Abstract
Rhizobium leguminosarum is a soil bacterium with the ability to form nitrogen-fixing nodules on the roots of leguminous plants. Soil-dwelling, free-living R. leguminosarum often encounters desiccation stress, which impacts its survival within the soil. The mechanisms by which soil bacteria resist the effects of desiccation stress have been described. However, the role of the cell envelope in the desiccation tolerance mechanisms of rhizobia is relatively uncharacterized. Using a transposon mutagenesis approach, a mutant of R. leguminosarum bv. viciae was isolated that was highly sensitive to desiccation. The mutation is located in the ATP-binding protein of an uncharacterized ATP-binding cassette transporter operon (RL2975–RL2977). Exopolysaccharide accumulation was significantly lower in the mutant and the decrease in desiccation tolerance was attributed to the decreased accumulation of exopolysaccharide. In addition to desiccation sensitivity, the mutant was severely impaired in biofilm formation, an important adaptation used by soil bacteria for survival. This work has identified a novel transporter required for physiological traits that are important for the survival of R. leguminosarum in the rhizosphere environment.
Keywords: ABC transporter, desiccation, biofilm, rhizobium, exopolysaccharide, cell envelope
Introduction
Rhizobium leguminosarum is a Gram-negative bacterium notable for its ability to fix nitrogen within root nodules of leguminous plants (recently reviewed by Gibson et al., 2008). In the absence of a host plant, R. leguminosarum dwells within the rhizosphere. The rhizosphere environment can be highly variable and is prone to fluctuations in oxygen concentration, decreased water availability, temperature extremes and nutrient limitations (Deaker et al., 2004). Bacteria residing in the rhizosphere frequently encounter desiccation stress and require strategies to persist during times of desiccation. The negative cellular effects of desiccation are wide ranging and include protein denaturation, DNA degradation and loss of membrane integrity (Billi & Potts, 2002). Therefore, it is not surprising that the mechanisms required for desiccation resistance are complex and require multiple genetic loci (Billi & Potts, 2002; van de Mortel & Halverson, 2004; Humann et al., 2009). An increased tolerance to desiccation can enhance the survival of rhizobacteria within the soil considerably (Rokitko et al., 2003). This is of particular interest with nitrogen-fixing rhizobia because desiccation is a major cause of the poor on-seed survival rates of commercial inoculants and the subsequent poor performance of rhizobial inoculants in the field (Deaker et al., 2004). To further our understanding of how bacteria persist in the soil environment, and improve on-seed survival of rhizobial inoculants, a complete understanding of the mechanisms used for desiccation tolerance is necessary.
Studies with the soil bacterium Pseudomonas sp. have identified a variety of mechanisms used to increase their tolerance to desiccation. For instance, the production of highly hygroscopic secreted polysaccharides is increased to maintain a hydrated microenvironment relative to the surrounding environment (Roberson & Firestone, 1992; van de Mortel & Halverson, 2004). Also, fatty acid isomerization is biased in favor of the trans-configuration to maintain membrane fluidity during drying (van de Mortel & Halverson, 2004). In Salmonella enterica serovar Typhimurium, the outer membrane O antigen of lipopolysaccharide was found to be crucial for desiccation tolerance (Garmiri et al., 2008). In addition, cell surface components such as fimbriae and cellulose are important in desiccation tolerance in Salmonella (Gibson et al., 2006; White et al., 2006). These results suggest that the cell envelope, which includes the inner and outer membranes, cell wall and excreted surface polysaccharides, is an important structural target for increasing desiccation tolerance in bacteria.
In rhizobia, the role of the cell envelope in desiccation tolerance has recently been described. Gilbert et al. (2007), isolated a ctpA mutant in R. leguminosarum bv. viciae with altered cell envelope integrity and a corresponding decrease in desiccation tolerance. In Bradyrhizobium, mild desiccation stress resulted in an increase in the degree of saturation of the fatty acids found in membrane phospholipids (Boumahdi et al., 2001). Microarray data from Bradyrhizobium japonicum revealed that a number of exopolysaccharides and lipopolysaccharides biosynthetic and transport genes are upregulated in response to desiccation stress (Cytryn et al., 2007). In addition, a R. leguminosarum bv. viciae lipopolysaccharide mutant lacking the 27-hydroxyoctacosanoatic acid of lipid A is less tolerant to desiccation stress (Vanderlinde et al., 2009).
A transposon mutagenesis screen to isolate genes required for maintaining a functional cell envelope in R. leguminosarum bv. viciae 3841 identified a strain with a mutation in the ATP-binding component of a previously uncharacterized ATP-binding cassette (ABC) transporter (RL2975, RL2976 and RL2977; Young et al., 2006). The level of exopolysaccharide in the cell envelope of the mutant was decreased compared with the wild type and this has been connected to a decreased tolerance to desiccation. The mutant was also defective in biofilm formation on a polystyrene support. The main findings of this study demonstrate the important role for exopolysaccharide in desiccation tolerance in R. leguminosarum and identify a new genetic element required for proper biofilm formation.
Materials and methods
Strains, media and growth conditions
Table 1 lists the strains, plasmids and primers used in this study. Escherichia coli strains were cultured using Luria–Bertani medium (Sambrook et al., 1989), supplemented as necessary with the following concentrations of antibiotics (μg mL−1): gentamicin, 15; ampicillin, 100; spectinomycin, 100; and tetracycline, 10. Rhizobium leguminosarum cells were cultured using tryptone–yeast (TY) (Beringer, 1974) or Vincent's minimal media (VMM) (Vincent, 1970) with 10 mM mannitol as a carbon and energy source, supplemented as required with the following concentrations of antibiotics (μg mL−1): gentamicin, 30; neomycin, 100; tetracycline, 5; and streptomycin, 500.
Table 1.
Strains, plasmids and primers used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| TOP 10 | F– mcrAD(mrr0hsdRMS0mcrBC) f80lacZDM15DlacX74 recA1 araD139D(ara0leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| S17-1 | RP4 tra region, mobilizer strain recA derivative of MM294A with RP4-2 (Tc::Mu::Km::Tn7) integrated into the chromosome | Simon et al. (1983) |
| DH5α | endA1,hsdR17,supE44,thi-1,recA1,gyrA96,relA1,D(argF–lacZYA),DlacU169,f80lacZDM15 | Invitrogen |
| R. leguminosarum | ||
| 3841 | Spontaneous streptomycin-resistant derivative of R. leguminosarum bv. viciae strain 300 | Glenn et al. (1980), Johnston & Beringer (1975) |
| 17B | 3841 ABC transporter, RL2975–RL2977 mutant, Tn5 TGN mutant, Smr, Nmr, Gmr | This study |
| 38EV30 | 3841 ABC transporter, RL2976–RL2977 mutant, Smr, Gmr | |
| Plasmids | ||
| PTGN | Tn5 derivative, Gmr, Ampr promoter-less nptII, gfp | Tang et al. (1999) |
| pCR2.1 topo | Topo TA cloning vector, 4.0 kb, Kmr Apr | Invitrogen |
| pJQ200SK+ | Suicide vector, P15a ori, mob, sacB, Gmr | Quandt & Hynes (1993) |
| pCos879 | R. leguminosarum VF39SM cosmid with RL2975–RL2977 | Yost et al. (1998) |
| pEV25 | pCR2.1 TOPO with a 1.3-kb PCR fragment containing the internal fragment of RL2975 and RL2976 Ampr, Kmr | This study |
| pEV30 | 1.3-kb reeA and reeB fragment cloned from pEV25 into pJQ200SK using SstI/XbaI Gmr | This study |
| Primers | ||
| GmTAIL-1 | 5′-CAACGCGCTTGGTGCTAATGTGAT-3′ | This study |
| GmTAIL2 | 5′-AGTTGGGCATACGGGAAGAAGTGA-3′ | This study |
| 17B1F | 5′-AGCTCGAGCATCCTGGTGCCGA-3′ | This study |
| 17B1R | 5′-AGCTCGAGTCCAGTCCAGCGAGAAAC-3′ | This study |
| 17BF | 5′-TCTGCGGCGGATGGACAA-3′ | This study |
| 17BR | 5′-GTGAGGATGATGGTGACG-3′ | This study |
| AD-1 | 5′-AGWGNAGWANCAWAGG-3′ | Liu & Huang (1998) |
Transposon mutagenesis
Mutagenesis was performed using the mini-Tn5 derivative found on plasmid pTGN (Tang et al., 1999). Biparental matings of the E. coli mobilizer strain S17-1 containing the pTGN vector and R. leguminosarum 3841 were performed at 30 °C for 24 h on VMM plates supplemented with 1 mM proline. Transconjugants were selected on VMM with streptomycin and neomycin, and subsequently screened for inability to grow on the solid complex medium TY. We have previously observed that mutants with defects in the cell envelope are unable to grow on complex solid media (Gilbert et al., 2007; Vanderlinde et al., 2009). Therefore, we used the lack of growth on TY as a selection to enrich for isolation of transposon mutants with defective cell envelopes. Genomic DNA was isolated from TY-negative isolates and used as a template to identify the transposon insertion site. The transposon insertion site was identified using the thermal asymmetric interlaced (TAIL)-PCR protocol and primers described by Liu & Huang (1998). Briefly, the specific primer GmTAIL1 (Table 1; SigmaGenosys Canada, Oakville, ON) that binds within the Gm cassette of the transposon and the arbitrary degenerate primer AD-1 described by Liu & Huang (1998) were used in the primary reaction, which was followed by a secondary reaction with the specific nested primer GmTAIL2 (Table 1), the arbitrary primer and 1 μL of primary reaction as template. The transposon insertion site was identified by sequencing the TAIL-PCR product and using a blastn search (Altschul et al., 1997) and the Rhizobase database [Kazusa DNA Research Institute (http://bacteria.kazusa.or.jp/rhizobase/)].
Standard molecular techniques
Plasmid preps were performed using the alkaline lysis method (Sambrook et al., 1989). Restriction endonucleases and DNA-modifying enzymes were purchased from Invitrogen (Burlington, ON) and used according to the manufacturer's instructions. When necessary, PCR products were isolated from agarose gels using reagents and protocols from the QIAEX II gel extraction kit (Qiagen, Missisuaga, ON).
Mutagenesis of RL2976
Primers 17B1F and 17B1R (Table 1) were used to PCR amplify a 1293-bp fragment of RL2975 and RL2976. The fragment contained the 3′ end of RL2975 and the 5′ start of RL2976. The genes RL2975 and RL2976 are likely part of an operon, beginning with RL2975; therefore, the fragment lacked a promoter for RL2976 and a single crossover event using this fragment resulted in mutation of RL2976. PCR reactions used 1 U of Taq DNA polymerase (UBI, Calgary, AB) and the following reaction conditions: 1× reaction buffer, 2 mM MgSO4, 0.2 mM dNTPs and 0.2 μM of each primer. PCR amplification was performed using a Techne TC312 Thermocycler (Techne, Staffordshire, UK) at 94 °C for 5 min, followed by 30 cycles of: 95 °C for 30 s, 58 °C for 30 s and 72 °C for 75 s and a final extension at 72 °C for 5 min. PCR products were cloned into the pCR2.1 topo vector (Invitrogen) as per the manufacturer's instructions. Mutagenesis of RL2976 was accomplished by allelic exchange using pEV30 and the method described by Quandt & Hynes (1993). Briefly, the fragment of RL2975 and RL2976 was excised using SstI and XbaI restriction sites in the pCR2.1 topo vector and ligated into the same sites in the vector pJQ200SK, creating pEV30. The mobilizer strain S17-1 was transformed with pEV30, and biparental matings were performed overnight on VMM supplemented with 0.5 mM proline. Mutants were selected on the basis of gentamicin resistance. Correct insertional disruption of RL2976 in the putative mutants was confirmed by PCR.
Sequence analysis
DNA sequencing was performed by the University of Calgary Core DNA Services (Calgary, AB). DNA sequence data were analyzed using 4peaks software [version 1.7.2; A. Griekspoor and Tom Groothuis (http://mekentosj.com/4peaks/)]. Primers were designed using oligo 4.0 software (National Biosciences, Plymouth, MN). Predictions of transmembrane domains were made using tmpred (Hofmann & Stoffel, 1993). Analysis of conserved domains and Hidden Markov model (HMM) sequences were from the InterPro database (Mulder et al., 2007). Sequence alignments were performed using clustalw (Thompson et al., 1994), and phylogenetic trees were constructed using njplot (Perrière & Gouy, 1996).
Complementation analysis
The 371-bp fragment amplified by the primers 17BF and 17BR (Table 1) was used to probe an R. leguminosarum bv. viciae VF39SM cosmid library (Yost et al., 1998). Probe labeling, hybridization and detection were performed using reagents and protocols of the digoxigenin labeling and detection system (Roche Diagnostics, Lavel, QC). Cosmids detected by the 17B probe were further confirmed for the presence of the entire ABC transporter operon by PCR and DNA sequencing. The sequences of the RL2975–RL2977 homologues in VF39SM are available from the GenBank database under accession numbers GQ183604–GQ183606.
Desiccation, detergent and osmotic stress assays
Desiccation sensitivity assays used the filtration method described by Ophir & Gutnick (1994) as modified by Gilbert et al. (2007). Cells were grown in TY broth or VMM broth to the late log phase and diluted in water to a total volume of 100 mL. The diluted culture was filtered in duplicate as 2 × 50 mL aliquots according to the manufacturer's specifications with a Millipore Vacuum Manifold (Millipore Inc., Bedford, MA) using Microfil Filtration Funnels and S-Pak 0.45-μm TypeHA membranes (Millipore Inc.). Membranes were placed on either TY or VMM plates and incubated for 48 h at 30 °C. Following incubation, one filter was transferred to 10 mL of water, vortexed for 5 min to remove cells and the amount of exopolysaccharide was determined using the precipitation method described below. The second filter was cut in half and transferred to either an empty Petri dish (air) or a water–agar plate (12.5 g L−1 agar) (water), and incubated at ambient temperature and humidity for 24 h. Following incubation, membranes were transferred to a centrifuge bottle containing 2 mL of water and vortexed vigorously for 10 min to remove cells from the membranes. The number of viable bacteria was then determined using the spread plate technique and the percent survival was calculated as the ratio of CFU to CFU . For exopolysaccharide complementation experiments, exopolysaccharide was isolated from wild-type bacteria as described below, and resuspended in sterile distilled water (dH2O), to a concentration of ~0.4 g mL−1. After cells were filtered as described above, 5 mL of the exopolysaccharide solution was filtered on top. The remainder of the assay was carried out as described above.
Detergent sensitivity assays were performed as described by Gilbert et al. (2007). Osmotic stress assays were performed by inoculating strains into TY broth with 69.5 mM NaCl. Cultures were grown at 30 °C for 48 h and then the OD600 nm was measured. Results are reported as the ratio of OD600 nm with NaCl to the OD600 nm without NaCl.
Quantitative determination of secreted polysaccharides
The method for total exopolysaccharide quantification was a modification of those described by Ngwai et al. (2006). Briefly, cells were grown to the stationary phase in 25 mL of VMM. Cells were pelleted at 7710 g for 20 min at 4 °C, and washed with 25 mL of 1 M NaCl, 10 mM EDTA at pH 8.0. The exopolysaccharide was then precipitated from the combined culture by the addition of two volumes of ice-cold isopropanol. The precipitated polysaccharide was then spooled and dried in a sterile Petri dish at 37 °C overnight. Results are reported as the mass of exopolysaccharide produced per milligram of dry cell mass. The capsular polysaccharide (CPS) was extracted according to the method of Zevenhuizen (1984). Precipitated CPS was dried overnight at 37 °C, and results were reported as the mass of CPS produced per milligram of dry cell mass. The amount of total neutral polysaccharide was determined by combining the remaining isopropanol supernatants from the exopolysaccharide and CPS determinations and quantifying the amount of glucose using the anthrone-sulfuric acid assay as described by Laurentin & Edwards (2003).
Structural analysis of acidic exopolysaccharide
Exopolysaccharide was precipitated from culture supernatants using isopropanol as described above. Following drying, the precipitates were resuspended in 20 mL of dH2O and dialyzed against dH2O for 20 h using the Spectra/Por® 7 dialysis membrane with a 2000 molecular weight cut-off (Spectrum® Laboratories Inc., Rancho Dominguez, CA). The dialyzed samples were then lyophilized before further analysis.
Exopolysaccharide preparations were purified by gelfiltration chromatography using a Sephacryl S-400 HR matrix (GE Healthcare) with 50 mM ammonium formate, pH 6.8, used as the eluent. The eluting fractions were monitored using the Shimadzu Refractive Index Detector (RID-10A).
Glycosyl composition analysis was performed by the preparation and GC-MS analysis of trimethylsilyl methyl glycosides as described by York et al. (1985). Briefly, samples were subjected to methanolysis at 80 °C for 18 h in 1 M methanolic HCl. The resulting methyl glycosides were N-acetylated at 100 °C for 1 h, trimethylsilylated at 80 °C for 30 min and then analyzed on a Hewlett-Packard HP5890 gas chromatograph equipped with a mass selective detector 5970 MSD using a Supelco DB-1 fused silica capillary column (30 m × 0.25 mm ID; J & W Scientific, Folsom, CA).
Nuclear magnetic resonance (NMR) analysis was performed by exchanging the sample two times with 99.9% deuterium oxide (D2O) and finally dissolving in 100% D2O (Cambridge Isotope Laboratories Inc.), and 1D proton spectra were recorded using a Varian Inova 500 MHz NMR spectrometer (Varian, Palo Alto, CA) at 70 °C.
Capsule staining
Capsule stains were performed on stationary-phase cultures grown in VMM broth. Cells were negatively stained using nigrosin, followed by counter-staining with crystal violet (CV). Stained cells were then visualized under × 100 oil immersion using an Olympus BX51 light microscope (Olympus, PA).
Biofilm cultivation
Biofilms were grown either in microtiter plates, using methods adapted from O'Toole & Kolter (1998) and Fujishige et al. (2006), or in the Calgary Biofilm Device (CBD, commercially available as the MBEC-HTP Assay, Innovotech, Edmonton, AB, Canada), as described previously by Ceri et al. (1999). To begin, strains were streaked out twice on the appropriate selective medium. Colonies were collected from the second agar subculture using a sterile cotton swab and were suspended in double-distilled water (ddH2O) to match a 1.0 McFarland standard. This standardized bacterial suspension was then diluted 1 in 15 into VMM that contained 1% mannitol and 500 μg mL−1 streptomycin, which, when verified by viable cell counting, provided a starting inoculum of approximately 1 × 107 CFU mL−1. For microtiter plate biofilms, 150 μL of this standardized inoculum was added to each well of a flat-bottom, cell culture-treated Nunc® brand 96-well plate (VWR International Ltd, Mississauga, ON, Canada). These plates were sealed with Parafilm™ and incubated at 30 °C for 48 h on a gyrorotary shaker set to 50 r.p.m. Alternatively, 22 mL of the standardized inoculum was added to the trough of the CBD. These devices were also sealed with Parafilm™ and then incubated at 30 °C for 72 h on a rocking table set to 3.5 rocks min− as described previously (Ceri et al., 1999).
CBD biofilms were rinsed by placing the peg lid into a microtiter plate that contained 200 μL of ddH2O in each well. For viable cell counts, the peg lids were transferred into a second microtiter plate, also containing 200 μL of ddH2O in each well, into which biofilms were disrupted using an ultrasonic cleaner as described previously (Ceri et al., 1999). Cells from the disrupted biofilms were serially diluted 10-fold and plated for viable cell counting on VMM+1% mannitol agar. Spot plates were grown for 72 h before enumeration.
Tetrazolium reduction assays
Microtiter plate biofilms were rinsed once with 200 μL of ddH2O to remove loosely adherent biomass and then immersed in fresh medium containing a metabolic indicator. To do this, 150 μL of ddH2O, 25 μL of VMM+1% mannitol and 25 μL of CellTiter 96 AQueous One solution (Promega Corporation, Madison, WI) were added to each well of the rinsed microtiter plate. AQueous One solution contains the tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS), which is presumably reduced to a colored formazan product by NADH and NADPH from metabolically active cells (Berridge & Tan, 1993). These plates were wrapped in an aluminum foil and incubated at 30 °C for 18 h. Biofilm metabolic activity was assessed by reading the OD490 nm of these plates on a Thermomax microtiter plate reader with softmax pro data analysis software (Molecular Devices, Sunnyvale, CA) as described previously (Harrison et al., 2007).
CV staining for biomass
Microtiter plate biofilms were rinsed once with 200 μL of ddH2O to remove loosely adherent biomass and then stained with CV using a method adapted from Fujishige et al. (2006). To do this, 200 μL of 0.4% CV (VWR International Ltd) was added to each well and these plates were incubated for 15 min. The CV was then aspirated and the wells were then rinsed three times with 200 μL of ddH2O for 1 min. The CV was solubilized by adding 200 μL of 95% ethanol to each well. The amount of CV bound to the biomass was quantified by reading the OD550 nm on a microtiter plate reader (as described above).
Confocal laser scanning microscopy (CLSM)
Pegs were broken from the lid of the CBD using a pair of flamed needle nose pliers. Biofilms that had been grown on the surface of the peg, were stained with acridine orange or with the Live/Dead® bacterial cell viability kit as described previously by Harrison et al. (2006, 2007). Acridine orange, which fluoresces green, is a nucleic acid intercalator that stains biofilm cells as well as extracelluar nucleic acids, and may thus be used as a biomass indicator. In contrast, the Live/Dead® kit contains the membrane-permeable DNA intercalator Syto-9, which fluoresces green, and the membrane-impermeable DNA intercalator propidium iodide, which fluoresces red. In principle, cells with compromised membrane integrity are dead and appear red, whereas live cells, which exclude propidium iodide from the cytoplasm, appear green.
Fluorescently labeled biofilms were placed in two drops of ddH2O on the surface of a glass coverslip. These pegs were examined using a Leica DM IRE2 spectral confocal and multiphoton microscope with a Leica TCS SP2 acoustic optical beam splitter (Leica Microsystems, Richmond Hill, ON, Canada) as described previously (Harrison et al., 2007). For acridine orange-stained samples, biofilms were scanned using 476 nm excitation and fluorescence was collected in the green region of the spectrum. For Live/Dead® staining, samples were sequentially scanned, frame by frame, first at 488 nm (Syto-9) and then at 543 nm (propidium iodide). Fluorescence emission was then sequentially collected in the green and red regions of the spectrum, respectively. A ×63 water-immersion objective was used in all imaging experiments. Image capture and two-dimensional (2D) reconstruction of z-stacks were performed using leica confocal software (Leica Microsystems).
Results
Mutagenesis and bioinformatic analysis of a novel ABC transporter operon
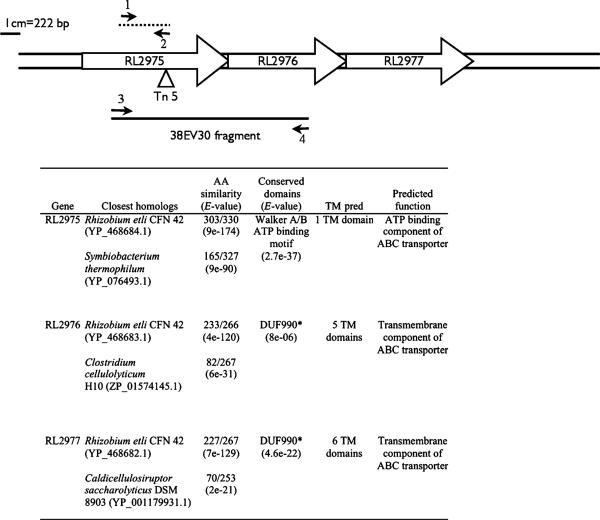
A R. leguminosarum 3841 mutant (17B) was isolated during a transposon (Tn5) mutagenesis screen for mutants with defective cell envelopes. The insertion site of the transposon was mapped to base pair 750 of a gene encoding a predicted ATP-binding component (RL2975) of a previously uncharacterized ABC transporter operon (Fig. 1). DNA sequence analysis suggests that the operon is comprised of three genes: RL2975 (1001 bp), RL2976 (800 bp) and RL2977 (848 bp), encoding for the ATP-binding protein and two transmembrane proteins, respectively. The start and stop codons of the three genes overlap, which may suggest transcriptional and translational coupling. Therefore, the transposon mutagenesis likely resulted in a polar mutation. The mutant 38EV30 is an RL2976 mutant constructed by homologous recombination as described in Materials and methods. This mutation disrupts RL2976 and RL2977; however, the gene encoding the ATP-binding protein, RL2975, should still be expressed.
Fig. 1.
Schematic of the RL2975—2977 ABC transporter operon. Small arrows indicate the positions of primers 17BF and 17BR (1 and 2) and 17B1F and 17B1R (3 and 4). Refer to Materials and methods for the primer sequences and amplification conditions used. The triangle indicates the transposon insertion site. The dashed line indicates the 17B probe fragment. Inset table summarizes the blastp analysis and predicted function of the three proteins encoded by the operon. *DUF990-domain of unknown function 990 is associated with ABC-2 transporters (Reizer et al., 1992). AA, amino acid; TM, transmembrane.
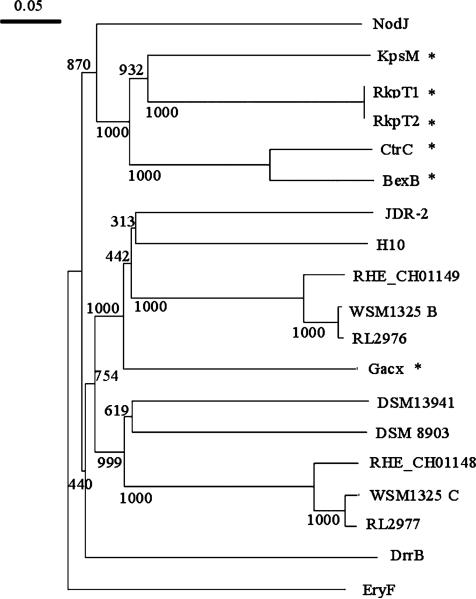
The proteins encoded by RL2975, RL2976 and RL2977 are predicted to contain 1, 5 and 6 transmembrane domains, respectively. Figures 1 and 2 illustrate that, with the exception of Rhizobium etli and R. leguminosarum bv. trifolii, this particular ABC transporter operon is not present in the genomes of other Rhizobiales sequenced to date. In fact, the next closest matches are to ABC transporters of unknown function in soil-dwelling Gram-positive bacteria such as Clostridium sp. and Paenibacillus sp. (Figs 1 and 2).
Fig. 2.
Neighbor-joining tree of RL2976 and RL2977 with the transmembrane components of well-characterized ABC-2 transporters and DUF990 proteins. Bootstrap analysis was performed with 1000 replicates, and values (out of 1000) are as displayed on tree. RHE_CH01148, Rhizobium etli CFN 42 (YP_468682.1); RL2977, Rhizobium leguminosarum bv. viciae 3841 (YP_768561.1); DrrB, Mycobacterium avium ssp. paratuberculosis K-10 (NP_960171.1); NodJ, R. leguminosarum bv. viciae 3841 (YP_770467.1); CtrC, Neisseria meningitidis Z2491 (NP_283043.1); BexB, Haemophilus influenzae (P22235); KpsM, Escherichia coli (AAC38078.1); RHE_CH01149, R. etli CFN 42 (YP_468683.1); RL2976, R. leguminosarum bv. viciae 3841 (YP_768560.1); RkpT1, Sinorhizobium meliloti 1021 (NP_437116); RkpT2, S. meliloti 1021 (NP_437102); JDR-2, Paenibacillus sp. JDR-2 (ZP_02846839); H10, Clostridium cellulolyticum H10 (ZP_01574145.1); WSM1325C, R. leguminosarum bv. trifolii WSM1325 (ZP_02293733); DSM 13941, Roseiflexus castenholzii DSM 13941 (YP_001431939); DSM 8903, Caldicellulosiruptor saccharolyticus DSM 8903 (YP_001179931.1); WSM1325B, R. leguminosarum bv. trifolii WSM1325 (ZP_02293734); EryF, R. leguminosarum bv. viciae 3841 (YP_764713); GacX, Streptomyces glaucescens (CAL64857.1). *Represents proteins where experimental data exists on their cellular function.
The two transmembrane proteins, encoded by RL2976 and RL2977, contain a conserved domain of unknown function (DUF990), which shares homology to a subgroup of ABC transporters classified as ABC-2 transporters (Reizer et al., 1992). The ABC-2 transporters are involved in the export of diverse substrates from the cell, such as sodium ions, signaling molecules and polysaccharides (Reizer et al., 1992). A few of these transporters have been well characterized and include the KpsMT, BexABC, CtrABC and RkpRST transporters, which are involved in exopolysaccharide export in E. coli, Haemophilus influenzae, Neisseria meningitidis and Sinorhizobium meliloti, respectively (Kroll et al., 1990; Smith et al., 1990; Frosch et al., 1991; Pavelka et al., 1991; Reizer et al., 1992; Kiss et al., 2001).
A clustalw alignment was performed with RL2977, the HMM sequence of the DUF990 domain and the HMM sequence of ABC-2 transporters (data not shown). An alignment comparing RL2976 with the DUF990 and ABC-2 HMM sequences yielded results similar to the RL2977 alignment. Based on the alignment results, both the proteins have amino acid sequences that are related to the DUF990 sequence with amino acid similarities of 53.4% and 55.7%, respectively, and have a weak similarity to the HMM ABC-2 transporter sequence, with amino acid similarities of 14.9% and 27.9%, respectively. A neighbor-joining phylogenetic tree with previously characterized members of the ABC-2 group and DUF990 proteins separated the previously characterized ABC-2 transporter sequences from the RL2976 and Rl2977 amino acid sequences and all of the other DUF990-containing proteins, suggesting that DUF990 proteins may represent a new uncharacterized subclass of ABC-2 transporters (Fig. 2).
Quantification and partial characterization of secreted polysaccharides in the 17B and 38EV30 mutants
Three types of polysaccharides constitute the majority of surface polysaccharides (SPS) in R. leguminosarum: exopolysaccharide, CPS and neutral polysaccharides (Skorupska et al., 2006). The exopolysaccharides and CPS have a similar or an identical basic structure, and are distinguished based on their location, and the distribution of noncarbohydrate residues such as pyruvate, 3-hydroxybutyrate and O-acetate (Skorupska et al., 2006). To determine the role of RL2975–RL2977 in SPS export, the amounts of exported exopolysaccharide, CPS and neutral polysaccharide from the mutant strains 17B and 38EV30 grown in VMM broth were quantified and compared with the wild-type strain. The amounts of exopolysaccharide produced by the 17B and 38EV30 mutants were 2.28±0.163 and 1.96±0.472 mg exopolysaccharide mg−1 dry cell weight, respectively, whereas the wild type, 3841, produced 6.20±0.849 mg exopolysaccharide mg−1 dry cell weight. The amount of CPS produced by wild type, 17B and 38EV30 was found to be 0.138±0.018, 0.092±0.010 and 0.082±0.038 mg CPS mg −1 dry cell weight, respectively. Finally, quantification of neutral polysaccharides determined that the wild type produced 0.054±0.0008 mg glucose mg−1 dry cell weight, 17B produced 0.059±0.004 mg glucose mg−1 dry cell weight and 38EV30 produced 0.044 mg glucose mg−1 dry cell weight. From these data, it is clear that while the mutants produce threefold lower amounts of exopolysaccharide in VMM broth, the amounts of CPS and neutral polysaccharides are not altered.
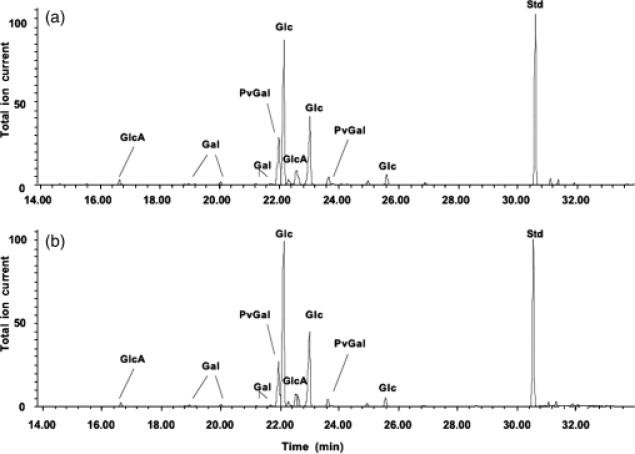
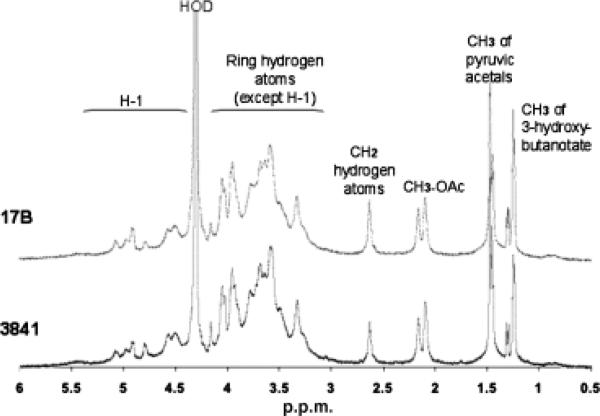
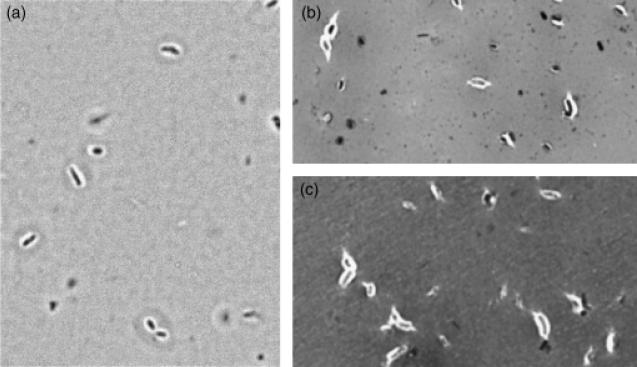
Gel-permeation chromatography was used to isolate the high molecular weight (HMW) acidic exopolysaccharide from both the wild type and the 17B mutant. Compositional analysis (Fig. 3) and NMR structural analysis (Fig. 4) indicated that the HMW acidic exopolysaccharide from the 17B mutant resembled that of the wild type and that the NMR spectrum was consistent with the published structure of R. leguminosarum HMW acidic exopolysaccharide (Amemura et al., 1983; O'Neill et al., 1991). These results suggest that loss of transporter function does not affect the structural composition of the HMW-secreted exopolysaccharide. Visualization of a capsule stain indicated that the transporter mutant still produced a capsule; however, the capsules produced by the mutant strains have a visibly altered structure (Fig. 5).
Fig. 3.
Compositional analysis of exopolysaccharides purified from Rhizobium leguminosarum 3841, wt (a) and 17B, mutant (b). Gas–liquid chromatography analysis of exopolysaccharide indicated a similar composition characteristics for HMW acidic exopolysaccharide. Abbreviations: GlcA, glucuronic acid; Gal, galactose; PvGal, 4,6-pyruvate acetal of galactose; Glc, glucose; Std, inositol used as a standard.
Fig. 4.
A500MHz 1H-NMR spectrometry comparative analysis of Rhizobium leguminosarum 3841, wild type and 17B, mutant exopolysaccharide.
Fig. 5.
Capsule stain of RL2975-77 transporter mutants grown in VMM broth. Cells are shown at ×1000 magnification. (a) wild-type 3841; (b) 17B; (c) 38EV30.
Desiccation, detergent and osmotic sensitivity of the mutants 17B and 38EV30
A desiccation sensitivity assay was used to demonstrate the importance of the ABC transporter for desiccation tolerance. The 17B and 38EV30 mutants were significantly reduced in desiccation tolerance when compared with the wild-type strain (Table 2). The mutants were not significantly sensitive to hyperosmotic stress, indicating that the sensitivity to decreased water availability is specific to desiccation stress. The mutants were as resistant as the wild type to the detergents deoxycholate or sarcosyl, suggesting that defects in outer membrane integrity are not responsible for the decrease in desiccation tolerance. Additionally, sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of lipopolysaccharide isolated from the 17B mutant indicated no obvious difference in the O-antigen structure compared with the wild type (data not shown).
Table 2.
Sensitivity to desiccation, osmotic stress and detergents
| Percent survival* |
||||
|---|---|---|---|---|
| Strains | Desiccation tolerance | Osmotic stress† | VMM+DOC‡ | VMM+SARC‡ |
| 3841 | 42±1.8 | 79.4±1.42 | 125±4.45 | 123±3.42 |
| 17B | 1.9±0.96** | 69.3±4.66 | 94.5±7.94 | 79.5±5.59 |
| 38EV30 | 6.45±2.6** | 79.7±6.07 | 101±20.6 | 117±23.6 |
All data presented are the average (±SD) percent survival of three independent trials.
Strains were grown in TY broth or TY supplemented with 69.5 mM NaCl for 2 days. ODs were measured at 600 nm.
Strains were grown in VMM broth or VMM supplemented with 75 μg mL–1 deoxycholate (DOC) or 50 μg mL–1 sarcosyl (SARC) for 2 days. ODs were measured at 600 nm.
Difference in percent growth between the wild type and the mutant is statistically significant at a P-value < 0.001 (Student's t-test).
Exopolysaccharide production is positively correlated with desiccation tolerance in R. leguminosarum
The importance of exopolysaccharide for desiccation tolerance has been reported in soil-dwelling bacteria such as Pseudomonas sp. (Roberson & Firestone, 1992; van de Mortel & Halverson, 2004); however, the relationship between exopolysaccharide production and desiccation tolerance has never been demonstrated for R. leguminosarum.To demonstrate that exopolysaccharide may play a role in desiccation tolerance in R. leguminosarum, the desiccation sensitivities of wild-type cells grown under conditions that produce low levels of exopolysaccharide (TY medium) and conditions that promote exopolysaccharide excretion (VMM medium) were compared (Table 3). The results confirm that conditions that promote exopolysaccharide secretion also significantly improve desiccation tolerance in R. leguminosarum.
Table 3.
Exopolysaccharide (EPS) production contributes to desiccation tolerance and restores tolerance to the transporter mutants
| VMM |
TY |
|||
|---|---|---|---|---|
| Strain | Amount EPS† produced (μg EPS mg–1 cell dry weight) | Desiccation tolerance‡ (percent survival) | Amount EPS† produced (μg EPS mg–1 cell dry weight) | Desiccation tolerance‡ (percent survival) |
| 3841 | 4.1±1.38 | 122±20.7 | 0.504±0.150* | 28±11.4* |
| 17B | 0.056±0.015** | 15±9.4** | 0.021±0.002** | 1.13±0.184** |
| 38EV30 | 0.115±0.035** | 26.8±7.21** | 0.029±0.007** | 1.20±0.516** |
| 17B+EPS | 1.80±0.593 | 95.9±23.8 | 0.97±0.478 | 35.1±6.41 |
| 38EV30+EPS | 2.75±0.565 | 104±34.0 | 0.74±0.175 | 47.2±4.72 |
Data presented are the average (±SD) of at least three independent trials.
Data presented are the average (±SD) percent survival of three independent trials.
Difference between 3841 (TY) and 3841 (VMM) is statistically significantly at a P-value < 0.002 (Student's t test).
Difference between the mutant and the wild type is statistically significant at a P-value < 0.003.
To refine further the relationship between desiccation sensitivity and exopolysaccharide production in 17B and 38EV30, the strains were grown on TY and VMM and exopolysaccharide was quantified and desiccation tolerance was measured (Table 3). Both mutant strains produce considerably less exopolysaccharide than the wild type and both strains have a significantly increased sensitivity to drying, suggesting that the decrease in desiccation tolerance in these mutants may be related to a decrease in exopolysaccharide. To support this hypothesis, exogenous exopolysaccharide was added to the mutants as described in Materials and methods. The additional exopolysaccharide restored the desiccation tolerance in these mutants to wild-type levels, confirming that the desiccation sensitivity in the 17B and 38EV30 mutants is likely related to a decrease in surface exopolysaccharide.
Biofilm formation
The transporter mutants have a propensity to flocculate heavily when grown in liquid VMM (data not shown). Changes in cell–cell adhesion can alter biofilm formation dynamics (Reisner et al., 2003). The ability of the 17B mutant to form biofilms on polystyrene microplates was investigated in order to determine whether the transporter mutation affected attachment of R. leguminosarum to solid surfaces. Biofilms of both the 17B mutant and R. leguminosarum 3841 were grown in microtiter plates and stained using CV (Table 4). These assays indicated that the surface-adherent biomass produced by the 17B mutant was significantly less than that produced by wild-type 3841. Similar results were obtained for the 38EV30 mutant (data not shown). Staining by CV is nonspecific and stains secreted polysaccharides as well as cellular biomass; therefore, metabolic staining with a tetrazolium salt (MTS) was used to assess the number of bacterial cells that had adhered to the surfaces of microtiter plates. In agreement with the CV assays, the total metabolic activity of the wild-type biofilm was, on average, fourfold higher than the 17B biofilm (Table 4).
Table 4.
Quantification of biofilm formation by the RL2975 (17B) mutant
| Strain | Crystal violet staining* (OD550nm) | Viable cell count* (log10 CFU peg–1) | MTS reduction assay* (OD490nm) |
|---|---|---|---|
| 3841 | 0.157±0.113 | 4.87±0.34 | 1.92±1.03 |
| 17B | 0.036±0.012** | 3.15±0.78** | 0.381±0.099** |
Results presented are the average (±SD) of 24 replicates (48 replicates for the viable cell count).
Difference between the wild type and the mutant strains is statistically significant at a P-value < 0.001 (Student's t-test).
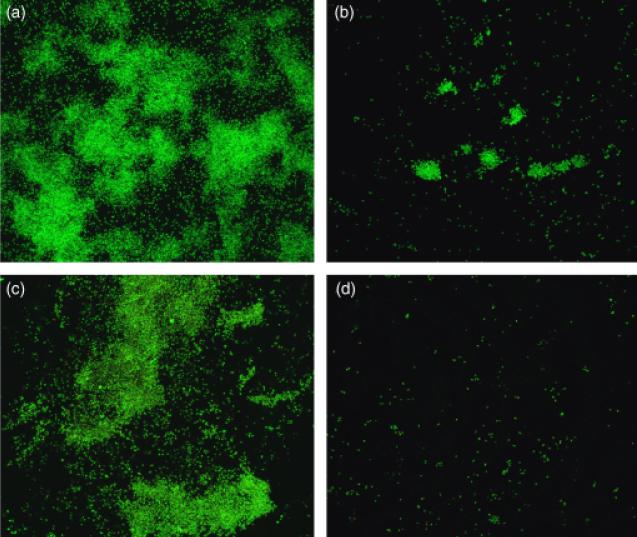
We further investigated the biofilm formation of the 17B mutant by growing biofilms of the mutant and wild-type strains in the CBD. The number of cells in the CBD biofilms was assessed by viable cell counting and CLSM using acridine orange and Live/Dead™ staining. The mean number of viable cells in the 17B biofilm was 52-fold lower in comparison with wild-type 3841 (Table 4). CLSM corroborated that the 17B mutant was impaired in biofilm formation. Both acridine orange and Live/Dead™ staining revealed that the wild-type strain 3841 could form micro-colonies at the air–liquid–surface interface of the CBD pegs (Fig. 6a and c). By contrast, only scattered cells of the 17B mutant had adhered to the pegs (Fig. 6b and d). Collectively, these data suggest that a functional RL2975–RL2977 transporter is essential for normal biofilm formation and subsequent maturation.
Fig. 6.
CLSM of biofilms produced by wild-type Rhizobium leguminosarum bv. viciae 3841 (a and c) and the 17B mutant (b and d). Biofilms of the wild type and mutant strains were stained with acridine orange (a and b) or using the Live/Dead™ cell viability kit (c and d).
Notably, when CBD biofilms were fixed before staining, a small number of very large cell aggregates remained attached to the peg surface (data not shown). These results, in addition to the flocculation observed in VMM broth culture, suggest that the mutants, while impaired in their ability to adhere (or remain attached) to solid substrata, are likely not impaired in their ability to form cell–cell contacts, and in fact may be prone to cell–cell adhesion relative to the wild type.
Attempted complementation of the 17B and 38EV30 mutants
Attempts to restore wild-type phenotypes to the 17B and 38EV30 mutants with a cosmid (pCos879) from an R. leguminosarum VF39SM genomic library (Yost et al., 1998) were unsuccessful. Rhizobium leguminosarum VF39SM is closely related to R. leguminosarum 3841, and DNA sequencing confirmed that the genes homologous to RL2975, RL2976 and RL2977, found on the VF39 cosmid (pCos879), coded for proteins that are 99%, 100% and 99% identical to the 3841 amino acid sequence, respectively. Therefore, DNA sequencing suggests that the genes should be functional in a 3841 background and the lack of complementation was attributed to a dominant-negative effect. Dominant-negative mutants of other ABC transporters have been reported (Bliss et al., 1996; Miyamoto et al., 2002). Mutations in the C-terminus of the ATP-binding protein KpsT of an E. coli ABC-2 transporter resulted in a dominant-negative mutation (Bliss et al., 1996). Because the transposon insertion site in the 17B mutant is located in the C-terminal portion of the ATP-binding component, it is possible that the lack of complementation is caused by a dominant-negative mutation. To circumvent the dominant-negative effect, we attempted to create a mutant where the genes RL2975–RL2977 are deleted; however, attempts have been unsuccessful to date. Although the phenotypes observed for the mutants were not restored by complementation, the two mutants, 17B and 38EV30, were isolated independently, and so it is unlikely that the observed defective phenotypes are due to either a secondary site mutation or an artifact of the transposon insertion.
Discussion
Reduced desiccation tolerance and defective biofilm formation have been reported previously for mutants with alterations to secreted polysaccharides in several bacterial species (Roberson & Firestone, 1992; Ophir & Gutnick, 1994; van de Mortel & Halverson, 2004; Russo et al., 2006; Balestrino et al., 2008). Therefore, we hypothesize that the threefold reduction of exopolysaccharide observed in the transporter mutants is at least partially responsible for the reported phenotypes of these mutants. This has been confirmed for the desiccation phenotype by restoring desiccation tolerance to wild-type levels with the addition of exogenous exopolysaccharide. The importance of exopolysaccharide for desiccation tolerance in soil-dwelling bacteria has been reported in Pseudomonas (Roberson & Firestone, 1992; van de Mortel & Halverson, 2004). Our results suggest that exopolysaccharide is also important for desiccation tolerance in Rhizobium. The ability of exopolysaccharide to absorb large amounts of water, creating a hydrating environment surrounding a cell, has been suggested as a possible mechanism for enhancing desiccation tolerance (Roberson & Firestone, 1992). Desiccation stress affects many cellular components (Billi & Potts, 2002) and adaptation is highly complex, likely requiring numerous genetic pathways. Humman et al. (2009) recently demonstrated that desiccation tolerance in the related S. meliloti requires genes involved in DNA repair and in the regulation of stress-induced pathways. Therefore, the ABC transporter characterized in this study is likely only one component to desiccation tolerance, and other genes involved in desiccation tolerance remain to be identified in R. leguminosarum.
Structural analysis of the secreted exopolysaccharide from the mutants was performed to help elucidate the possible role for the transporter in exopolysaccharide secretion. The structures of the HMW exopolysaccharide were identical between the mutants and the wild type, suggesting that the transporter does not transport a substrate that plays a role in proper HMW exopolysaccharide assembly. Mutation of pssC, a gene involved in exopolysaccharide biosynthesis, results in a 50% reduction in the amount of exopolysaccharide synthesized (van Workum et al., 1997). It is possible that the ABC transporter's substrate may be involved in regulating the expression of pssC, or other exopolysaccharide assembly-related genes. Future studies will attempt to identify the substrate of the RL2975–RL2977 transporter and its linkage to the regulation of exopolysaccharide production. These studies may also help elucidate the functions and substrates of transporters classified within the DUF990 subfamily of ABC-2-type transporters.
The 17B and 38EV30 mutants flocculate extensively in VMM broth. In Klebsiella pneumoniae, exopolysaccharide has been attributed to preventing cell-to-cell aggregation by shielding the intercellular binding of adhesin proteins (Schembri et al., 2004; Balestrino et al., 2008). In R. leguminosarum, cell–cell aggregation in exopolysaccharidedeficient mutants is thought to be due to the cell-to-cell binding of exposed cellulose microfibrils (Napoli et al., 1975; Laus et al., 2005). Furthermore, a mutant strain of R. leguminosarum that overproduces cellulose has been observed to flocculate heavily (Ausmees et al., 1999). The genome of R. leguminosarum 3841 contains genes that putatively code for cellulose synthesis (Young et al., 2006). It is possible that intercellular shielding of the cellulose microfibrils is absent in the 17B and 38EV30 mutants as a result of the decreased level of exopolysaccharide, contributing to increased cell-to-cell aggregation.
Russo et al. (2006) have shown that exopolysaccharide plays an important role in the biofilm formation in R. leguminosarum. Biofilm formation is severely impaired in the 17B and 38EV30 mutants. Future experiments will attempt to determine whether the decreased exopolysaccharide production in 17B and 38EV30 directly contributes to the defective biofilm phenotype. Observations made from CLSM imaging of fixed and nonfixed biofilms suggested that the mutant is specifically deficient in surface adhesion. These results are similar to those found by Balestrino et al. (2008), who reported that exopolysaccharide-deficient mutants of K. pneumoniae were also impaired in the initial attachment phase of biofilm formation. In K. pneumoniae, attachment to the solid substrate occurred at the poles of the cells, and so it is possible that the alterations in the capsule structure at the poles of the 17B and 38EV30 mutants (Fig. 5) are interfering with surface adhesion. Biofilm formation can occur during plant root colonization by rhizobacteria (Fujishige et al., 2006; Danhorn & Fuqua, 2007; Santaella et al., 2008). Given the defective biofilm phenotype of 17B and 38EV30, future experiments will determine the importance of the transporter encoded by RL2975–RL2977 for root colonization of host legume plants.
Acknowledgements
This research has been supported by Natural Sciences and Engineering Research Council (NSERC) grants to C.K.Y. and R.J.T. E.M.V. was supported by a Postgraduate Scholarship from NSERC. J.J.H. was supported by a Canada Graduate Scholarship from NSERC as well as by a PhD studentship from the Alberta Heritage Foundation for Medical Research. CLSM was facilitated by a grant from the Canadian Foundation for Innovation to Dr Howard Ceri at the University of Calgary. Analysis of the exopolysaccharide was supported, in part, by National Institutes of Health Grant GM39583 to R.W.C., and Department of Energy Grant DE-FG02-98ER20307 to the Complex Carbohydrate Research Center.
Footnotes
Present address: Joe J. Harrison, Department of Microbiology, University of Washington School of Medicine, Seattle 98195-7242, WA, USA.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemura A, Harada T, Abe M, Higashi S. Structural studies on the extracellular acidic polysaccharide from Rhizobium trifolii 4S. Carbohyd Res. 1983;115:165–174. [Google Scholar]
- Ausmees N, Jonsson H, Hoglund S, Ljunggren H, Lindberg M. Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii. Microbiology. 1999;145:1253–1262. doi: 10.1099/13500872-145-5-1253. [DOI] [PubMed] [Google Scholar]
- Balestrino D, Ghigo JM, Charbonnel N, Haagensen JAJ, Forestier C. The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ Microbiol. 2008;10:685–701. doi: 10.1111/j.1462-2920.2007.01491.x. [DOI] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:189–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Berridge MV, Tan AS. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Mtt) – subcellular-localization, substrate dependence, and involvement of mitochondrial electron-transport in Mtt reduction. Arch Biochem Biophys. 1993;303:474–482. doi: 10.1006/abbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- Billi D, Potts M. Life and death of dried prokaryotes. Res Microbiol. 2002;153:7–12. doi: 10.1016/s0923-2508(01)01279-7. [DOI] [PubMed] [Google Scholar]
- Bliss J, Garon C, Silver R. Polysialic acid export in Escherichia coli K1: the role of KpsT, the ATP-binding component of an ABC transporter, in chain translocation. Glycobiology. 1996;6:445–452. doi: 10.1093/glycob/6.4.445. [DOI] [PubMed] [Google Scholar]
- Boumahdi M, Mary P, Hornez JP. Changes in fatty acid composition and degree of unsaturation of (brady)rhizobia as a response to phases of growth, reduced water activities and mild desiccation. Antonie van Leeuwenhoek. 2001;79:73–79. doi: 10.1023/a:1010291818304. [DOI] [PubMed] [Google Scholar]
- Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cytryn EJ, Sangurdekar DP, Streeter JG, et al. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J Bacteriol. 2007;189:6751–6762. doi: 10.1128/JB.00533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn T, Fuqua C. Biofilm formation by plant-associated bacteria. Ann Rev Microbiol. 2007;61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- Deaker R, Roughley RJ, Kennedy IR. Legume seed inoculation technology – a review. Soil Biol Biochem. 2004;36:1275–1288. [Google Scholar]
- Frosch M, Edwards U, Bousset K, Krauße B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in Gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- Fujishige N, Kapadia N, De Hoff P, Hirsch A. Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol. 2006;56:195–206. doi: 10.1111/j.1574-6941.2005.00044.x. [DOI] [PubMed] [Google Scholar]
- Garmiri P, Coles KE, Humphrey TJ, Cogan TA. Role of outer membrane lipopolysaccharides in the protection of Salmonella enterica serovar Typhimurium from desiccation damage. FEMS Microbiol Lett. 2008;281:155–159. doi: 10.1111/j.1574-6968.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- Gibson DL, White AP, Snyder SD, et al. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol. 2006;188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert K, Vanderlinde EM, Yost CK. Mutagenesis of the carboxy terminal protease CtpA decreases desiccation tolerance in Rhizobium leguminosarum. FEMS Microbiol Lett. 2007;272:65–72. doi: 10.1111/j.1574-6968.2007.00735.x. [DOI] [PubMed] [Google Scholar]
- Glenn AR, Poole PS, Hudman JF. Succinate uptake by free-living and bacteroid forms of Rhizobium leguminosarum. J Gen Microbiol. 1980;119:267–271. [Google Scholar]
- Harrison JJ, Ceri H, Yerly J, Stremick CA, Hu YP, Martinuzzi R, Turner RJ. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary biofilm device. Biol Proced Online. 2006:194–215. doi: 10.1251/bpo127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JJ, Ceri H, Yerly J, Rabiei M, Hu YP, Martinuzzi R, Turner RJ. Metal ions may suppress or enhance cellular differentiation in Candida albicans and Candida tropicalis biofilms. Appl Environ Microb. 2007;73:4940–4949. doi: 10.1128/AEM.02711-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase – a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Humann JL, Ziemkiewicz HT, Yurgel SN, Kahn ML. Regulatory and DNA repair genes contribute to the desiccation resistance of Sinorhizobium meliloti Rm1021. Appl Environ Microb. 2009;75:446–453. doi: 10.1128/AEM.02207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AWB, Beringer JE. Identification of Rhizobium strains in pea root nodules using genetic markers. J Gen Microbiol. 1975;87:343–350. doi: 10.1099/00221287-87-2-343. [DOI] [PubMed] [Google Scholar]
- Kiss E, Kereszt A, Barta F, Stephens S, Reuhs BL, Kondorosi A, Putnoky P. The rkp-3 gene region of Sinorhizobium meliloti Rm41 contains strain-specific genes that determine K-antigen structure. Mol Plant Microbe In. 2001;14:1395–1403. doi: 10.1094/MPMI.2001.14.12.1395. [DOI] [PubMed] [Google Scholar]
- Kroll JS, Loynds B, Brophy LN, Moxon ER. The bex lovus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- Laurentin A, Edwards C. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal Biochem. 2003;315:143–145. doi: 10.1016/s0003-2697(02)00704-2. [DOI] [PubMed] [Google Scholar]
- Laus M, van Brussel A, Kijne J. Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Mol Plant Microbe In. 2005;18:533–538. doi: 10.1094/MPMI-18-0533. [DOI] [PubMed] [Google Scholar]
- Liu YG, Huang N. Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol Biol Rep. 1998;16:175–181. [Google Scholar]
- Miyamoto A, Matsuyama S, Tokuda H. Dominant negative mutant of a lipoprotein-specific molecular chaperone, LolA, tightly associates with LolCDE. FEBS Lett. 2002;528:193–196. doi: 10.1016/s0014-5793(02)03305-7. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Dazzo F, Hubbell D. Production of cellulose microfibrils in Rhizobium. Appl Microbiol. 1975;30:123–131. doi: 10.1128/am.30.1.123-131.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwai YB, Adachi Y, Ogawa Y, Hara H. Characterization of biofilm-forming abilities of antibiotic-resistant Salmonella typhimurium DT104 on hydrophobic abiotic surfaces. J Microbiol Immunol Infect. 2006;39:278–291. [PubMed] [Google Scholar]
- O'Neill MA, Darvill AG, Albersheim P. The degree of esterification and points of substitution by O-acetyl and O-(3-hydroxybutanoyl) groups in the acidic extracellular polysaccharides secreted by Rhizobium leguminosarum biovars viciae, trifolii, and phaseoli are not related to host range. J Biol Chem. 1991;266:9549–9555. [PubMed] [Google Scholar]
- Ophir T, Gutnick D. A role for expolysaccharide in the protection of microorganisms from desiccation. Appl Environ Microb. 1994;60:740–745. doi: 10.1128/aem.60.2.740-745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Pavelka MS, Jr, Wright LF, Silver RP. Identification of the two genes, kpsM and kpsT, in region 3 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1991;173:4603–4610. doi: 10.1128/jb.173.15.4603-4610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière G, Gouy M. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- Reisner A, Haagensen JAJ, Schembri MA, et al. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol. 2003;48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- Reizer J, Reizer A, Saier JR. A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci. 1992;1:1326–1332. doi: 10.1002/pro.5560011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson EB, Firestone MK. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl Environ Microb. 1992;58:1284–1291. doi: 10.1128/aem.58.4.1284-1291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokitko PV, Romanovskaya VA, Malashenko YR, et al. Soil drying as a model for the action of stress factors on natural bacterial populations. Microbiology. 2003;72:756–761. [PubMed] [Google Scholar]
- Russo DM, Williams A, Edwards A, Posadas DM, Finnie C, Dankert M, Downie JA, Zorreguieta A. Proteins exported via the PrsD–PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J Bacteriol. 2006;188:4474–4486. doi: 10.1128/JB.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Santaella C, Schue M, Berge O, Heulin T, Achouak W. The exopolysaccharide of Rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environ Microbiol. 2008;10:2150–2163. doi: 10.1111/j.1462-2920.2008.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri MA, Dalsgaard D, Klemm P. Capsule shields the function of short bacterial adhesins. J Bacteriol. 2004;186:1249–1257. doi: 10.1128/JB.186.5.1249-1257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering – transposon mutagenesis in Gram negative bacteria. Bio-Technology. 1983;1:784–791. [Google Scholar]
- Skorupska A, Janczarek M, Marczak M, Mazur A. Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb Cell Fact. 2006;5:7–26. doi: 10.1186/1475-2859-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AN, Boulmois GJ, Roberts IS. Molecular analysis of the Escherichia coli K5 kps locus: identification and characterization of an inner-membrane capsular polysaccharide transport system. Mol Microbiol. 1990;4:1863–1869. doi: 10.1111/j.1365-2958.1990.tb02035.x. [DOI] [PubMed] [Google Scholar]
- Tang X, Lu BF, Pan SQ. A bifunctional transposon mini-Tn5gfp-km which can be used to select for promoter fusions and report gene expression levels in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1999;179:37–42. doi: 10.1111/j.1574-6968.1999.tb08704.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W – improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel M, Halverson LJ. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol Microbiol. 2004;52:735–750. doi: 10.1111/j.1365-2958.2004.04008.x. [DOI] [PubMed] [Google Scholar]
- Vanderlinde EM, Muszynski A, Harrison JJ, et al. Rhizobium leguminosarum biovar viciae 3841, deficient in 27-hydroxyoctacosanoate-modified lipopolysaccharide is impaired in desiccation tolerance, biofilm formation, and motility. Microbiology. 2009;155:3055–3069. doi: 10.1099/mic.0.025031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Workum WAT, Canter Cremers HCJ, Wijfjes AHM, van der Kolk C, Wijffelman CA, Kijne JW. Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol Plant Microbe In. 1997;10:290–301. doi: 10.1094/MPMI.1997.10.2.290. [DOI] [PubMed] [Google Scholar]
- Vincent JM. A Manual for the Practical Study of Root-nodule Bacteria (IBP Handbook no. 15) Blackwell Scientific; Oxford: 1970. [Google Scholar]
- White AP, Gibson DL, Kim W, Kay WW, Surette MG. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J Bacteriol. 2006;188:3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. Isolation and characterization of plant cell wall components. Method Enzymol. 1985;118:3–40. [Google Scholar]
- Yost CK, Rochepeau P, Hynes MF. Rhizobium leguminosarum contains a group of genes that appear to code for methyl-accepting chemotaxis proteins. Microbiology. 1998;144:1945–1956. doi: 10.1099/00221287-144-7-1945. [DOI] [PubMed] [Google Scholar]
- Young JPW, Crossman LC, Johnston AWB, et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006;7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zevenhuizen LPTM. Gel-forming capsular polysaccharide of fast-growing rhizobia: occurrence and rheological properties. Appl Microbiol Biot. 1984;20:393–399. [Google Scholar]