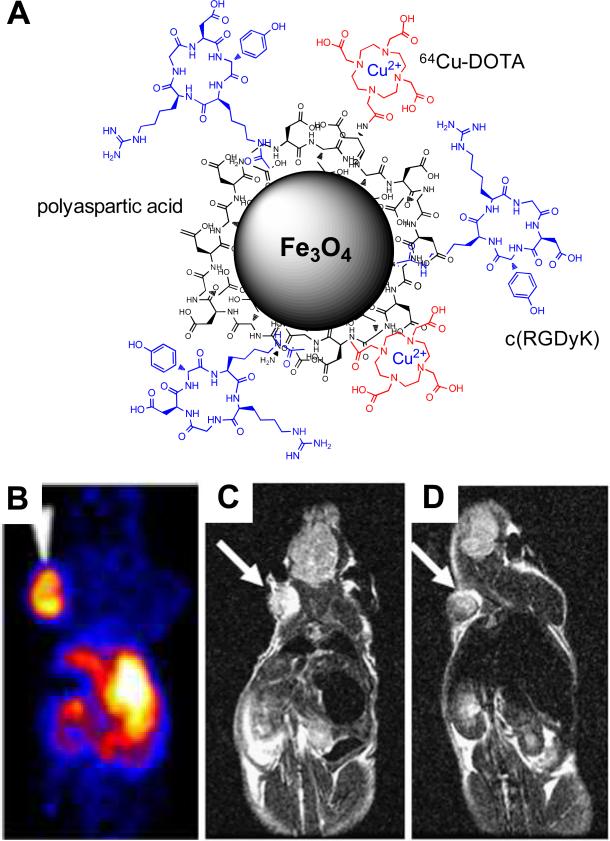
5.2. Iron Oxide Nanoparticle-Peptide Probes
The term iron oxide usually refers to maghemite and magnetite (Fe2O3, Fe3O4), which are strong magnetic materials (92 emug−1 for magnetite and 78 emug−1 for maghemite at 300K)217, and are generally regarded as biocompatible and biodegradable.215 When made in nanoscale, these IONPs become superparamagnetic, i.e., a state of possessing zero magnetic moment when no external magnetic field is present. With the presence of an external magnetic field, such as in a MRI system, these small magnets are readily magnetized, turning into sensitive probes that attenuate the local signals on a T2 or T2* weighted map.223 These features have made them a major class of contrast agents in MRI, which compensate the intrinsic low sensitivity of the latter.28,215,225,226 Due to their superior magnetic property, ease of modification, and biocompatibility, IONPs are one of the most studied nanoparticles in the field of molecular imaging, and several IONP formulations have been approved by the FDA for MRI and are widely used in clinics.28 Recently, IONPs have been used extensively to label and track the migration of cells, including stem cells, immune cells, and many other cell types, in numerous models.227 Conventional IONP formulations rely on nonspecific passive targeting in vivo, and it is difficult to deliver a sufficient dose of such imaging agents to a target region (e.g., tumor). To overcome this limitation, a number of targeting molecules, including peptides, have been used to modify the surface of IONPs and allow the targeting of specific tissues in vivo. IONPs targeting and imaging integrins that are expressed in tumors cells have been developed.228 RGD peptide analogs, [Gly-Ser-Ser-Lys(FITC)-Gly-Gly-Gly]-c(RGD), have been chemically conjugated to the primary amines on amino cross-linked iron oxide (CLIO) nanoparticles using disuccinimidyl suberimidate. The NPs demonstrate integrin-mediated cellular uptake in BT-20 tumor cells in vitro and localize in BT-20 tumors in vivo. In another example, c(RGDyK) peptide was conjugated to 4-methylcatechol (MC)-coated ultrasmall IONPs (4.5 nm) via a Mannich reaction.229 The resulting c(RGDyK)/MC-IONPs have an overall diameter of 8.4 nm and are stable under physiological conditions. The NPs have shown 5-fold higher cellular uptake in integrin-positive U87MG cells compared to integrin-negative cells and accumulate preferentially in tumors when intravenously administered in an U87MG tumor-bearing mice model. Chlorotoxin (CTX), a 36-amino acid peptide that has a strong affinity for tumors of neuroectodermal origin230,231, has also been introduced into nanoparticles. The nanoparticles comprised IONP-coated with PEG or PEG-grafted chitosan copolymer, to which CTX was conjugated.232,233 The probes showed higher internalization and localization properties in mice bearing 9L xenograft tumors and in transgenic ND2:SmoA1 mice, respectively. The specificities of luteinizing hormone-releasing hormone (LHRH)-bound IONPs were tested in human breast cancer cells in vitro and in a xenograft model.234 LHRH is a decapeptide comprised of naturally occurring amino acids, Glu-His-Trp-Ser-Tyr-Leu-Arg-Pro-Gly-NH2, and over 50% of human breast cancers express receptor binding sites for LHRH.235
MRI provides three-dimensional tomographic images and higher spatial resolution; however, a limitation of MRI is the inherently low sensitivity of the scanner. In order to combine the benefits of MRI with those of other imaging modalities, including PET and optical imaging, growing attention has focused on the development of dual-imaging probes.6 Such probes are designed to combine the high spatial and temporal resolution and tissue penetration of MRI with the high sensitivity of PET and the optical imaging methods. An RGD-peptide-based MRI/PET probe has been developed by conjugation of 64Cu-DOTA and c(RGDyK) on the surface of polyaspartic acid-coated IONPs (Fig. 17).212 When administered intravenously in U87MG tumor-bearing mice, both PET and MRI have demonstrated integrin-specific delivery by nanoparticles. Nanoparticles for MRI and optical imaging have also been frequently studied. A tumor-specific contrast agent targeting the underglycosylated mucin-1 antigen (uMUC-1) tumor antigen has been reported for the application of MRI-optical dual-imaging.236 uMUC-1 provides a unique advantage for imaging and monitoring the therapeutic response of breast cancer, because it is found on more than 90% of breast cancers and is predictive of chemotherapeutic response.237 The probe consists of IONPs for MRI, modified with Cy5.5 for optical imaging, and conjugated to uMCU-1-targeting peptide, EPPT (Cys-AHA-Ala-Arg-Glu-Pro-Pro-Thr-Arg-Thr-Phe-Ala-Tyr-Trp-Gly-Lys(FITC). The probe shows specificity for uMUC-1 in vitro, and the dependence of tumor-specific accumulation on the expression of uMUC-1 in a uMUC-1-positive BT-20 tumor-bearing mice model has been confirmed by both MRI and optical imaging. Similarly, different peptide-based IONPs, such as c(RGDyK), amino-terminal fragment of peptide of urokinase-type plasminogn activator (uPA) and CTX conjugated IONPs, are simply-labeled NIR fluorophores that have been tested in vivo for their dual-imaging abilities.233,238,239 All the nanoparticles have the potential to be molecularly targeted and to serve as dual-modality imaging probes for in vivo applications. Besides peptide-conjugated IONPs, the recent development of various IONP-based platforms for imaging and therapy has been described and reviewed elsewhere.6,215,240
Figure 17.
(A) Schematic diagram of 64Cu-DOTA-IONPs-c(RGDyK) probe for PETG and MRI imaging. (B) microPET image of U87MG tumor-bearing mouse at 4 h after intravenous injection of the probe. T2-weighted MR images of mice (the arrow indicates the tumor) C) before and D) 4 h after intravenous injection of the probe. Modified with permission from ref. 212. Copyright 2008, the Society of Nuclear Medicine, Inc.
5.3. Quantum-dot-labeled Peptide Probes
QDs are made from semiconductor materials a few nanometers in diameter. Interest in the use of QDs for bioapplications arises from their unique optical properties, including photo-stability, broad absorption cross sections, wide absorption spectra, and narrow emission spectra.241 Many efforts to employ QDs as replacements for currently utilized organic dyes have been made.151,224,242,243 In the case of IONPs, pyrolysis based synthesis permits precise control over many optical aspects of QD characteristics by accurately tuning their size and composition. For instance, CdSe QDs with various sizes have emission spectra that can span most of the visible region, while the CdS and ZnSe QDs have emission spectra in the blue and near-ultraviolet regions.244 However, the application of these particles in living subjects may encounter problems of limited tissue penetration depth and high background. To address this issue, formulas, which include InP-, InAs-, and Cu-doped InP, with emission in the NIR range are under intensive study.245-248 Another motive for having such transitions is to avoid the utilization of some extremely toxic metals, such as Cd, Pb, and Hg. Although In, Cu and As are also toxic, generally, the human body has better tolerance of those metals.249 Several biocompatible QDs have been applied for labeling cells and long-term multicolor cell imaging.250 A variety of targeting peptides have been explored to label QDs for better targeting and specificity. For example, a cellular nuclear localizing sequence (NLS) (Pro-Pro-Lys-Lys-Lys-Arg-Lys-Val)251, cell-penetrating Tat peptides (Arg-Arg-Arg-Gln-Arg-Arg-Lys-Lys-Arg-Gly-Tyr)252, neuropeptide AST1 (Ala-Pro-Ser-Gly-Ala-Gln-Arg-Leu-Tyr-Gly-Phe-Gly-Leu-NH2)253, and neurotoxins such as CTX and dendrotoxin-1254, have been successfully labeled on QDs and have provided improved stability and targetability. However, so far, their use has been limited to in vitro. Most of the in vivo applications have been by non-targeted QDs, for instance, in vivo cell trafficking, vasculature imaging, sentinel lymph node mapping, and neural imaging.2 Molecularly-targeted imaging applications of QDs are limited mainly by their size, short half-lives, surface charge, surface functionality, and toxicity.255 There are very few reports in the literature of peptide-associated QDs with successful in vivo applications. For the first time, surfaces of QDs were decorated by peptides with affinity for tumor vasculatures, such as GFE (Cys-Gly-Phe-Glu-Cys-Val-Arg-Gln-Cys-Pro-Glu-Arg-Cys, which binds to membrane dipeptidase on the endothelia cells in lung blood vessels), F3 (Lys-Asp-Glu-Pro-Gln-Arg-Arg-Ser-Ala-Arg-Leu-Ser-Ala-Lys-Pro-Ala-Pro-Pro-Lys-Pro-Glu-Pro-Lys-Pro-Lys-Lys-Ala-Pro-Ala-Lys-Lys, which binds to blood vessels and tumor cells in various tumors), and LyP-1 (Cys-Gly-Asn-Lys-Arg-Thr-Arg-Gly-Cys, which recognizes lymphatic vessels and tumor cells in certain tumors).256 Each peptide-conjugated QD revealed tumor specificity in in vitro histology. However, the QD probes were not detected in living animals. Successful in vivo targeted imaging of tumor vasculature has been achieved by conjugating RGD peptide analogs to poly(ethylene glycol) coated QD705 (Fig. 18).224,257 Conjugation of thiolated RGD peptide to PEG-coated QDs was achieved through a heterobifunctional linker, 4-maleimidobutyric acid N-hydroxysuccinimide ester. A competitive cell binding assay showed that QD-peptide conjugates retain specificity to integrin receptor, and in vivo imaging of tumor has been successfully achieved in a U87MG xenograft model. In addition, this probe was further developed as a dual-modality probe for both PET and optical imaging by conjugating [64Cu-DOTA] on the surface of the QD-peptide.258 A similar strategy has been applied to develop cyclic Asn-Gly-Arg (cNGR) analog-conjugated paramagnetic QDs (pQDs) for in vivo imaging of tumor angiogenic activity.259,260 The probe was prepared by non-covalent conjugation of two different biotinylated peptide and DTPA analogs, Ac-c(NGR)/Gly-Gly-Lys(biotin)-NH2 and biotinylated poly-L-lysine-DTPA, with streptavidin-coated PEG-conjugated QDs. Subsequently constructed cNGR-pQDs allowed in vivo quantification and accurate localization of angiogenic activity in an LS1747 colorectal adenocarcinoma mouse model.259 More recently, zwitterionic QDs with an overall diameter of 5.5 nm have been coupled with RGD. Those particles demonstrated good tumor targeting and, owing to their extremely compact size, showed renal clearance within 4 h post-injection. The results of this study suggest that targeted nanoparticles, if properly designed, can be eliminated through renal clearance.261
Figure 18.
(A) Schematic diagram of QD705-RGD. (B) Staining of live U87MG cells with QD705-RGD (left) and QD705 (right). (C) In vivo NIR fluorescence imaging of U87MG tumor-bearing mice at 6 h after intravenous injection of QD705-RGD (left) or QD705 (right). Arrows indicate tumors. Modified with permission from ref. 224. Copyright 2009, American Chemical Society.
QDs have fulfilled their promise as a new class of probes for molecular imaging, while the exploration of peptide conjugates for in vivo targeting is still at an early stage. QDs modified with other macromolecules, including antibodies, antigens, aptamers, and targeting proteins for in vivo uses, have been reported and have been reviewed elsewhere 255,262,263.
5.4. Other Nanomaterial-labeled Peptide Probes
Recently, a number of molecular imaging applications have been proposed that are based on the unique Raman signatures and photoacoustic properties of carbon nanotubes. Development of Raman spectroscopy could potentially solve the common issues encountered in optical imaging, including background autofluoresence, limited tissue penetration depth, and photo-bleaching. Moreover, compared with fluorescence optical imaging, which has a limited number of wavelengths in the NIR region, Raman spectroscopy can differentiate the spectral fingerprint of many molecules and, therefore, encourages multiplexing imaging.264 The surface of single-walled carbon nanotubes (SWNTs) were coated with phospholipids bearing PEG linked c(RGD) peptides and thein vivo biodistribution and tumor targeting of the SWNTs were investigated.265,266 Effectively, PEGylated SWNTs were stable in serum and demonstrated desirable pharmacokinetics in vivo. Highly efficient targeting of integrin-positive U87MG tumors in mice has been achieved by the multivalent effect of decorated c(RGD) peptide and confirmed by Raman and PET imaging. Photoacoustic imaging provides optical contrast with improved spatial resolution and tissue penetration compared to purely optical imaging techniques. Besides the Raman characteristics, SWNTs have proven capable of producing very strong photoacoustic signals in a dose-dependent manner. Therefore, SWNT-c(RGD) was also applied in in vivo photoacoustic and Raman imaging, and it demonstrated an 8-fold higher tumor specificity in the tumor compared with mice injected with plain SWNT (Fig. 19).267
Figure 19.
(A) Schematic diagrams of single-walled carbon nanotubes (plain SWNT) and SWNT–RGD. (B) Photographs of the tumors in mice and the corresponding photoacoustic subtraction images (green) shown as horizontal slices through the tumors. After the photoacoustic scan, the tumors were excised and scanned using a Raman microscope (red). Modified with permission from ref. 267. Copyright 2008, Macmillan Publishers Limited.
Recent advances in biocompatible polymer chemistry have provided a number of different polymer structures, such as dendrimers, branched, amphiphilic, graft, and block-co-polymers, and all of these unique platforms have been associated with targeting peptide and investigated as imaging probes.30 Phage-selected atherosclerotic plaque-selective peptide with the sequence of Cys-Arg-Lys-Arg-Leu-Asp-Arg-Asn-Cys (AP) has been associated with self-assembled polymeric nanoparticles for atherosclerotic lesion optical imaging.268 AP binds to atherosclerotic plaque in vivo through interleukin (IL)/4 receptors on endothelial cells, macrophages, and smooth muscle cells.269 The peptide was chemically conjugated on the surface of Cy5.5 fluorophore-labeled hydrophobically modified glycol chitosan (HGC) nanoparticles (diameter 250 nm) by thiol-maleimide reaction. AP-conjugated HGCs bound avidly to IL-4 receptors and demonstrated improved specificity to atherosclerotic lesions in an Ldlr-/- atherosclerotic mouse model compared to binding in a normal mouse.269 Other examples of polymeric nanoparticles are surface-localized tumor vasculature targeting F3 peptides and encapsulated imaging agents.270 The core of the polymeric nanoparticle was synthesized from polyacrylamide, which was embedded with drug and/or imaging agents such as IONPs and fluorophores. PEG linker and a targeting peptide, F3, were attached to target these nanoparticles to cancer cells. Peptide labeled on the nanoparticles (40 nm) successfully targeted receptors on MDA-MB/435 cells in vitro and serial MRI confirmed its tumor specificity in rat 9L gliomas following intravenous injection. A biodegradable positron-emitting dendritic nanoprobe was developed for angiogenesis imaging using the RGD motif as a peptide ligand.271 A biodegradable heterobifunctional dendritic core, tyrosine-conjugated pentaerythritol, was synthesized and functionalized with heterobifunctional PEG chains. Cyclic RGD peptides were conjugated at the terminal ends of the PEG chains and tyrosine group moieties were radio-labeled with 76Br. The dendritic nanoprobe showed a 50-fold enhanced receptor binding affinity with respect to the monovalent RGD peptide in vitro and revealed highly specific accumulation of 76Br-labeled dendritic nanoparobes in muscles undergoing angiogenesis in a hindlimb ischemia mouse model for angiogenesis. Recently, a new protease activatable strategy has been designed by a combination of a polymeric nanoparticle platform and activatable dark-quenched peptide (Fig. 20).272 The probe consists of strongly quenched MMP-specific NIR activatable peptide probes (Cy5.5-Gly-Pro-Leu-Gly-Val-Arg-Gly-Lys(BHQ-3)-Gly-Gly) on the surface of tumor-homing polymeric nanoparticles (a self-assembled glycol chitosan-5β-cholanic conjugates) as a carrier. Chemically labeled activatable peptide probes on the surface of tumor-homing nanoparticles demonstrated a higher specificity and sensitivity of the probe in MMP-positive disease models (MMP-positive SCC-7 xenograft tumor and a colon cancer mouse model) compared to the peptide probe without nanoparticles, since the nanoparticles can deliver the probe effectively to the tumor region by the enhanced permeability retention (EPR) effect,273 and because the nanoparticles can be strongly dual-quenched by both the dye-dark quencher and NIR dye-dye self-quenching mechanisms.
Figure 20.
(A) Schematic diagrams of MMPs activatable polymeric nanoparticle-based probe. (B) Structures of the probe. (C) In vivo NIR fluorescence images of subcutaneous MMP-positive SCC7 tumor-bearing mice after intravenous injection of the probe with or without the MMP inhibitor. Only tumors injected with the probe without inhibitor were clearly visualized. (D) Upper: photo image of colon tumors from an A/J mouse treated with azoxymethane (AOM), lower: NIR fluorescence image of MMP-positive colon tumors after intravenous injection of the probe.
Superior and unique characteristics of inorganic and polymeric nanoparticles have generally been blunted by low specificity and low stability under physiological conditions. Simple decoration of these nanomaterials with targeting peptides may not overcome many of the current limitations. However, nanoparticles can provide researchers with potential targeting tools. Most of the examples introduced in this section are associated with well-known RGD peptides. However, the probe design platforms are flexible and tunable for a wide array of applications by substituting new peptide ligands. Most of the studies on peptide-nanoparticle-conjugate based imaging probes are still in the proof-of-concept stage. None of the above mentioned examples has advanced into clinical trials. Two of the major concerns, as mentioned above, are the acute and chronic toxicity of the nanoparticle materials. Other complications include potential immunogenicity and relatively poor batch-to-batch reproducibility of the nanomaterials.
6. CONCLUSIONS
The potential for preclinical and clinical applications of molecular imaging is tremendous. These powerful methods will offer invaluable opportunities to explore complex disease-related biological processes at the molecular level in vivo. The emergence of current molecular imaging technologies is not only dependent on the progress of imaging systems, but, more importantly, on the pool of available molecular imaging probes. Undoubtedly, the specific sequences of targeting peptides will play an essential role in the design of appropriate molecular imaging probes. Peptide-based imaging has now become an established approach in nuclear imaging, and its application is expanding to other imaging modalities.
In this article, we introduced and discussed numerous peptide-based molecular imaging probes with respect to their design strategies and applications. Beginning with the success of octreotide in the clinic, significant research effort has been made over the past two decades toward the development and evaluation of novel peptide-based probes. However, many of the sophisticated probes reported in animal models have failed to reach clinical reality and are still in the investigative stage. Significant challenges remain to be overcome that are largely responsible for the failure of classic peptide-based probe diagnostics to gain a significant foothold in the clinic. There is, for example, a need to discover novel synthetic peptides that are metabolically stable and capable of binding to specific receptors subtype in vivo. Various peptide-based probes have demonstrated good tumor uptake; however, they have frequently shown nonspecific uptake in normal organs, such as the liver and kidneys. An ideal probe should have favorable pharmacokinetics resulting in rapid target localization and clearance to produce a sufficient ‘target-to-background’ ratio of imaging signals. Another important issue is the availability of new techniques for efficient and reproducible labeling of peptides. Maintaining in vivo integrity of peptides labeled with imaging moieties, including chelators, short half-life radioisotopes and nanoparticles, needs to be guaranteed. Specific structural modification is also preferred to ensure that the presence of the imaging moiety has minimal impact on the biological activity of peptides. Development of optimized peptide-based probes is fundamentally different from traditional drug development. A major challenge is to harmonize many different factors to develop more sensitive, specific, and stable peptide-based diagnostics. The development and application of peptide probes has a long way to go. Despite these challenges, it is important to keep in mind that interdisciplinary research at the interface of imaging science, molecular peptide biology, and nanotechnology can provide numerous unexplored possibilities, and lead to novel probes that will become valuable preclinical and clinical tools, ideally in the near future.
Acknowledgment
This project was supported, in part, by the Intramural Research Program, NIBIB, NIH. We thank Dr. Henry S. Eden for English editing and proof-reading of the manuscript.
7. Abbreviations
- Aba
4-aminobutyric acid
- Ac
Acetyl
- ACPP
activatable cell-penetrating peptide
- Ahx
6-aminohexanoic acid
- Ava
5-aminovaleric acid
- α-MSH
alpha-melanocyte stimulating hormone
- BBN
bombesin
- BHQ-3
black hole quencher-3
- BODIPY
dipyrromethene boron difluoride
- BS3
bis[sulfosuccinimidyl] suberate
- c
cyclo
- c(RGDfK)
cyclo(Arg-Gly-Asp-D-Phe-Lys)
- c(RGDyK)
cyclo(Arg-Gly-Asp-D-Tyr-Lys)
- CB-TE2A
4,11-bis(carboxymethyl)/1,4,8,11-tetraazabicyclo[6.6.2]hexadecane
- CCK
cholecystokinin
- CCK-2r
CCK/2 receptor
- CCMSH
[Cys3,4,10,D-Phe7]-MSH3-13
- cMBP
c-Met binding peptide
- c-MET
mesenchymal-epithelial transition factor
- Css
disulfide bond between cysteine
- CTX
chlorotoxin
- CXCR4
chemokine receptor
- Cy
Cyanine
- Cypate
bispropylcarboxymethylinodocyanine
- DMSA
2,3-dimercaptosuccinic acid
- DOTA
1,4,7,10-tetraazacyclodecane-1,4,7,10-tetraacetic acid
- DOTA-NOC
[DOTA-Nal3]-octreotide
- DOTA-TATE
[DOTA-D-Phe1,Tyr3]-octreotide
- DOTA-TOC
[DOTA-Tyr3]-octreotide
- DTPA
diethylene triamine pentaacetic acid
- EDC
1-ethyl-3-(3-dimethylaminopropyl
- EDTA
ethylenediaminetetraacetic acid
- eIND
emergency investigational new drug
- EPR
enhanced permeability retention
- [18F]FAZA
[18F]fluoroazomycinarabinofuranoside
- [18F]FBEM
N-(2-aminoethyl)maleimide
- [18F]FDG
2-[18F]fluoro-2-deoxy-D-glucose
- [18F]FDGMHO
[18F]FDG-maleimidehexyloxime
- [18F]FLT
3’-deoxy-3’-[18F]fluorothymidine
- [18F]SFB
N-succinimidyl-4-[18F]fluorobenzoate
- [18F]SFBS
4-[18F]fluorobenzylamine succinimidyl ester
- [18F]SFMB
N-succinimidyl-4-([18F]fluoromethyl)benzoate)
- [18F]SFMB
3-[18F]fluoro-5-nitrobenzimidate
- [18F]SPB
4-[18F]fluorophenacyl bromide
- FITC
fluorescein isothiocyanate
- FPRGD2
NH2-PEG3-Glu-[c(RGDyK)]2
- FRET
fluorescence resonance energy transfer
- FRGD2
Glu-[c(RGDyK)]2
- GIST
gastrointestinal stromal tumor
- GLP/1
glucagon-like peptide-1
- GPCr
G-protein-coupled receptor
- GRP
gastrin-releasing peptide
- GRPr
GRP receptor
- HGC
hydrophobically modified glycol chitosan
- HYNIC
6-Hydrazinopyridine-3-carboxylic acid
- ICG
indocyanine green
- IONPs
iron oxide nanoparticles
- ITCC
indotricarbocyanie
- LHRH
luteinizing hormone-releasing hormone
- MAG2
mercapto Ac-Gly-Gly
- MAG3
mercapto Ac-Gly-Gly-Gly
- MG
minigastrin
- MMP
matrix metalloproteinase
- MRI
magnetic resonance imaging
- MTC
medullary thyroid carcinoma
- NDP
[Met4,D-Phe7]-MSH
- NGR
Asn-Gly-Arg
- NHS
N-hydroxysuccinimide
- NIR
near-infrared
- NOTA
2,2',2”-(1,4,7-triazacyclononane-1,4,7-triyl)triacetic acid
- NPY
nueropeptide Y
- NT
neurotensin
- OA
osteoarthritis
- OBOC
one-bead one-copound
- OctreoScan
[111In-DTPA]-octreotide
- PAMAM
polyamidoamine
- PDAC
pancreatic ductal adenocarcinoma
- PEG
poly(ethylene glycol)
- PET
positron emission tomography
- PLGA
poly(lactic-co-glycolic acid)
- pQDs
paramagnetic QDs
- PS
phosphatidylserine
- PVP
poly(vinylpyrrolidone)
- qABPs
quenched activity-based probes
- QDs
quantum dots
- QY
quantum yield
- RAFT
regioselectively addressable functionalized template
- RGD
Arg-Gly-Asp
- SAA
7-amino-L-glycero-L-galacto-2,6-anhydro-7-deoxyheptanoic acid
- SATA
N-hydroxysuccinimide ester
- SCLC
small cell lung cancers
- SPECT
single photon emission computed tomography
- SST
somatostatin
- SSTrs
human somatostatin receptors
- sulfo-SMCC
sulfosuccinimidyl 4-[N-maleimidomethyl]cyclohexane-1-carboxylate
- SWNTs
single-walled carbon nanotubes
- TAMRA
5-carboxytetramethylrhodamine
- TETA
triethylenetetramine
- uMUC-1
underglycosylated mucin-1 antigen
- uPA
urokinase-type plasminogn activator
- VCAM-1
vascular cell adhesion molecule-1
- VIP
vasoactive intestinal peptide
- ε
molar extinction coefficient
Biographies
Xiaoyuan Chen
 Xiaoyuan Chen was born in Jiangsu, China in 1974. Dr. Chen received his B.S. in 1993
and M.S. in 1996 from Nanjing University, China. He then moved to the United States
and obtained his Ph.D. in chemistry from the University of Idaho in 1999. After two
quick postdocs at Syracuse University and Washington University in St. Louis, he
joined the University of Southern California as an Assistant Professor of Radiology.
He then moved to Stanford University in 2004 to help build up the Molecular Imaging
Program at Stanford (MIPS) under the directorship of Prof. Sanjiv Sam Gambhir. He
was promoted to Associate Professor in 2008, and in the summer of 2009 he joined the
intramural research program of the National Institute of Biomedical Imaging and
Bioengineering (NIBIB), at the National Institutes of Health, as a tenured senior
investigator and lab chief. He took over the Positron Emission Tomography
Radiochemistry Group and expanded it into the Laboratory of Molecular Imaging and
Nanomedicine (LOMIN). He also has joint appointments with the NIH Clinical Center
and the National Institute of Standards and Technology (NIST). Dr. Chen has
published over 180 papers, 4 books and 15 book chapters. He sits on the editorial
board of 11 peer-reviewed journals and is a regular reviewer for over 50 journals.
The Laboratory of Molecular Imaging and Nanomedicine (LOMIN) specializes in
synthesizing molecular imaging probes for positron emission tomography (PET),
single-photon emission computed tomography (SPECT), magnetic resonance imaging
(MRI), optical (bioluminescent, fluorescent and Raman) imaging, contrast enhanced
ultrasound, photoacoustic imaging, andmultimodality imaging. His lab aims to develop
a molecular imaging toolbox for better understanding of biology, for early diagnosis
of disease, guiding drug discovery-development, and monitoring therapeutic
responses. The laboratory has a special emphasis on high sensitivity nanosensors for
biomarker detection and theranostic nanomedicine.
Xiaoyuan Chen was born in Jiangsu, China in 1974. Dr. Chen received his B.S. in 1993
and M.S. in 1996 from Nanjing University, China. He then moved to the United States
and obtained his Ph.D. in chemistry from the University of Idaho in 1999. After two
quick postdocs at Syracuse University and Washington University in St. Louis, he
joined the University of Southern California as an Assistant Professor of Radiology.
He then moved to Stanford University in 2004 to help build up the Molecular Imaging
Program at Stanford (MIPS) under the directorship of Prof. Sanjiv Sam Gambhir. He
was promoted to Associate Professor in 2008, and in the summer of 2009 he joined the
intramural research program of the National Institute of Biomedical Imaging and
Bioengineering (NIBIB), at the National Institutes of Health, as a tenured senior
investigator and lab chief. He took over the Positron Emission Tomography
Radiochemistry Group and expanded it into the Laboratory of Molecular Imaging and
Nanomedicine (LOMIN). He also has joint appointments with the NIH Clinical Center
and the National Institute of Standards and Technology (NIST). Dr. Chen has
published over 180 papers, 4 books and 15 book chapters. He sits on the editorial
board of 11 peer-reviewed journals and is a regular reviewer for over 50 journals.
The Laboratory of Molecular Imaging and Nanomedicine (LOMIN) specializes in
synthesizing molecular imaging probes for positron emission tomography (PET),
single-photon emission computed tomography (SPECT), magnetic resonance imaging
(MRI), optical (bioluminescent, fluorescent and Raman) imaging, contrast enhanced
ultrasound, photoacoustic imaging, andmultimodality imaging. His lab aims to develop
a molecular imaging toolbox for better understanding of biology, for early diagnosis
of disease, guiding drug discovery-development, and monitoring therapeutic
responses. The laboratory has a special emphasis on high sensitivity nanosensors for
biomarker detection and theranostic nanomedicine.
Seulki Lee
 Seulki Lee was born in Seoul, Korea in 1977. He received his B.S. in polymer
engineering in 2000 from the Sungkyunkwan University, and M.S. and Ph.D. degrees
from the Department of Materials Science and Engineering at Gwangju Institute of
Science and Technology in Korea. His Ph.D. project involved the design and
development of novel therapeutic peptide/protein drugs using various
biopolymer-based nanomedicine technologies. Under the supervision of Dr. Youngro
Byun, he completed his Ph.D. program in 2006, and then focused his training and
research on molecular imaging at the Biomedical Research Center, Korea Institute of
Science and Technology under the mentorship of Dr. Ick Chan Kwon. During that
period, he developed various smart molecular probes for real-time imaging of
cancer-related biological processes. He then moved to the United States where he
joined the Molecular Imaging Program at Stanford (MIPS) in 2008 under the
supervision of Dr. Xiaoyuan Chen. Subsequently, in the summer of 2009, he joined Dr.
Xiaoyuan Chen's new Laboratory of Molecular Imaging and Nanomedicine (LOMIN) at the
National Institute of Biomedical Imaging and Bioengineering (NIBIB), at the National
Institutes of Health. Dr. Lee has published over 50 papers and 2 book chapters.
Currently, he is a NIH-NIST fellow and leader of the Theranostic Nanomedicine Group
in the LOMIN. With a background in nanomedicine and molecular imaging, his main
research interests are smart nanoplatforms for future diagnosis and therapy of
various diseases with the emphasis on theranostics.
Seulki Lee was born in Seoul, Korea in 1977. He received his B.S. in polymer
engineering in 2000 from the Sungkyunkwan University, and M.S. and Ph.D. degrees
from the Department of Materials Science and Engineering at Gwangju Institute of
Science and Technology in Korea. His Ph.D. project involved the design and
development of novel therapeutic peptide/protein drugs using various
biopolymer-based nanomedicine technologies. Under the supervision of Dr. Youngro
Byun, he completed his Ph.D. program in 2006, and then focused his training and
research on molecular imaging at the Biomedical Research Center, Korea Institute of
Science and Technology under the mentorship of Dr. Ick Chan Kwon. During that
period, he developed various smart molecular probes for real-time imaging of
cancer-related biological processes. He then moved to the United States where he
joined the Molecular Imaging Program at Stanford (MIPS) in 2008 under the
supervision of Dr. Xiaoyuan Chen. Subsequently, in the summer of 2009, he joined Dr.
Xiaoyuan Chen's new Laboratory of Molecular Imaging and Nanomedicine (LOMIN) at the
National Institute of Biomedical Imaging and Bioengineering (NIBIB), at the National
Institutes of Health. Dr. Lee has published over 50 papers and 2 book chapters.
Currently, he is a NIH-NIST fellow and leader of the Theranostic Nanomedicine Group
in the LOMIN. With a background in nanomedicine and molecular imaging, his main
research interests are smart nanoplatforms for future diagnosis and therapy of
various diseases with the emphasis on theranostics.
Jin Xie
 Jin Xie was born in Yangzhou, China in 1980. He received his B.S. degree in
Chemistry from Nanjing University, China in 2003. In 2004, he moved to the U.S. and
started pursuing his Ph.D. in the department of Chemistry at Brown University under
the supervision of Dr. Shouheng Sun. His Ph.D. work focused on synthesis, surface
modification and bioapplication of magnetic nanoparticles. After completing his
Ph.D. studies in 2008, he joined the Molecular Imaging Program at Stanford (MIPS) as
a postdoctoral research fellow, and worked with Dr. Xiaoyuan Chen on developing
nanoparticle-based imaging probes. Then, in the summer of 2009, he moved with Dr.
Chen to National Institute of Biomedical Imaging and Bioengineering (NIBIB) at
National Institutes of Health (NIH), where he has continued his research in the
field of molecular imaging. His current research interests include the development
and evaluation of nanoparticle or protein based imaging probes and drug delivery
vehicles. Especially, he is interested in developing “all-in-one”
theranostic agents integrative of multiple functions that allow simultaneous
imaging, therapy, and therapeutic response monitoring.
Jin Xie was born in Yangzhou, China in 1980. He received his B.S. degree in
Chemistry from Nanjing University, China in 2003. In 2004, he moved to the U.S. and
started pursuing his Ph.D. in the department of Chemistry at Brown University under
the supervision of Dr. Shouheng Sun. His Ph.D. work focused on synthesis, surface
modification and bioapplication of magnetic nanoparticles. After completing his
Ph.D. studies in 2008, he joined the Molecular Imaging Program at Stanford (MIPS) as
a postdoctoral research fellow, and worked with Dr. Xiaoyuan Chen on developing
nanoparticle-based imaging probes. Then, in the summer of 2009, he moved with Dr.
Chen to National Institute of Biomedical Imaging and Bioengineering (NIBIB) at
National Institutes of Health (NIH), where he has continued his research in the
field of molecular imaging. His current research interests include the development
and evaluation of nanoparticle or protein based imaging probes and drug delivery
vehicles. Especially, he is interested in developing “all-in-one”
theranostic agents integrative of multiple functions that allow simultaneous
imaging, therapy, and therapeutic response monitoring.
8. References
- 1.Hoffman JM, Gambhir SS. Radiology. 2007;244:39. doi: 10.1148/radiol.2441060773. [DOI] [PubMed] [Google Scholar]
- 2.Cai W, Chen X. Small. 2007;3:1840. doi: 10.1002/smll.200700351. [DOI] [PubMed] [Google Scholar]
- 3.Wu AM, Olafsen T. Cancer J. 2008;14:191. doi: 10.1097/PPO.0b013e31817b07ae. [DOI] [PubMed] [Google Scholar]
- 4.Zaccaro L, Del Gatto A, Pedone C, Saviano M. Curr. Med. Chem. 2009;16:780. doi: 10.2174/092986709787549307. [DOI] [PubMed] [Google Scholar]
- 5.Schottelius M, Wester HJ. Methods. 2009;48:161. doi: 10.1016/j.ymeth.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, Chen X. Mol. Imaging. 2009;8:87. [PubMed] [Google Scholar]
- 7.Lee S, Park K, Kim K, Choi K, Kwon IC. Chem. Commun. (Camb) 2008:4250. doi: 10.1039/b806854m. [DOI] [PubMed] [Google Scholar]
- 8.Aloj L, Morelli G. Curr. Pharm. Des. 2004;10:3009. doi: 10.2174/1381612043383511. [DOI] [PubMed] [Google Scholar]
- 9.Reubi JC, Maecke HR. J. Nucl. Med. 2008;49:1735. doi: 10.2967/jnumed.108.053041. [DOI] [PubMed] [Google Scholar]
- 10.Okarvi SM. Med. Res. Rev. 2004;24:357. doi: 10.1002/med.20002. [DOI] [PubMed] [Google Scholar]
- 11.Hruby VJ. Nat Rev Drug Discov. 2002;1:847. doi: 10.1038/nrd939. [DOI] [PubMed] [Google Scholar]
- 12.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. Mol. Pharm. 2007;4:631. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 13.Petrenko V. Expert Opin. Drug Deliv. 2008;5:825. doi: 10.1517/17425247.5.8.825. [DOI] [PubMed] [Google Scholar]
- 14.Krohn KA. Nucl. Med. Biol. 2001;28:477. doi: 10.1016/s0969-8051(01)00216-5. [DOI] [PubMed] [Google Scholar]
- 15.Mankoff DA, Link JM, Linden HM, Sundararajan L, Krohn KA. J. Nucl. Med. 2008;49(Suppl. 2):149S. doi: 10.2967/jnumed.107.045963. [DOI] [PubMed] [Google Scholar]
- 16.Reubi JC. Endocr. Rev. 2003;24:389. doi: 10.1210/er.2002-0007. [DOI] [PubMed] [Google Scholar]
- 17.Cai W, Niu G, Chen X. Curr. Pharm. Des. 2008;14:2943. doi: 10.2174/138161208786404308. [DOI] [PubMed] [Google Scholar]
- 18.Reubi JC. Curr. Top. Med. Chem. 2007;7:1239. doi: 10.2174/156802607780960546. [DOI] [PubMed] [Google Scholar]
- 19.Korner M, Stockli M, Waser B, Reubi JC. J. Nucl. Med. 2007;48:736. doi: 10.2967/jnumed.106.038679. [DOI] [PubMed] [Google Scholar]
- 20.Miao Y, Quinn TP. Front. Biosci. 2007;12:4514. doi: 10.2741/2406. [DOI] [PubMed] [Google Scholar]
- 21.Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Science. 1991;251:767. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 22.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Nature. 1991;354:84. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 23.Zuckermann RN, Kerr JM, Siani MA, Banville SC, Santi DV. Proc. Natl. Acad. Sci. U. S. A. 1992;89:4505. doi: 10.1073/pnas.89.10.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam KS, Liu R, Miyamoto S, Lehman AL, Tuscano JM. Acc. Chem. Res. 2003;36:370. doi: 10.1021/ar0201299. [DOI] [PubMed] [Google Scholar]
- 25.Newton J, Deutscher SL. Handb. Exp. Pharmacol. 2008:145. doi: 10.1007/978-3-540-77496-9_7. [DOI] [PubMed] [Google Scholar]
- 26.Brown KC. Curr. Opin. Chem. Biol. 2000;4:16. doi: 10.1016/s1367-5931(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 27.Benedetti E, Morelli G, Accardo A, Mansi R, Tesauro D, Aloj L. BioDrugs. 2004;18:279. doi: 10.2165/00063030-200418050-00001. [DOI] [PubMed] [Google Scholar]
- 28.Corot C, Robert P, Idee JM, Port M. Adv. Drug. Deliv. Rev. 2006;58:1471. doi: 10.1016/j.addr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Frangioni JV. Curr. Opin. Chem. Biol. 2003;7:626. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Kim J-H, Park K, Nam HY, Lee S, Kim K, Kwon IC. Prog. Polym. Sci. 2007;32:1031. [Google Scholar]
- 31.Fass L. Mol. Oncol. 2008;2:115. doi: 10.1016/j.molonc.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ametamey SM, Honer M, Schubiger PA. Chem. Rev. 2008;108:1501. doi: 10.1021/cr0782426. [DOI] [PubMed] [Google Scholar]
- 33.Hermanson GT. Bioconjugate techniques. Academic Press; San Diego: 1996. [Google Scholar]
- 34.Sosabowski JK, Mather SJ. Nat. Protoc. 2006;1:972. doi: 10.1038/nprot.2006.175. [DOI] [PubMed] [Google Scholar]
- 35.De Leon-Rodriguez LM, Kovacs Z, Dieckmann GR, Sherry AD. Chemistry. 2004;10:1149. doi: 10.1002/chem.200305389. [DOI] [PubMed] [Google Scholar]
- 36.Lewis MR, Jia F, Gallazzi F, Wang Y, Zhang J, Shenoy N, Lever SZ, Hannink M. Bioconjug. Chem. 2002;13:1176. doi: 10.1021/bc025591s. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Edwards DS. Chem. Rev. 1999;99:2235. doi: 10.1021/cr980436l. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Edwards DS, Looby RJ, Poirier MJ, Rajopadhye M, Bourque JP, Carroll TR. Bioconjug. Chem. 1996;7:196. doi: 10.1021/bc9500958. [DOI] [PubMed] [Google Scholar]
- 39.Kasina S, Sanderson JA, Fitzner JN, Srinivasan A, Rao TN, Hobson LJ, Reno JM, Axworthy DB, Beaumier PL, Fritzberg AR. Bioconjug. Chem. 1998;9:108. doi: 10.1021/bc970047i. [DOI] [PubMed] [Google Scholar]
- 40.Gano L, Patricio L, Marques E, Cantinho G, Pena H, Martins T, Hnatowich DJ. Nucl. Med. Biol. 1998;25:395. doi: 10.1016/s0969-8051(97)00225-4. [DOI] [PubMed] [Google Scholar]
- 41.Kelly JD, Forster AM, Higley B, Archer CM, Booker FS, Canning LR, Chiu KW, Edwards B, Gill HK, McPartlin M, Nagle KR, Latham I. A.l, Pickett RD, Storey AE, Webbon PM. J. Nucl. Med. 1993;34:222. [PubMed] [Google Scholar]
- 42.Higley B, Smith FW, Smith T, Gemmell HG, Das Gupta P, Gvozdanovic DV, Graham D, Hinge D, Davidson J, Lahiri A. J. Nucl. Med. 1993;34:30. [PubMed] [Google Scholar]
- 43.Miller PW, Long NJ, Vilar R, Gee AD. Angew. Chem. Int. Ed. Engl. 2008;47:8998. doi: 10.1002/anie.200800222. [DOI] [PubMed] [Google Scholar]
- 44.Labroo VM, Hebel D, Kirk KL, Cohen LA, Lemieux C, Schiller PW. Int. J. Pept. Protein Res. 1991;37:430. doi: 10.1111/j.1399-3011.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- 45.Cai W, Zhang X, Wu Y, Chen X. J. Nucl. Med. 2006;47:1172. [PMC free article] [PubMed] [Google Scholar]
- 46.Shiue CY, Wolf AP, Hainfeld JF. J. Labelled Compd. Radiopharm. 1998;26:287. [Google Scholar]
- 47.Wuest F, Berndt M, Bergmann R, van den Hoff J, Pietzsch J. Bioconjug. Chem. 2008;19:1202. doi: 10.1021/bc8000112. [DOI] [PubMed] [Google Scholar]
- 48.Li ZB, Wu Z, Chen K, Chin FT, Chen X. Bioconjug. Chem. 2007;18:1987. doi: 10.1021/bc700226v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glaser M, Arstad E. Bioconjug. Chem. 2007;18:989. doi: 10.1021/bc060301j. [DOI] [PubMed] [Google Scholar]
- 50.Weckbecker G, Lewis I, Albert R, Schmid HA, Hoyer D, Bruns C. Nat. Rev. Drug Discov. 2003;2:999. doi: 10.1038/nrd1255. [DOI] [PubMed] [Google Scholar]
- 51.Susini C, Buscail L. Ann. Oncol. 2006;17:1733. doi: 10.1093/annonc/mdl105. [DOI] [PubMed] [Google Scholar]
- 52.de Jong M, Breeman WA, Kwekkeboom DJ, Valkema R, Krenning EP. Acc. Chem. Res. 2009;42:873. doi: 10.1021/ar800188e. [DOI] [PubMed] [Google Scholar]
- 53.Kwekkeboom DJ, Krenning EP. Semin. Nucl. Med. 2002;32:84. doi: 10.1053/snuc.2002.31022. [DOI] [PubMed] [Google Scholar]
- 54.Rufini V, Calcagni ML, Baum RP. Semin. Nucl. Med. 2006;36:228. doi: 10.1053/j.semnuclmed.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 55.Cwikla JB, Mikolajczak R, Pawlak D, Buscombe JR, Nasierowska-Guttmejer A, Bator A, Maecke HR, Walecki J. J. Nucl. Med. 2008;49:1060. doi: 10.2967/jnumed.107.046961. [DOI] [PubMed] [Google Scholar]
- 56.Li WP, Lewis JS, Kim J, Bugaj JE, Johnson MA, Erion JL, Anderson CJ. Bioconjug. Chem. 2002;13:721. doi: 10.1021/bc015590k. [DOI] [PubMed] [Google Scholar]
- 57.Schottelius M, Wester HJ, Reubi JC, Senekowitsch-Schmidtke R, Schwaiger M. Bioconjug. Chem. 2002;13:1021. doi: 10.1021/bc0200069. [DOI] [PubMed] [Google Scholar]
- 58.Meisetschlager G, Poethko T, Stahl A, Wolf I, Scheidhauer K, Schottelius M, Herz M, Wester HJ, Schwaiger M. J. Nucl. Med. 2006;47:566. [PubMed] [Google Scholar]
- 59.Ginj M, Chen J, Walter MA, Eltschinger V, Reubi JC, Maecke HR. Clin. Cancer Res. 2005;11:1136. [PubMed] [Google Scholar]
- 60.Cescato R, Erchegyi J, Waser B, Piccand V, Maecke HR, Rivier JE, Reubi JC. J. Med. Chem. 2008;51:4030. doi: 10.1021/jm701618q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waser B, Tamma ML, Cescato R, Maecke HR, Reubi JC. J. Nucl. Med. 2009;50:936. doi: 10.2967/jnumed.108.061457. [DOI] [PubMed] [Google Scholar]
- 62.Wadas TJ, Eiblmaier M, Zheleznyak A, Sherman CD, Ferdani R, Liang K, Achilefu S, Anderson CJ. J. Nucl. Med. 2008;49:1819. doi: 10.2967/jnumed.108.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 64.Brooks PC, Clark RA, Cheresh DA. Science. 1994;264:569. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 65.Horton MA. Int. J. Biochem. Cell Biol. 1997;29:721. doi: 10.1016/s1357-2725(96)00155-0. [DOI] [PubMed] [Google Scholar]
- 66.Dijkgraaf I, Beer AJ, Wester HJ. Front. Biosci. 2009;14:887. doi: 10.2741/3284. [DOI] [PubMed] [Google Scholar]
- 67.Cai W, Rao J, Gambhir SS, Chen X. Mol. Cancer Ther. 2006;5:2624. doi: 10.1158/1535-7163.MCT-06-0395. [DOI] [PubMed] [Google Scholar]
- 68.Schottelius M, Laufer B, Kessler H, Wester HJ. Acc. Chem. Res. 2009;42:969. doi: 10.1021/ar800243b. [DOI] [PubMed] [Google Scholar]
- 69.Haubner R, Wester HJ, Weber WA, Mang C, Ziegler SI, Goodman SL, Senekowitsch-Schmidtke R, Kessler H, Schwaiger M. Cancer Res. 2001;61:1781. [PubMed] [Google Scholar]
- 70.Beer AJ, Grosu AL, Carlsen J, Kolk A, Sarbia M, Stangier I, Watzlowik P, Wester HJ, Haubner R, Schwaiger M. Clin. Cancer Res. 2007;13:6610. doi: 10.1158/1078-0432.CCR-07-0528. [DOI] [PubMed] [Google Scholar]
- 71.Beer AJ, Niemeyer M, Carlsen J, Sarbia M, Nahrig J, Watzlowik P, Wester HJ, Harbeck N, Schwaiger M. J. Nucl. Med. 2008;49:255. doi: 10.2967/jnumed.107.045526. [DOI] [PubMed] [Google Scholar]
- 72.Schnell O, Krebs B, Carlsen J, Miederer I, Goetz C, Goldbrunner RH, Wester HJ, Haubner R, Popperl G, Holtmannspotter M, Kretzschmar HA, Kessler H, Tonn JC, Schwaiger M, Beer AJ. Neuro Oncol. 2009 doi: 10.1215/15228517-2009-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kenny LM, Coombes RC, Oulie I, Contractor KB, Miller M, Spinks TJ, McParland B, Cohen PS, Hui AM, Palmieri C, Osman S, Glaser M, Turton D, Al-Nahhas A, Aboagye EO. J. Nucl. Med. 2008;49:879. doi: 10.2967/jnumed.107.049452. [DOI] [PubMed] [Google Scholar]
- 74.Morrison MS, Ricketts SA, Barnett J, Cuthbertson A, Tessier J, Wedge SR. J. Nucl. Med. 2009;50:116. doi: 10.2967/jnumed.108.056077. [DOI] [PubMed] [Google Scholar]
- 75.Indrevoll B, Kindberg GM, Solbakken M, Bjurgert E, Johansen JH, Karlsen H, Mendizabal M, Cuthbertson A. Bioorg. Med. Chem. Lett. 2006;16:6190. doi: 10.1016/j.bmcl.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 76.Zhang X, Xiong Z, Wu Y, Cai W, Tseng JR, Gambhir SS, Chen X. J. Nucl. Med. 2006;47:113. [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Z, Li ZB, Cai W, He L, Chin FT, Li F, Chen X. Eur. J. Nucl. Med. Mol. Imaging. 2007;34:1823. doi: 10.1007/s00259-007-0427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu S, Liu Z, Chen K, Yan Y, Watzlowik P, Wester HJ, Chin FT, Chen X. Mol. Imaging Biol. 2009 doi: 10.1007/s11307-009-0284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, Park R, Shahinian AH, Tohme M, Khankaldyyan V, Bozorgzadeh MH, Bading JR, Moats R, Laug WE, Conti PS. Nucl. Med. Biol. 2004;31:179. doi: 10.1016/j.nucmedbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 80.Liu Z, Niu G, Shi J, Liu S, Wang F, Chen X. Eur. J. Nucl. Med. Mol. Imaging. 2009;36:947. doi: 10.1007/s00259-008-1045-1. [DOI] [PubMed] [Google Scholar]
- 81.Shi J, Kim YS, Zhai S, Liu Z, Chen X, Liu S. Bioconjug. Chem. 2009;20:750. doi: 10.1021/bc800455p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi J, Wang L, Kim YS, Zhai S, Liu Z, Chen X, Liu S. J. Med. Chem. 2008;51:7980. doi: 10.1021/jm801134k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li ZB, Cai W, Cao Q, Chen K, Wu Z, He L, Chen X. J. Nucl. Med. 2007;48:1162. doi: 10.2967/jnumed.107.039859. [DOI] [PubMed] [Google Scholar]
- 84.Kolb H, Walsh J, Liang Q, Zhao T, Gao D, Secrest J, Gomez L, Scott P. J. NUCL. MED. MEETING ABSTRACTS. 2009;50:329. [Google Scholar]
- 85.Cho HJ, Lee JD, Park JY, Yun M, Kang WJ, Walsh JC, Kolb H, Zhang JJ. J. NUCL. MED. MEETING ABSTRACTS. 2009;50:1910. [Google Scholar]
- 86.Smith CJ, Volkert WA, Hoffman T. J. Nucl. Med. Biol. 2005;32:733. doi: 10.1016/j.nucmedbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 87.Van de Wiele C, Dumont F, Dierckx RA, Peers SH, Thornback JR, Slegers G, Thierens H. J. Nucl. Med. 2001;42:1722. [PubMed] [Google Scholar]
- 88.Van de Wiele C, Phonteyne P, Pauwels P, Goethals I, Van den Broecke R, Cocquyt V, Dierckx RA. J. Nucl. Med. 2008;49:260. doi: 10.2967/jnumed.107.047167. [DOI] [PubMed] [Google Scholar]
- 89.Ferro-Flores G, Arteaga de Murphy C, Rodriguez-Cortes J, Pedraza-Lopez M, Ramirez-Iglesias MT. Nucl. Med. Commun. 2006;27:371. doi: 10.1097/01.mnm.0000202863.52046.7f. [DOI] [PubMed] [Google Scholar]
- 90.Hoffman TJ, Gali H, Smith CJ, Sieckman GL, Hayes DL, Owen NK, Volkert WA. J. Nucl. Med. 2003;44:823. [PubMed] [Google Scholar]
- 91.Prasanphanich AF, Nanda PK, Rold TL, Ma L, Lewis MR, Garrison JC, Hoffman TJ, Sieckman GL, Figueroa SD, Smith CJ. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12462. doi: 10.1073/pnas.0705347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang H, Schuhmacher J, Waser B, Wild D, Eisenhut M, Reubi JC, Maecke HR. Eur. J. Nucl. Med. Mol. Imaging. 2007;34:1198. doi: 10.1007/s00259-006-0347-4. [DOI] [PubMed] [Google Scholar]
- 93.Biddlecombe GB, Rogers BE, de Visser M, Parry JJ, de Jong M, Erion JL, Lewis JS. Bioconjug. Chem. 2007;18:724. doi: 10.1021/bc060281l. [DOI] [PubMed] [Google Scholar]
- 94.Zhang X, Cai W, Cao F, Schreibmann E, Wu Y, Wu JC, Xing L, Chen X. J. Nucl. Med. 2006;47:492. [PubMed] [Google Scholar]
- 95.Nock BA, Nikolopoulou A, Galanis A, Cordopatis P, Waser B, Reubi JC, Maina T. J. Med. Chem. 2005;48:100. doi: 10.1021/jm049437y. [DOI] [PubMed] [Google Scholar]
- 96.Maina T, Nock BA, Zhang H, Nikolopoulou A, Waser B, Reubi JC, Maecke HR. J. Nucl. Med. 2005;46:823. [PubMed] [Google Scholar]
- 97.Cescato R, Maina T, Nock B, Nikolopoulou A, Charalambidis D, Piccand V, Reubi JC. J. Nucl. Med. 2008;49:318. doi: 10.2967/jnumed.107.045054. [DOI] [PubMed] [Google Scholar]
- 98.Dimitrakopoulou-Strauss A, Hohenberger P, Haberkorn U, Macke HR, Eisenhut M, Strauss LG. J. Nucl. Med. 2007;48:1245. doi: 10.2967/jnumed.106.038091. [DOI] [PubMed] [Google Scholar]
- 99.Li ZB, Wu Z, Chen K, Ryu EK, Chen X. J. Nucl. Med. 2008;49:453. doi: 10.2967/jnumed.107.048009. [DOI] [PubMed] [Google Scholar]
- 100.Liu Z, Yan Y, Chin FT, Wang F, Chen X. J. Med. Chem. 2009;52:425. doi: 10.1021/jm801285t. [DOI] [PubMed] [Google Scholar]
- 101.Liu Z, Niu G, Wang F, Chen X. Eur. J. Nucl. Med. Mol. Imaging. 2009;36:1483. doi: 10.1007/s00259-009-1123-z. [DOI] [PubMed] [Google Scholar]
- 102.Liu Z, Li ZB, Cao Q, Liu S, Wang F, Chen X. J. Nucl. Med. 2009;50:1168. doi: 10.2967/jnumed.108.061739. [DOI] [PubMed] [Google Scholar]
- 103.Liu ZF, Yan YJ, Liu SL, Wang F, Chen XY. Bioconjug. Chem. 2009;20:1016. doi: 10.1021/bc9000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reubi JC, Schaer JC, Waser B. Cancer Res. 1997;57:1377. [PubMed] [Google Scholar]
- 105.Reubi JC, Waser B. Int. J. Cancer. 1996;67:644. doi: 10.1002/(SICI)1097-0215(19960904)67:5<644::AID-IJC9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 106.Koopmans KP, Neels ON, Kema IP, Elsinga PH, Links TP, de Vries EG, Jager PL. Crit. Rev. Oncol. Hematol. 2009;71:199. doi: 10.1016/j.critrevonc.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 107.Baldwin GS, Shulkes A. Curr. Top. Med. Chem. 2007;7:1232. doi: 10.2174/156802607780960492. [DOI] [PubMed] [Google Scholar]
- 108.Aloj L, Caraco C, Panico M, Zannetti A, Del Vecchio S, Tesauro D, De Luca S, Arra C, Pedone C, Morelli G, Salvatore M. J. Nucl. Med. 2004;45:485. [PubMed] [Google Scholar]
- 109.Aloj L, Panico M, Caraco C, Del Vecchio S, Arra C, Affuso A, Accardo A, Mansi R, Tesauro D, De Luca S, Pedone C, Visentin R, Mazzi U, Morelli G, Salvatore M. Cancer Biother. Radiopharm. 2004;19:93. doi: 10.1089/108497804773391739. [DOI] [PubMed] [Google Scholar]
- 110.D'Andrea LD, Testa I, Panico M, Di Stasi R, Caraco C, Tarallo L, Arra C, Barbieri A, Romanelli A, Aloj L. Biopolymers. 2008;90:707. doi: 10.1002/bip.21041. [DOI] [PubMed] [Google Scholar]
- 111.Behr TM, Jenner N, Behe M, Angerstein C, Gratz S, Raue F, Becker W. J. Nucl. Med. 1999;40:1029. [PubMed] [Google Scholar]
- 112.von Guggenberg E, Behe M, Behr TM, Saurer M, Seppi T, Decristoforo C. Bioconjug. Chem. 2004;15:864. doi: 10.1021/bc0300807. [DOI] [PubMed] [Google Scholar]
- 113.Mather SJ, McKenzie AJ, Sosabowski JK, Morris TM, Ellison D, Watson SA. J. Nucl. Med. 2007;48:615. doi: 10.2967/jnumed.106.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Good S, Walter MA, Waser B, Wang X, Muller-Brand J, Behe MP, Reubi JC, Maecke HR. Eur. J. Nucl. Med. Mol. Imaging. 2008;35:1868. doi: 10.1007/s00259-008-0803-4. [DOI] [PubMed] [Google Scholar]
- 115.Tatro JB, Wen Z, Entwistle ML, Atkins MB, Smith TJ, Reichlin S, Murphy JR. Cancer Res. 1992;52:2545. [PubMed] [Google Scholar]
- 116.Chen J, Giblin MF, Wang N, Jurisson SS, Quinn TP. Nucl. Med. Biol. 1999;26:687. doi: 10.1016/s0969-8051(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 117.Chen J, Cheng Z, Owen NK, Hoffman TJ, Miao Y, Jurisson SS, Quinn TP. J. Nucl. Med. 2001;42:1847. [PubMed] [Google Scholar]
- 118.McQuade P, Miao Y, Yoo J, Quinn TP, Welch MJ, Lewis JS. J. Med. Chem. 2005;48:2985. doi: 10.1021/jm0490282. [DOI] [PubMed] [Google Scholar]
- 119.Cantorias MV, Figueroa SD, Quinn TP, Lever JR, Hoffman TJ, Watkinson LD, Carmack TL, Cutler CS. Nucl. Med. Biol. 2009;36:505. doi: 10.1016/j.nucmedbio.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 120.Miao Y, Gallazzi F, Guo H, Quinn TP. Bioconjug. Chem. 2008;19:539. doi: 10.1021/bc700317w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo H, Yang J, Gallazzi F, Prossnitz ER, Sklar LA, Miao Y. Bioconjug. Chem. 2009;20:2162. doi: 10.1021/bc9003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J, Guo H, Gallazzi F, Berwick M, Padilla RS, Miao Y. Bioconjug. Chem. 2009;20:1634. doi: 10.1021/bc9001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vincent JP. Cell. Mol. Neurobiol. 1995;15:501. doi: 10.1007/BF02071313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reubi JC, Waser B, Friess H, Buchler M, Laissue J. Gut. 1998;42:546. doi: 10.1136/gut.42.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Souaze F, Dupouy S, Viardot-Foucault V, Bruyneel E, Attoub S, Gespach C, Gompel A, Forgez P. Cancer Res. 2006;66:6243. doi: 10.1158/0008-5472.CAN-06-0450. [DOI] [PubMed] [Google Scholar]
- 126.de Visser M, Janssen PJ, Srinivasan A, Reubi JC, Waser B, Erion JL, Schmidt MA, Krenning EP, de Jong M. Eur. J. Nucl. Med. Mol. Imaging. 2003;30:1134. doi: 10.1007/s00259-003-1189-y. [DOI] [PubMed] [Google Scholar]
- 127.Maes V, Garcia-Garayoa E, Blauenstein P, Tourwe D. J. Med. Chem. 2006;49:1833. doi: 10.1021/jm051172f. [DOI] [PubMed] [Google Scholar]
- 128.Garcia-Garayoa E, Blauenstein P, Bruehlmeier M, Blanc A, Iterbeke K, Conrath P, Tourwe D, Schubiger PA. J. Nucl. Med. 2002;43:374. [PubMed] [Google Scholar]
- 129.Buchegger F, Bonvin F, Kosinski M, Schaffland AO, Prior J, Reubi JC, Blauenstein P, Tourwe D, Garcia Garayoa E, Bischof Delaloye A. J. Nucl. Med. 2003;44:1649. [PubMed] [Google Scholar]
- 130.Reubi JC, Laderach U, Waser B, Gebbers JO, Robberecht P, Laissue JA. Cancer Res. 2000;60:3105. [PubMed] [Google Scholar]
- 131.Thakur ML, Marcus CS, Saeed S, Pallela V, Minami C, Diggles L, Le Pham H, Ahdoot R, Kalinowski EA. J. Nucl. Med. 2000;41:107. [PubMed] [Google Scholar]
- 132.Thakur ML, Aruva MR, Gariepy J, Acton P, Rattan S, Prasad S, Wickstrom E, Alavi A. J. Nucl. Med. 2004;45:1381. [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang K, Aruva MR, Shanthly N, Cardi CA, Rattan S, Patel C, Kim C, McCue PA, Wickstrom E, Thakur ML. J. Nucl. Med. 2008;49:112. doi: 10.2967/jnumed.107.043703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kieffer TJ, Habener JF. Endocr. Rev. 1999;20:876. doi: 10.1210/edrv.20.6.0385. [DOI] [PubMed] [Google Scholar]
- 135.Bullock BP, Heller RS, Habener JF. Endocrinology. 1996;137:2968. doi: 10.1210/endo.137.7.8770921. [DOI] [PubMed] [Google Scholar]
- 136.Gotthardt M, Fischer M, Naeher I, Holz JB, Jungclas H, Fritsch HW, Behe M, Goke B, Joseph K, Behr TM. Eur. J. Nucl. Med. Mol. Imaging. 2002;29:597. doi: 10.1007/s00259-002-0761-1. [DOI] [PubMed] [Google Scholar]
- 137.Wild D, Behe M, Wicki A, Storch D, Waser B, Gotthardt M, Keil B, Christofori G, Reubi JC, Macke HR. J. Nucl. Med. 2006;47:2025. [PubMed] [Google Scholar]
- 138.Mukai E, Toyoda K, Kimura H, Kawashima H, Fujimoto H, Ueda M, Temma T, Hirao K, Nagakawa K, Saji H, Inagaki N. Biochem. Biophys. Res. Commun. 2009;389:523. doi: 10.1016/j.bbrc.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 139.Wild D, Macke H, Christ E, Gloor B, Reubi JC. N. Engl. J. Med. 2008;359:766. doi: 10.1056/NEJMc0802045. [DOI] [PubMed] [Google Scholar]
- 140.Korner M, Reubi JC. Peptides. 2007;28:419. doi: 10.1016/j.peptides.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 141.Korner M, Waser B, Reubi JC. Clin. Cancer Res. 2008;14:5043. doi: 10.1158/1078-0432.CCR-07-4551. [DOI] [PubMed] [Google Scholar]
- 142.Langer M, La Bella R, Garcia-Garayoa E, Beck-Sickinger AG. Bioconjug. Chem. 2001;12:1028. doi: 10.1021/bc015514h. [DOI] [PubMed] [Google Scholar]
- 143.Zwanziger D, Khan IU, Neundorf I, Sieger S, Lehmann L, Friebe M, Dinkelborg L, Beck-Sickinger AG. Bioconjug. Chem. 2008;19:1430. doi: 10.1021/bc7004297. [DOI] [PubMed] [Google Scholar]
- 144.Raman D, Baugher PJ, Thu YM, Richmond A. Cancer Lett. 2007;256:137. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hanaoka H, Mukai T, Tamamura H, Mori T, Ishino S, Ogawa K, Iida Y, Doi R, Fujii N, Saji H. Nucl. Med. Biol. 2006;33:489. doi: 10.1016/j.nucmedbio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 146.Egeblad M, Werb Z. Nat. Rev. Cancer. 2002;2:161. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 147.Scherer RL, McIntyre JO, Matrisian LM. Cancer Metastasis Rev. 2008;27:679. doi: 10.1007/s10555-008-9152-9. [DOI] [PubMed] [Google Scholar]
- 148.Kuhnast B, Bodenstein C, Haubner R, Wester HJ, Senekowitsch-Schmidtke R, Schwaiger M, Weber WA. Nucl. Med. Biol. 2004;31:337. doi: 10.1016/j.nucmedbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 149.Hanaoka H, Mukai T, Habashita S, Asano D, Ogawa K, Kuroda Y, Akizawa H, Iida Y, Endo K, Saga T, Saji H. Nucl. Med. Biol. 2007;34:503. doi: 10.1016/j.nucmedbio.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 150.Licha K, Olbrich C. Adv. Drug. Deliv. Rev. 2005;57:1087. doi: 10.1016/j.addr.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 151.Biju V, Itoh T, Anas A, Sujith A, Ishikawa M. Anal. Bioanal. Chem. 2008;391:2469. doi: 10.1007/s00216-008-2185-7. [DOI] [PubMed] [Google Scholar]
- 152.Ntziachristos V. Annu. Rev. Biomed. Eng. 2006;8:1. doi: 10.1146/annurev.bioeng.8.061505.095831. [DOI] [PubMed] [Google Scholar]
- 153.Berlier JE, Rothe A, Buller G, Bradford J, Gray DR, Filanoski BJ, Telford WG, Yue S, Liu J, Cheung CY, Chang W, Hirsch JD, Beechem JM, Haugland RP. J. Histochem. Cytochem. 2003;51:1699. doi: 10.1177/002215540305101214. [DOI] [PubMed] [Google Scholar]
- 154.Weissleder R, Ntziachristos V. Nat. Med. 2003;9:123. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 155.Nakayama A, Bianco AC, Zhang CY, Lowell BB, Frangioni JV. Mol. Imaging. 2003;2:37. doi: 10.1162/15353500200303103. [DOI] [PubMed] [Google Scholar]
- 156.Hintersteiner M, Enz A, Frey P, Jaton AL, Kinzy W, Kneuer R, Neumann U, Rudin M, Staufenbiel M, Stoeckli M, Wiederhold KH, Gremlich HU. Nat. Biotechnol. 2005;23:577. doi: 10.1038/nbt1085. [DOI] [PubMed] [Google Scholar]
- 157.Zhao W, Carreira EM. Chemistry. 2006;12:7254. doi: 10.1002/chem.200600527. [DOI] [PubMed] [Google Scholar]
- 158.Ho NH, Weissleder R, Tung CS. Tetrahedron. 2006;62:578. [Google Scholar]
- 159.Bright FV. Anal. Chem. 1988;60:1031A. doi: 10.1021/ac00169a001. [DOI] [PubMed] [Google Scholar]
- 160.Wu P, Brand L. Anal. Biochem. 1994;218:1. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 161.Johansson MK, Cook RM. Chemistry. 2003;9:3466. doi: 10.1002/chem.200304941. [DOI] [PubMed] [Google Scholar]
- 162.Edwards DR, Murphy G. Nature. 1998;394:527. doi: 10.1038/28961. [DOI] [PubMed] [Google Scholar]
- 163.Christensen JG, Burrows J, Salgia R. Cancer Lett. 2005;225:1. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 164.Kim EM, Park EH, Cheong SJ, Lee CM, Jeong HJ, Kim DW, Lim ST, Sohn MH. Bioconjug. Chem. 2009;20:1299. doi: 10.1021/bc8005539. [DOI] [PubMed] [Google Scholar]
- 165.Becker A, Hessenius C, Licha K, Ebert B, Sukowski U, Semmler W, Wiedenmann B, Grotzinger C. Nat. Biotechnol. 2001;19:327. doi: 10.1038/86707. [DOI] [PubMed] [Google Scholar]
- 166.Kostenich G, Livnah N, Bonasera TA, Yechezkel T, Salitra Y, Litman P, Kimel S, Orenstein A. Lung Cancer. 2005;50:319. doi: 10.1016/j.lungcan.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 167.Kostenich G, Oron-Herman M, Kimel S, Livnah N, Tsarfaty I, Orenstein A. Int. J. Cancer. 2008;122:2044. doi: 10.1002/ijc.23353. [DOI] [PubMed] [Google Scholar]
- 168.Pu Y, Wang WB, Tang GC, Zeng F, Achilefu S, Vitenson JH, Sawczuk I, Peters S, Lombardo JM, Alfano RR. Technol. Cancer Res. Treat. 2005;4:429. doi: 10.1177/153303460500400410. [DOI] [PubMed] [Google Scholar]
- 169.Ma L, Yu P, Veerendra B, Rold TL, Retzloff L, Prasanphanich A, Sieckman G, Hoffman TJ, Volkert WA, Smith CJ. Mol. Imaging. 2007;6:171. [PubMed] [Google Scholar]
- 170.Chen X, Conti PS, Moats RA. Cancer Res. 2004;64:8009. doi: 10.1158/0008-5472.CAN-04-1956. [DOI] [PubMed] [Google Scholar]
- 171.Wang W, Ke S, Wu Q, Charnsangavej C, Gurfinkel M, Gelovani JG, Abbruzzese JL, Sevick-Muraca EM, Li C. Mol. Imaging. 2004;3:343. doi: 10.1162/15353500200404148. [DOI] [PubMed] [Google Scholar]
- 172.Achilefu S, Bloch S, Markiewicz MA, Zhong T, Ye Y, Dorshow RB, Chance B, Liang K. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7976. doi: 10.1073/pnas.0503500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Waldeck J, Hager F, Holtke C, Lanckohr C, von Wallbrunn A, Torsello G, Heindel W, Theilmeier G, Schafers M, Bremer C. J. Nucl. Med. 2008;49:1845. doi: 10.2967/jnumed.108.052514. [DOI] [PubMed] [Google Scholar]
- 174.Cheng Z, Wu Y, Xiong Z, Gambhir SS, Chen X. Bioconjug. Chem. 2005;16:1433. doi: 10.1021/bc0501698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu Y, Cai W, Chen X. Mol. Imaging. Biol. 2006;8:226. doi: 10.1007/s11307-006-0041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Boturyn D, Coll JL, Garanger E, Favrot MC, Dumy P. J. Am. Chem. Soc. 2004;126:5730. doi: 10.1021/ja049926n. [DOI] [PubMed] [Google Scholar]
- 177.Jin ZH, Josserand V, Foillard S, Boturyn D, Dumy P, Favrot MC, Coll JL. Mol. Cancer. 2007;6:41. doi: 10.1186/1476-4598-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Cancer Res. 2009;69:2435. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tait JF. J. Nucl. Med. 2008;49:1573. doi: 10.2967/jnumed.108.052803. [DOI] [PubMed] [Google Scholar]
- 180.Thapa N, Kim S, So IS, Lee BH, Kwon IC, Choi K, Kim IS. J. Cell. Mol. Med. 2008;12:1649. doi: 10.1111/j.1582-4934.2008.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Thapa N, Hong HY, Sangeetha P, Kim IS, Yoo J, Rhee K, Oh GT, Kwon IC, Lee BH. J. Control. Release. 2008;131:27. doi: 10.1016/j.jconrel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 182.Hong HY, Choi JS, Kim YJ, Lee HY, Kwak W, Yoo J, Lee JT, Kwon TH, Kim IS, Han HS, Lee BH. J. Control. Release. 2008;131:167. doi: 10.1016/j.jconrel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 183.Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Nat. Med. 2008;14:454. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang W, Shao R, Wu Q, Ke S, McMurray J, Lang FF, Jr., Charnsangavej C, Gelovani JG, Li C. Mol. Imaging Biol. 2009;11:424. doi: 10.1007/s11307-009-0219-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kelly KA, Waterman P, Weissleder R. Neoplasia. 2006;8:1011. doi: 10.1593/neo.06610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Newton JR, Kelly KA, Mahmood U, Weissleder R, Deutscher SL. Neoplasia. 2006;8:772. doi: 10.1593/neo.06331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yao N, Xiao W, Wang X, Marik J, Park SH, Takada Y, Lam KS. J. Med. Chem. 2009;52:126. doi: 10.1021/jm801062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xiao W, Yao N, Peng L, Liu R, Lam KS. Eur. J. Nucl. Med. Mol. Imaging. 2009;36:94. doi: 10.1007/s00259-008-0920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tung CH. Biopolymers. 2004;76:391. doi: 10.1002/bip.20139. [DOI] [PubMed] [Google Scholar]
- 190.Elias DR, Thorek DL, Chen AK, Czupryna J, Tsourkas A. Cancer Biomark. 2008;4:287. doi: 10.3233/cbm-2008-4602. [DOI] [PubMed] [Google Scholar]
- 191.Pham W, Choi Y, Weissleder R, Tung CH. Bioconjug. Chem. 2004;15:1403. doi: 10.1021/bc049924s. [DOI] [PubMed] [Google Scholar]
- 192.Johansson MK, Cook RM, Xu J, Raymond KN. J. Am. Chem. Soc. 2004;126:16451. doi: 10.1021/ja0452368. [DOI] [PubMed] [Google Scholar]
- 193.Lee S, Park K, Lee SY, Ryu JH, Park JW, Ahn HJ, Kwon IC, Youn IC, Kim K, Choi K. Bioconjug. Chem. 2008;19:1743. doi: 10.1021/bc800264z. [DOI] [PubMed] [Google Scholar]
- 194.Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17867. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chen J, Liu TW, Lo PC, Wilson BC, Zheng G. Bioconjug. Chem. 2009;20:1836. doi: 10.1021/bc900207k. [DOI] [PubMed] [Google Scholar]
- 196.Vaux DL, Korsmeyer SJ. Cell. 1999;96:245. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 197.Riedl SJ, Shi Y. Nat. Rev. Mol. Cell. Biol. 2004;5:897. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 198.Maxwell D, Chang Q, Zhang X, Barnett EM, Piwnica-Worms D. Bioconjug. Chem. 2009;20:702. doi: 10.1021/bc800516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Barnett EM, Zhang X, Maxwell D, Chang Q, Piwnica-Worms D. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9391. doi: 10.1073/pnas.0812884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang Z, Fan J, Cheney PP, Berezin MY, Edwards WB, Akers WJ, Shen D, Liang K, Culver JP, Achilefu S. Mol. Pharm. 2009;6:416. doi: 10.1021/mp800264k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Nat. Chem. Biol. 2007;3:668. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 202.Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Nat. Chem. Biol. 2005;1:203. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- 203.Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Nat. Med. 2009;15:967. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kim JH, Park K, Nam HY, Lee S, Kim K, Kwon IC. Prog. Polym. Sci. 2007;32:1031. [Google Scholar]
- 205.Bremer C, Tung CH, Weissleder R. Nat. Med. 2001;7:743. doi: 10.1038/89126. [DOI] [PubMed] [Google Scholar]
- 206.Ntziachristos V, Tung CH, Bremer C, Weissleder R. Nat. Med. 2002;8:757. doi: 10.1038/nm729. [DOI] [PubMed] [Google Scholar]
- 207.Deguchi JO, Aikawa M, Tung CH, Aikawa E, Kim DE, Ntziachristos V, Weissleder R, Libby P. Circulation. 2006;114:55. doi: 10.1161/CIRCULATIONAHA.106.619056. [DOI] [PubMed] [Google Scholar]
- 208.Chen J, Tung CH, Allport JR, Chen S, Weissleder R, Huang PL. Circulation. 2005;111:1800. doi: 10.1161/01.CIR.0000160936.91849.9F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Galande AK, Hilderbrand SA, Weissleder R, Tung CH. J. Med. Chem. 2006;49:4715. doi: 10.1021/jm051001a. [DOI] [PubMed] [Google Scholar]
- 210.Hilderbrand SA, Kelly KA, Weissleder R, Tung CH. Bioconjug. Chem. 2005;16:1275. doi: 10.1021/bc0501799. [DOI] [PubMed] [Google Scholar]
- 211.Scherer RL, VanSaun MN, McIntyre JO, Matrisian LM. Mol. Imaging. 2008;7:118. [PMC free article] [PubMed] [Google Scholar]
- 212.Lee HY, Li Z, Chen K, Hsu AR, Xu C, Xie J, Sun S, Chen X. J. Nucl. Med. 2008;49:1371. doi: 10.2967/jnumed.108.051243. [DOI] [PubMed] [Google Scholar]
- 213.Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. J. Med. Chem. 2006;49:6087. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 214.Mulder WJ, Koole R, Brandwijk RJ, Storm G, Chin PT, Strijkers GJ, de Mello Donega C, Nicolay K, Griffioen AW. Nano Lett. 2006;6:1. doi: 10.1021/nl051935m. [DOI] [PubMed] [Google Scholar]
- 215.Xie J, Huang J, Li X, Sun S, Chen X. Curr. Med. Chem. 2009;16:1278. doi: 10.2174/092986709787846604. [DOI] [PubMed] [Google Scholar]
- 216.Li ZB, Cai W, Chen X. J. Nanosci. Nanotechnol. 2007;7:2567. doi: 10.1166/jnn.2007.628. [DOI] [PubMed] [Google Scholar]
- 217.Gupta AK, Gupta M. Biomaterials. 2005;26:3995. doi: 10.1016/j.biomaterials.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 218.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat. Mater. 2005;4:435. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 219.Bahr JL, Tour JM. J. Mater. Chem. 2002;12:1952. [Google Scholar]
- 220.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 221.Choi JS, Jun YW, Yeon SI, Kim HC, Shin JS, Cheon J. J. Am. Chem. Soc. 2006;128:15982. doi: 10.1021/ja066547g. [DOI] [PubMed] [Google Scholar]
- 222.Jun YW, Huh YM, Choi JS, Lee JH, Song HT, Kim S, Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. J. Am. Chem. Soc. 2005;127:5732. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 223.Lee JH, Huh YM, Jun Y, Seo J, Jang J, Song HT, Kim S, Cho EJ, Yoon HG, Suh JS, Cheon J. Nat. Med. 2007;13:95. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 224.Cai W, Shin DW, Chen K, Gheysens O, Cao Q, Wang SX, Gambhir SS, Chen X. Nano Lett. 2006;6:669. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 225.Ito A, Shinkai M, Honda H, Kobayashi T. J. Biosci. Bioeng. 2005;100:1. doi: 10.1263/jbb.100.1. [DOI] [PubMed] [Google Scholar]
- 226.Jun YW, Seo JW, Cheon A. Acc. Chem. Res. 2008;41:179. doi: 10.1021/ar700121f. [DOI] [PubMed] [Google Scholar]
- 227.Budde MD, Frank JA. J. Nucl. Med. 2009;50:171. doi: 10.2967/jnumed.108.053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Montet X, Montet-Abou K, Reynolds F, Weissleder R, Josephson L. Neoplasia. 2006;8:214. doi: 10.1593/neo.05769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Xie J, Chen K, Lee HY, Xu C, Hsu AR, Peng S, Chen X, Sun S. J. Am. Chem. Soc. 2008;130:7542. doi: 10.1021/ja802003h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lyons SA, O'Neal J, Sontheimer H. Glia. 2002;39:162. doi: 10.1002/glia.10083. [DOI] [PubMed] [Google Scholar]
- 231.Veiseh M, Gabikian P, Bahrami SB, Veiseh O, Zhang M, Hackman RC, Ravanpay AC, Stroud MR, Kusuma Y, Hansen SJ, Kwok D, Munoz NM, Sze RW, Grady WM, Greenberg NM, Ellenbogen RG, Olson JM. Cancer Res. 2007;67:6882. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- 232.Sun C, Veiseh O, Gunn J, Fang C, Hansen S, Lee D, Sze R, Ellenbogen RG, Olson J, Zhang M. Small. 2008;4:372. doi: 10.1002/smll.200700784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Veiseh O, Sun C, Fang C, Bhattarai N, Gunn J, Kievit F, Du K, Pullar B, Lee D, Ellenbogen RG, Olson J, Zhang M. Cancer Res. 2009;69:6200. doi: 10.1158/0008-5472.CAN-09-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Leuschner C, Kumar CS, Hansel W, Soboyejo W, Zhou J, Hormes J. Breast Cancer Res. Treat. 2006;99:163. doi: 10.1007/s10549-006-9199-7. [DOI] [PubMed] [Google Scholar]
- 235.Chatzistamou L, Schally AV, Nagy A, Armatis P, Szepeshazi K, Halmos G. Clin Cancer Res. 2000;6:4158. [PubMed] [Google Scholar]
- 236.Medarova Z, Rashkovetsky L, Pantazopoulos P, Moore A. Cancer Res. 2009;69:1182. doi: 10.1158/0008-5472.CAN-08-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Moore A, Medarova Z, Potthast A, Dai G. Cancer Res. 2004;64:1821. doi: 10.1158/0008-5472.can-03-3230. [DOI] [PubMed] [Google Scholar]
- 238.Yang L, Peng XH, Wang YA, Wang X, Cao Z, Ni C, Karna P, Zhang X, Wood WC, Gao X, Nie S, Mao H. Clin. Cancer Res. 2009;15:4722. doi: 10.1158/1078-0432.CCR-08-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chen K, Xie J, Xu H, Behera D, Michalski MH, Biswal S, Wang A, Chen X. Biomaterials. 2009;30:6912. doi: 10.1016/j.biomaterials.2009.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Peng XH, Qian X, Mao H, Wang AY, Chen ZG, Nie S, Shin DM. Int. J. Nanomedicine. 2008;3:311. doi: 10.2147/ijn.s2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. Nat. Biotechnol. 2004;22:969. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 243.Green M. Angew. Chem. Int. Ed. Engl. 2004;43:4129. doi: 10.1002/anie.200301758. [DOI] [PubMed] [Google Scholar]
- 244.Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Curr. Opin. Biotechnol. 2002;13:40. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 245.Xie R, Peng X. J. Am. Chem. Soc. 2009;131:10645. doi: 10.1021/ja903558r. [DOI] [PubMed] [Google Scholar]
- 246.Xie R, Battaglia D, Peng X. J. Am. Chem. Soc. 2007;129:15432. doi: 10.1021/ja076363h. [DOI] [PubMed] [Google Scholar]
- 247.Xie R, Peng X. Angew. Chem. Int. Ed. Engl. 2008;47:7677. doi: 10.1002/anie.200802867. [DOI] [PubMed] [Google Scholar]
- 248.Choi HS, Ipe BI, Misra P, Lee JH, Bawendi MG, Frangioni JV. Nano Lett. 2009;9:2354. doi: 10.1021/nl900872r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zimmer JP, Kim SW, Ohnishi S, Tanaka E, Frangioni JV, Bawendi MG. J. Am. Chem. Soc. 2006;128:2526. doi: 10.1021/ja0579816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nat. Biotechnol. 2003;21:47. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 251.Chen F, Gerion D. Nano Lett. 2004;4:1827. [Google Scholar]
- 252.Ruan G, Agrawal A, Marcus AI, Nie S. J. Am. Chem. Soc. 2007;129:14759. doi: 10.1021/ja074936k. [DOI] [PubMed] [Google Scholar]
- 253.Anas A, Okuda T, Kawashima N, Nakayama K, Itoh T, Ishikawa M, Biju V. ACS Nano. 2009;3:2419. doi: 10.1021/nn900663r. [DOI] [PubMed] [Google Scholar]
- 254.Orndorff RL, Rosenthal SJ. Nano Lett. 2009;9:2589. doi: 10.1021/nl900789e. [DOI] [PubMed] [Google Scholar]
- 255.Smith AM, Duan H, Mohs AM, Nie S. Adv. Drug Deliv. Rev. 2008;60:1226. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Akerman ME, Chan WC, Laakkonen P, Bhatia SN, Ruoslahti E. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12617. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cai W, Chen X. Nat. Protoc. 2008;3:89. doi: 10.1038/nprot.2007.478. [DOI] [PubMed] [Google Scholar]
- 258.Cai W, Chen K, Li ZB, Gambhir SS, Chen X. J. Nucl. Med. 2007;48:1862. doi: 10.2967/jnumed.107.043216. [DOI] [PubMed] [Google Scholar]
- 259.Oostendorp M, Douma K, Hackeng TM, Dirksen A, Post MJ, van Zandvoort MA, Backes WH. Cancer Res. 2008;68:7676. doi: 10.1158/0008-5472.CAN-08-0689. [DOI] [PubMed] [Google Scholar]
- 260.Mulder WJ, Castermans K, van Beijnum JR, Oude Egbrink MG, Chin PT, Fayad ZA, Lowik CW, Kaijzel EL, Que I, Storm G, Strijkers GJ, Griffioen AW, Nicolay K. Angiogenesis. 2009;12:17. doi: 10.1007/s10456-008-9124-2. [DOI] [PubMed] [Google Scholar]
- 261.Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Nat. Nanotechnol. 2009;5:42. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Rhyner MN, Smith AM, Gao X, Mao H, Yang L, Nie S. Nanomed. 2006;1:209. doi: 10.2217/17435889.1.2.209. [DOI] [PubMed] [Google Scholar]
- 263.Gao X, Dave SR. Adv. Exp. Med. Biol. 2007;620:57. doi: 10.1007/978-0-387-76713-0_5. [DOI] [PubMed] [Google Scholar]
- 264.Keren S, Zavaleta C, Cheng Z, de la Zerda A, Gheysens O, Gambhir SS. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5844. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Liu Z, Cai W, He L, Nakayama N, Chen K, Sun X, Chen X, Dai H. Nat. Nanotechnol. 2007;2:47. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 266.Zavaleta C, de la Zerda A, Liu Z, Keren S, Cheng Z, Schipper M, Chen X, Dai H, Gambhir SS. Nano Lett. 2008;8:2800. doi: 10.1021/nl801362a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.De la Zerda A, Zavaleta C, Keren S, Vaithilingam S, Bodapati S, Liu Z, Levi J, Smith BR, Ma TJ, Oralkan O, Cheng Z, Chen X, Dai H, Khuri-Yakub BT, Gambhir SS. Nat. Nanotechnol. 2008;3:557. doi: 10.1038/nnano.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Park K, Hong HY, Moon HJ, Lee BH, Kim IS, Kwon IC, Rhee K. J. Control. Release. 2008;128:217. doi: 10.1016/j.jconrel.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 269.Hong HY, Lee HY, Kwak W, Yoo J, Na MH, So IS, Kwon TH, Park HS, Huh S, Oh GT, Kwon IC, Kim IS, Lee BH. J. Cell. Mol. Med. 2008;12:2003. doi: 10.1111/j.1582-4934.2008.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Reddy GR, Bhojani MS, McConville P, Moody J, Moffat BA, Hall DE, Kim G, Koo YE, Woolliscroft MJ, Sugai JV, Johnson TD, Philbert MA, Kopelman R, Rehemtulla A, Ross BD. Clin. Cancer Res. 2006;12:6677. doi: 10.1158/1078-0432.CCR-06-0946. [DOI] [PubMed] [Google Scholar]
- 271.Almutairi A, Rossin R, Shokeen M, Hagooly A, Ananth A, Capoccia B, Guillaudeu S, Abendschein D, Anderson CJ, Welch MJ, Frechet JM. Proc. Natl. Acad. Sci. U. S. A. 2009;106:685. doi: 10.1073/pnas.0811757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lee S, Ryu JH, Park K, Lee A, Lee SY, Youn IC, Ahn CH, Yoon SM, Myung SJ, Moon DH, Chen X, Choi K, Kwon IC, Kim K. Nano Lett. 2009;9:4412. doi: 10.1021/nl902709m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Matsumura Y, Maeda H. Cancer Res. 1986;46:6387. [PubMed] [Google Scholar]