Abstract
Promoter hypermethylation mediated by DNA methyltransferases (DNMTs) is the main reason for epigenetic inactivation of tumor suppressor genes (TSGs). Previous studies showed that DNMT1 and DNMT3B play an important role in CpG island methylation in tumorigenesis. Little is known about the role of DNMT3A in this process, especially in hepatocellular carcinoma (HCC). In the present study, increased DNMT3A expression in 3 out of 6 HCC cell lines and 16/25 (64%) HCC tissues implied that DNMT3A is involved in hepatocellular carcinogenesis. Depletion of DNMT3A in HCC cell line SMMC-7721 inhibited cell proliferation and decreased the colony formation (about 65%). Microarray data revealed that 153 genes were upregulated in DNMT3A knockdown cells and that almost 71% (109/153) of them contain CpG islands in their 5′ region. 13 of them including PTEN, a crucial tumor suppressor gene in HCC, are genes involved in cell cycle and cell proliferation. Demethylation of PTEN promoter was observed in DNMT3A-depleted cells implying that DNMT3A silenced PTEN via DNA methylation. These results provide insights into the mechanisms of DNMT3A to regulate TSGs by an epigenetic approach in HCC.
1. Introduction
Aberrant methylation of tumor suppressor genes (TSGs), which result in their silencing, is a common phenomenon in tumorigenesis [1, 2]. DNA methyltransferases (DNMTs) DNMT1, DNMT3A, and DNMT3B have been identified as functional DNA methylation enzymes in eukaryotic cells [3, 4]. DNMT3A and DNMT3B function as de novo methyltransferases that are responsible for de novo methylation, whereas DNMT1 functions as a maintenance enzyme for retaining methylation patterns. CpG islands residing within the promoter or the 5′ end of genes are normally unmethylated in normal cells, and abnormal methylation of these CpG islands can efficiently repress transcription of the associated gene [5]. Silencing of TSGs by epigenetic alterations at promoter CpG islands is a common event found in many types of human cancers, including hepatocellular carcinoma (HCC). Overexpression of DNMTs has been reported in numerous studies on various malignancies including prostate, colorectal, and breast tumors. These findings suggest that DNMTs may contribute to tumorigenesis through epigenetic silencing of genes [6–11]. The targeting of individual DNMTs to a gene is believed to play a critical role in determining its state of methylation. RNA interference-mediated knockdown or specific inhibition by antisense oligo-nucleotides of DNMT1 resulted in the reduction of promoter methylation in TSGs, such as RASSF1A, p16INK4a, and CDH1 [12, 13]. Satellite DNA is the target of methylation by DNMT3B. Interestingly, DNMT3B is also responsible for immunodeficiency, centromere instability, and facial anomalies (ICF) syndrome [14, 15]. Moreover, recent studies have shown that DNMT3B expression contributes to a CpG island methylator phenotype and that it epigenetically silences CXCL12 [16], RECK [17], and RASSF1A [18] through DNMT3B-mediated promoter methylation.
The function of DNMT3A is obscure and has not been associated with disease. DNMT3A is ubiquitously overexpressed in various types of tumors [19–22] implying that it may be involved in and play an important role in tumorigenesis. However, very little is known about DNMT3A's ability to regulate gene expression, especially in regulating tumor suppressor genes. Recent results showed that DNMT3A expression mediates the repression of SFRP5 [23] and plakoglobin transcription [24] by direct hypermethylation of their promoter, indicating that DNMT3A's methylation activities are gene specific. However, the target genes of DNMT3A remain unknown. In this study, we attempted to identify tumor-related genes that are potentially regulated by DNMT3A and further discuss the prospect of gene expression regulation by DNMT3A. In this report, we also investigated the role of DNMT3A in the aberrant methylation and inactivation of genes in human tumor cells as well as its role in the maintenance of the transformed phenotype.
2. Materials and Methods
2.1. HCC Cell Lines
2.1.1. HCC Cell Lines and Tissue Samples from Patients
HCC-derived cell lines (QGY-7701, Bel-7402, Bel-7404, Bel-7405, SMMC-7721) para-carcinoma cell line QSG-7701, and immortalized human normal hepatocyte cell line HL-7702 were obtained from TCC Cell Bank (Shanghai, China) and cultured with standard medium: RPMI 1640 (GIBCO-BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (GIBCO-BRL) in a 5% CO2 humidified chamber at 37°C. Tumor tissues and adjacent noncancerous tissue from 25 HCCs were obtained from the First Affiliated Hospital, Nanjing Medical University, from May 2008 to December 2009. All patients were confirmed histopathologically and samples were obtained with informed consent from patients.
2.2. DNMT3A RNA Interference Constructs and Transfection
Three candidate siRNAs against human DNMT3A were designed for targeting different coding regions of the DNMT3A isoforms (Genbank No. NM_175629.1, NM_153759.2, NM_022552.3). Synthesized 64nt siRNA template oligonucleotides were constructed into BglII and HindIII (TAKARA) sites of pSUPER-EGFP (gift from Professor Dianqing Wu at UCHC, USA), which has a green fluorescent protein marker and resistance to the G418, and named as pMT3A. The scramble sequences of DNMT3A siRNA were used as control and inserted into pSUPER-EGFP to form sMT3A. SMMC-7721 cells were transfected with recombinant plasmids using Lipofectamine 2000 according to the manufacturer's protocols (Invitrogen Corp., Carlsbad, CA) and were selected with medium containing 200 mg/mL G418 (Gibco, Grand Island, NY) for 2 months after the initial transfection. The cells stably harboring the targeting vector were monitored with green fluorescent protein (GFP) expression. SMMC-7721 cells were transfected with pMT3A and labeled as 7721–pMT3A cell lines; these were transfected with sMT3A and were designated as 7721–sMT3A cell lines or control.
2.3. Western Blot Analysis
Cells (5 × 106) were harvested and the total proteins were prepared. Cell precipitate was resuspedded in 200 ul ice-cold lysis buffer (10 mmol/L Tris-Hcl (pH 8.0), containing 150 mmol/L NaCl, 1 mmol/L phenylmethylsulfonyl fluoride, and 1% Triton X-100). An equal amount of cellular proteins was loaded on 7% polyacrylamide gels (PAGE) in a 1×SDS-PAGE buffer (1 g/L SDS, 3 g/L Tris-base, and 14.4 g/L glycine). Proteins were transferred to a polyvinylpyrrolidine difluoride membrane electrophoretically and incubated overnight at 4 in block buffer (5% nonfat milk in Tris-Base Tween-20 (TBST). Membranes were incubated for 1 hour at room temperature with DNMT3A-specific monoclonal antibodies (1 : 5000, Imagenex) followed by six five-minute washes with TBST. After washing, the membrane was incubated with antimouse horseradish peroxidase-conjugated secondary antibodies (1 : 80000, Sigma), and the immune complexes were detected with enhanced chemiluminescence (Pierce). The membranes were probed with an antiactin antibody (Biomedical Technologies, Stoughton, MA) to confirm equal loading of each protein samples. β-actin protein levels were used as a control to ensure equal protein loading (β-actin, 1 : 8000, Sigma). Each experiment was repeated at least 3 times. The intensity ratios of the 2 bands from the same sample were determined and calculated by Gel-Pro Analyzer 3.0.
2.4. Microarray Hybridizations and Induced Genes Analysis
Affymetrix Human Genome U133A 2.0 Array (Affymetrix, Santa Clara, CA), encompassing 18,400 transcripts and variants, including 14,500 well-characterized human genes on one array was processed. Microarray hybridization was performed in the SBC (Shanghai, China) using standard affymetrix procedures. Raw microarray data were acquired using GCOS1.2 software of affymetrix. The data were preprocessed using robust multiarray analysis (RMA) with a log base 2 (log2) transformation. Genes identified as differentially expressed genes, which is exhibiting, upregulated more than 2-fold to the control. Gene ontology of the differentially expressed genes was performed through the GO Mining Tool (http://www.affymetrix.com/analysis/query/go_analysis.affx). The potential promoter regions spanning 2000 bp upstream and 5'UTR around the transcription start site of 153 upregulated genes. Subsequently, CpG islands were identified within the core promoter region by CpG Island Searcher program (http://www.methdb.de/) in DNA Methylation Database (http://www.uscnorris.com/cpgisland2).
2.5. Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Real-Time Quantitative RT-PCR (qPCR) Analysis
Total RNA was isolated using TRIzol reagent (Invitrogen, NY). cDNA was synthesized from 1 μg of total RNA using Oligo(dT)18 as primers and reverse transcriptase following reverse-transcribed with Reverse Transcription System (Promega,) according to the kit's instructions. The expression levels of differentially expressed genes of microarray after DNMT3A RNAi were quantified using qRT-PCR analysis. PCR reactions containing 10 ng cDNA, SYBR green (1 : 20,000 dilutions) were included in each reaction to allow quantification of RNA levels using the ABI 7300 detection system (Applied Biosystems). To normalize the input load of cDNA among samples, either β-actin was quantified and used as an endogenous standard. The relative levels of expression of each target genes among different samples were calculated accordingly (ABI PRISM 7300 Detection System, USA). The conditions and primers designed for detected genes were listed in Table 1. Each duplex PCR was repeated with at least 3 different cDNA preparations and 2 independent qPCR reactions for each cDNA.
Table 1.
Primers sequences and related amplicons for quantitative real-time RT-PCR or RT-PCR.
| Genes | Primers (5′–3′) | Anneal temperature (°C) | Amplicon (bp) |
|---|---|---|---|
| β-actin | Forward: AAAGACCTGTACGCCAACAC | 61 | 220 |
| Reverse: GTCATACTCCTGCTTGCTGAT | |||
| DNMT3A | Forward: TATTGATGAGCGCACAAGAGAGC | 65 | 111 |
| Reverse: GGGTGTTCCAGGGTAACATTGAG | |||
| DNMT3B | Forward: GACTTGGTGATTGGCGGAA | 64 | 271 |
| Reverse: GGCCCTGTGAGCAGCAGA | |||
| DNMT1 | Forward: CCGAGTTGGTGATGGTGTGTAC | 61 | 325 |
| Reverse: AGGTTGATGTCTGCGTGGTAGC | |||
| PTEN-q | Forward: CCGTTACCTGTGTGTGGTGATATC | 60 | 100 |
| Reverse: AATGTATTTACCCAAAAGTGAAACATT |
2.6. Methylation-Specific PCR (MSP), Bisulfite Sequencing, and Treatment of 5-aza-dC
Genomic DNA was extracted according to standard method [25]. Bisulfite treatment was performed by the Methylamp DNA Modification Kit (Epigentek) following the manufacturer's instruction. Modified DNA was amplified to determine the methylation status of the promoter region of gene by MSP. The primer sequences and PCR conditions for amplification above were shown in Table 2. PCR products for the bisulfite sequencing were gel-purified, subcloned into a pEASY-T3 vector system (TransGen Biotech). At least ten colonies were sequenced to assess the degree of methylation in each CpG site. For demethylation analysis, cells were plated and treated with 100 μmol/L 5-aza-dC (Sigma) for up to 4 days. The total RNA was extracted from exponentially growing cultures on the 3rd day after treatment.
Table 2.
Primers sequences for MSP and bisulfite sequencing analysis.
| Gene | Primers (5′–3′) | Anneal temperature (°C) |
|---|---|---|
| PTEN-Ma | Forward: TTTTTTTTCGGTTTTTCGAGGC | 59 |
| Reverse: CAATCGCGTCCCAACGCCG | ||
| PTEN-UMb | Forward: TTTTGAGGTGTTTGGGTTTTTGGT | 59 |
| Reverse: ACACAATCACATCCCAACACCA | ||
| PTEN-BGSc | Forward: ATTGGGTATGTTTAGTAGAGTT | 60 |
| Reverse: CAACTCTCAAACTTCCATCATA |
a: Methylated primer sequences used for MSP. b: Unmethylated primer sequences used for MSP. c: Primer sequences used for bisulfite genomic sequencing.
2.7. Cell Proliferation and Colony Formation Assay
Cell proliferation of transfected stable SMMC-7721 cells with construct pMT3A and its control sMT3A were measured by Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan). The absorbance at 450 nm was measured to represent the cell viability, which was measured to assess the cell proliferation of those cell lines. All experiments were independently repeated at least three times.
1 × 103 cells were suspended in 1.5 ml of 0.3%-melted agar in RPMI 1640 medium (Invitrogen, Carlsbad, CA. USA) with 5% FCS, and plated in a standard 6-well cell culture plate containing a solidified layer of 3 ml 0.6% agarose gel in the same medium. After 14 days of incubation at 37°C in 5% CO2, colony growth in soft agar was assayed by visually counting the colonies. All evaluations were performed in a blinded manner. The experiments were independently performed three times.
2.8. Statistical Analysis
Data were presented as means ± standard error (SE). Data were analyzed using Graphpad Prism 5 software (GraphPad Software, San Diego, CA). Mean difference was determined by Student t-tests. Statistical significant differences were defined as P < .05. Standard statistical tests were performed using SPSS (version 13.0).
3. Results
3.1. Increased Expression of DNMT3A in HCC Cell Lines and HCC Tissues
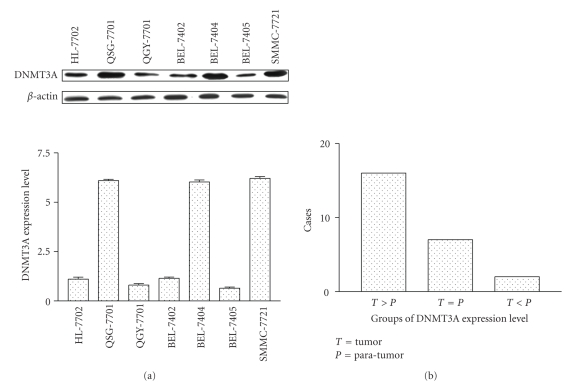
The expression level of DNMT3A was measured in HCC cell lines by western blot and was normalized to β-actin. There was lower expressed DNMT3A in human normal liver cells (HL-7702) and differentially expressed in HCC cell lines (Figure 1(a)). Increased expression of DNMT3A was found in 3 out of 6 HCC cell lines, especially in SMMC-7721 and BEL-7402. Specifically, there was a 6.1- and 5.8-fold increase in DNMT3A expression in SMMC-7721 and BEL-7402, compared to HL-7702, respectively. The mRNA expression level of the DNMT3A was detected by qPCR in tumor tissues and noncancerous liver tissues from 25 HCC patients. Increased expression of DNMT3A was detected in 16 out of 25 cases (64%) of HCCs (Figure 1(b)). These data suggested that DNMT3A may play a role in the progression of HCC tumorigenesis, and this finding needs to be confirmed by a larger study.
Figure 1.
DNMT3A expression is analyzed in HCC cell lines, HCC tumor tissue, and adjacent nontumor samples. (a) DNMT3A protein expression level was detected in HCC cell lines by western blot. Upper panel, DNMT3A was examined by western blot in HCC cell lines. Bottom panel, the intensity ratios of both target gene band and control from the same sample were determined and calculated by Gel-Pro Analyzer 3.0. DNMT3A was increased more in 3 HCC cell lines than in immortalized normal liver cell line HL-7702. (b) Real-time quantitative RT-PCR analysis of DNMT3A expression in 25 pairs of HCC specimens and adjacent noncancerous liver tissues, normalized to β-actin.
3.2. DNMT3A siRNA Construct Suppresses DNMT3A Specifically and Stably in HCC Cell Line
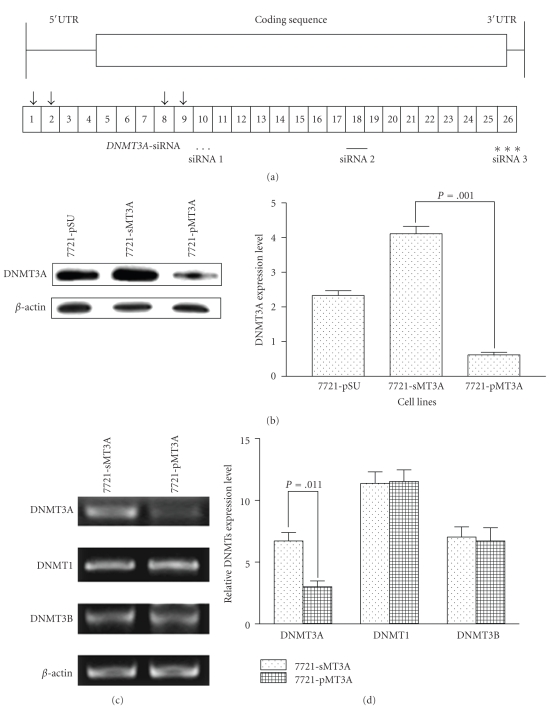
Knockdown of transcripts using siRNA is a powerful tool for studying gene function. We designed three target siRNAs (Figure 2(a)), which can suppress all of the different DNMT3A transcripts, and one nonspecific scramble siRNA, which was used as control (see Table 1 in Supplementary Material available online at doi:10.1155/2010/737535).The candidate siRNA-2 effectively suppressed DNMT3A expression in levels of protein (P = .001)and encoding mRNA (P = .011) (Figures 2(b), 2(c), 2(d)). No significant inhibition on DNMT1 and DNMT3B was detected in DNMT3A RNAi treatment cells (Figure 2(d)). Expression of endogenousβ-actin was used as a control to normalize the expression level of DNMT1, DNMT3A, and DNMT3B.
Figure 2.
DNMT3A gene expression is inhibited by siRNA of DNMT3A in SMMC-7721 cells. (a) Schematic representation of DNMT3A mRNA structure and siRNA positions is showed. (b) Western blot analysis efficiency of DNMT3A siRNA, β-actin was used as reference control. Lanes 1–3 indicated the cell line transfected with plasmid pSUPER-EGFP, sMT3A, and pMT3A, respectively. Right: the intensity ratios of both target gene band and control from the same sample were determined and calculated by Gel-Pro Analyzer 3.0. (c) RT-PCR evaluated the affect of DNMT3A-specific siRNA on other DNMTs expression. (d) Real-time quantitative RT-PCR confirmed the efficiency of DNMT3A-specific siRNA and the affect on other DNMTs mRNA expression.
3.3. Depletion of DNMT3A Inhibits Cell Proliferation and Colony Formation in SMMC-7721 Cells
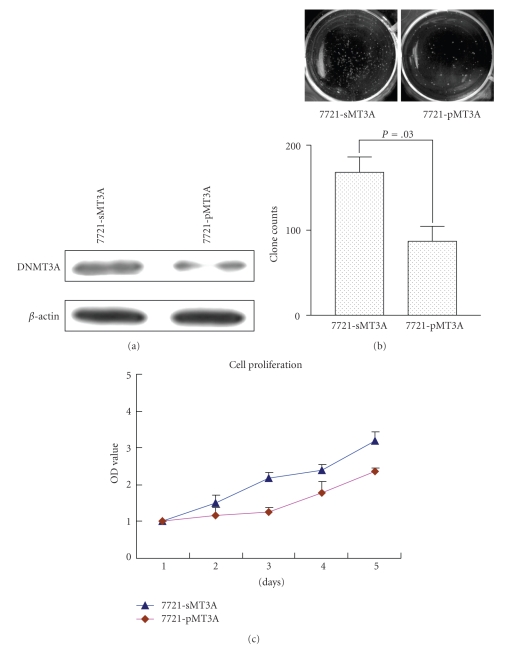
To explore the functional consequences of DNMT3A depletion in HCC, we evaluated the role of DNMT3A-specific siRNA treatment on cell proliferation and colony formation in HCC cells. Transfected clones were picked up and expanded for an additional 2 months and analyzed for DNMT3A in protein levels. DNMT3A expression was stably suppressed in SMMC-7721 (Figure 3(a)). Compared to control 7721-sMT3A cells, 7721-pMT3A formed significantly fewer colonies (about 65%) in soft agar assay (Figure 3(b)) and with lower cell proliferation ability in CCK-8 cell assay (Figure 3(c)).
Figure 3.
Depletion of DNMT3A suppressed HCC cell growth and colony formation ability. (a) Expression of DNMT3A was stably suppressed in SMMC-7721. 7721-sMT3A, which is transfected with scramble vector sMT3A, 7721-pMT3A, which is transfected with candidate DNMT3A siRNA constructs. (b) Colony formation of SMMC-7721 cells was inhibited when transfected with DNMT3A siRNA. The scramble vector served as a control. The number of colonies was counted from three independent experiments. The histogram shows the colony formation efficiency, where the numbers represent the average value of three independent experiments.(c) Cell growth curve of SMMC-7721 cells treated with the DNMT3A siRNA. The cells transfected with the scramble vector served as control. The experiments were repeated at least three times. The result represents the average value of triplicate wells +/- the standard deviation.
3.4. Gene Expression Profiling Induced by DNMT3A Knockdown
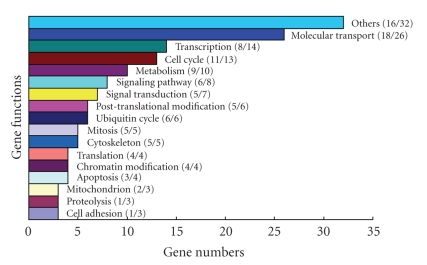
To elucidate the effects of increased DNMT3A expression in HCC tumorigenesis, Affymetrix HG U133A oligonucleotide microarray was used to evaluate induced gene expression profiling in DNMT3A depletion cells compared with control. The data revealed that 153 genes were upregulated in DNMT3A knockdown cell (Supplementary Table 2).These genes were categorized to numerous biological processes by using the GO Mining Tool [26]. All genes were combined into 17 superordinate categories (Figure 4). Three categories contained the largest number of affected genes: molecular transport (26 genes), transcription regulation (14 genes), and cell cycle (13 genes). Since a CpG island (CGI) near the transcription initiation site is usually important for gene repression by DNA methylation, we searched for the presence of CGIs in each gene by using CpG Island Searcher in the DNA Methylation Database, referring to the criteria established by Takai and Jones [27]. We found that almost 71% (109/153) of genes contain CpG islands in their 5′ region. This indicated that these genes might be regulated by DNA methylation. In an effort to provide further experimental evidence that the gene expression changes observed on arrays were valid, quantitative real-time PCR was used to analyze several cancer-related genes and confirmed the data from the array.
Figure 4.
Superordinate categories of biological processes identified by gene ontology (GO) mapping in depletion DNMT3A HCC cell lines. All genes induced by inhibiting DNMT3A were combined into 17 superordinate categories. Three categories contained the largest number of affected genes: molecular transport, transcription regulation, and cell cycle.
3.5. Depletion of DNMT3A Induced PTEN via Demethylation of CpG Sites in Its Promoter Region
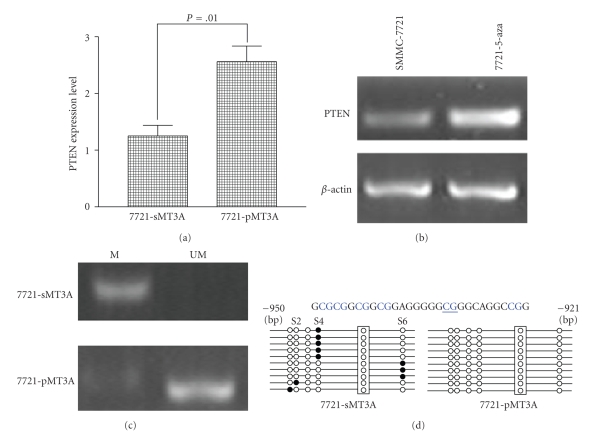
In DNMT3A depletion cells, tumor suppressor gene PTEN was activated (Figure 5(a)). As 5-Aza-2′deoxycytidine, a demethylation agent could restore PTEN expression (Figure 5(b)); we supposed that activated PTEN in DNMT3A depletion cells is maybe involved in demethylation. To explore whether the repression of PTEN expression is due to DNA methylation; we examined the status of methylation of its promoter in DNMT3A depletion cells 7721-pMT3A and control by MSP method. The result showed that restoring of the PTEN was significantly associated with its promoter demethylation in the 7721-pMT3A cell line (Figure (5(c))). Subsequently, a 331 bp (−1128/−798) fragment that contained part of the PTEN promoter and its 5′ untranslated region (5′ UTR) (Figure 5(d)) is chosen for further methyaltion analysis by bisulfite sequencing. Ten individual colonies were sequenced to identify methylated cytosine residues. We found that all six CpG sites were unmethylated in 7721-pMT3A. In contrast, different CpG sites (S1, S2, S4, or S6) were methylated in the 7721-sMT3A cell line. The results indicated that DNMT3A knockdown changed the methylation status of certain CpG sites within the 5′ UTR and promoter of the PTEN gene. These data suggested that PTEN expression is regulated in a methylation-dependent manner and that PTEN may be preferentially methylated by DNMT3A.
Figure 5.
Restoring of PTEN expression by depletion of DNMT3A in HCC cells is partly due to methylation of its promoter. (a) Quantitative-PCR analysis of PTEN expression level in DNMT3A-depleted cell line. (b) 5-Aza-2′deoxycytidine restored endogenous PTEN mRNA expression in SMMC-7721 cells. (c) Methylated status evaluation of PTEN promoter region determined by MSP. The presence of visible PCR product in lanes marked “U” indicates the unmethylated gene, whereas the presence of product in lanes marked “M” indicates the methylated gene. (d) Bisulfite sequencing of the selected region of the PTEN promoter in DNMT3A-depleted SMMC-7721 cells. Top, sequences within the fragment include 6 CpG sites (S1, S2, S3, S4, S5, S6). Bottom, the methylation pattern of 6 CpG sites in PTEN promoter region is shown. Ten random clones from DNMT3A-depleted SMMC-7721cell line and control were sequenced to examine the methylated status. Open and closed circles indicate unmethylated and methylated sites, respectively.
4. Discussion
HCC is a severe disease with a very poor prognosis and is the fourth largest cause of cancer-related deaths worldwide [26, 28]. DNA hypermethylation of tumor suppressor genes is frequently observed in HCCs [25, 29–31]. The process of aberrant DNA methylation is catalyzed by DNA methyltransferases (DNMTs). Moreover, DNMTs expression is increased during early tumorigenesis. In hepatocarcinogenesis, increased expression of DNMTs mRNA is correlated with a progressive increase in the number of methylated genes from normal liver, chronic hepatitis/cirrhosis to HCC [32]. One hypothesis is that abnormal expression of the three functional DNMTs may be important in regulating tumor suppressor gene expression during hepatocellular tumorigenesis. DNMT1 and DNMT3B were both shown to be important for cancer cell survival and tumorigenesis [33]. However, the relationship between DNMT3A and tumorigenesis is still uncertain. Previous studies suggested that DNMT3A is developmentally regulated and was shown to be important in establishing maternal imprints in mice [34, 35]. Recently, researchers found that elevated DNMT3A activity promotes polyposis in Apc (Min) mice [23] and that knockdown of DNMT3A dramatically inhibits melanoma growth and metastasis in mouse melanoma models [36]. Several studies showed that increased expression of DNMT3A in HCC implied that DNMT3A was involved in hepatocarcinogenesis. In order to evaluate the effect of DNMT3A depletion, we examined the characteristics of SMMC-7721 cells in vitro, using proliferation and colony-forming. The results showed that cell proliferation and colony-forming ability did differ between the WT and DNMT3A-depleted SMMC-7721 cells. DNMT3A depletion produced primarily antiproliferative effects. These results were observed in another HCC cell line QSG-7701 (Supplementary). Microarray analysis revealed that inhibition of DNMT3A induced expression of various genes that are involved in cell cycle regulation, transcription regulation, and signal transduction and are thus implicated in the inhibition of HCC cell growth. Whether depletion of DNMT3A is capable of inhibiting the growth of hepatocellular carcinoma in vivo, mice malignant hepatocellular carcinoma models should be established in the further study.
Our previous studies showed that depleting DNMT1 and DNMT3B induced expression of different sets of TSGs in HCC cell lines [37, 38]. Here, DNMT3A depletion significantly altered the expression of tumor-related genes on the array. However, the regulation of these genes by DNMT3A-mediated methylation or other epigenetic mechanisms is unknown. In addition to analyzing the influence of DNMT3A depletion on the broad changes in gene expression by cDNA arrays, we sought to elucidate the potential mechanism by which DNMT3A may regulate TSGs. To this end, we began an analysis of CGI sites contained within the promoter regions of altered genes by using CpG Island Searcher. Almost 71% (109/153) of induced genes contain CpG islands in their 5′ region, suggesting that DNA methylation plays an important role in the regulation of gene expression by DNMT3A. Compared with control, phosphatase and tensin homolog (PTEN), a tumor suppressor gene in HCC, is significantly induced by DNMT3A knockdown. PTEN is silenced by DNA methylation at high frequencies in HCC, and promoter methylation and silencing of PTEN is reported to be associated with HCC [39, 40]. One of the mechanisms of PTEN inactivation is epigenetic silencing, which has been reported in various tumors including hematologic malignancies and some solid tumors [41–44]. In this study we found that PTEN can be restored due to demethylation of its promoter by demethylating agents in SMMC-7721 cells. PTEN is known for its essential role in regulating the PI3K/AKT signaling pathway [45]. PTEN loss leads to activation of AKT kinases, which contributes to cell survival, growth, proliferation, and invasion [46, 47]. The methylation of the PTEN promoter and its associated loss in activity is observed in HCC. These results demonstrate that depletion of DNMT3A affects cell proliferation and that this effect may be a result of the reexpression of the formerly silenced PTEN. In the present study, we found that DNMT3A may play a role in hepatocellular carcinogenesis by its role in regulating the expression of some tumor suppressor genes such as PTEN. However, it is necessary to further study additional DNMT3A mechanisms of regulation of TSGs and its function in tumorigenesis.
5. Conclusion
Our results provide evidence that DNMT3A depletion significantly inhibited cell growth and colony formation in HCC cells. The most likely mechanism for this inhibition of tumorigenesis includes the reactivation of epigenetically silenced tumor suppressor genes, such as the PTEN gene.
Supplementary Material
Supplementary materials include two tables, one is about candidate DNMT3A siRNA sequences (19nt) for knocking down DNMT3A in HCC cell lines, the other is about 153 genes up-regulated in DNMT3A knockdown cell.
Supplementary materials also provided the depletion of DNMT3A in QSG-7701 cells to perform the same experiments, including RNA interference, cell proliferation, colony formation and PTEN expression.
Acknowledgments
This work was supported by The National Natural Science Foundation of China, no. 30470950 and no. 30971605. The authors are grateful to Professor Dianqing WU in UConn Health Center USA for providing siRNA expression vector. They also thank Dr. Sen Lu, and Dr. Chuanjun Wen for their assistance in collecting samples.
References
- 1.Jones PA. DNA methylation and cancer. Oncogene. 2002;21(35):5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- 2.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 3.Bestor TH. The DNA methyltransferases of mammals. Human Molecular Genetics. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genetics. 1998;19(3):219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 5.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in Genetics. 2000;16(4):168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 6.Oh BK, Kim H, Park HJ, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. International Journal of Molecular Medicine. 2007;20(1):65–73. [PubMed] [Google Scholar]
- 7.Park H-J, Yu E, Shim Y-H. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Letters. 2006;233(2):271–278. doi: 10.1016/j.canlet.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 8.Choi MS, Shim Y-H, Hwa JY, et al. Expression of DNA methyltransferases in multistep hepatocarcinogenesis. Human Pathology. 2003;34(1):11–17. doi: 10.1053/hupa.2003.5. [DOI] [PubMed] [Google Scholar]
- 9.Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clinical Cancer Research. 2003;9(12):4415–4422. [PubMed] [Google Scholar]
- 10.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Molecular Carcinogenesis. 2002;33(3):163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 11.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Research. 1999;59(10):2302–2306. [PubMed] [Google Scholar]
- 12.Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene re-expression in human lung and breast cancer cells . Cancer Research. 2004;64(9):3137–3143. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- 13.Robert M-F, Morin S, Beaulieu N, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nature Genetics. 2003;33(1):61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 14.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 15.Xu G-L, Bestor TH, Bourc’his D, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402(6758):187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 16.Sowińska A, Jagodzinski PP. RNA interference-mediated knockdown of DNMT1 and DNMT3B induces CXCL12 expression in MCF-7 breast cancer and AsPC1 pancreatic carcinoma cell lines. Cancer Letters. 2007;255(1):153–159. doi: 10.1016/j.canlet.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Chang H-C, Cho C-Y, Hung W-C. Silencing of the metastasis suppressor RECK by RAS oncogene is mediated by DNA methyltransferase 3b-induced promoter methylation. Cancer Research. 2006;66(17):8413–8420. doi: 10.1158/0008-5472.CAN-06-0685. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Bhutani M, Pathak AK, et al. ΔDNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Research. 2007;67(22):10647–10652. doi: 10.1158/0008-5472.CAN-07-1337. [DOI] [PubMed] [Google Scholar]
- 19.Kautiainen TL, Jones PA. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. The Journal of Biological Chemistry. 1986;261(4):1594–1598. [PubMed] [Google Scholar]
- 20.Robertson KD, Uzvolgyi E, Liang G, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Research. 1999;27(11):2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagai M, Nakamura A, Makino R, Mitamura K. Expression of DNA (5-cytosin)-methyltransferases (DNMTs) in hepatocellular carcinomas. Hepatology Research. 2003;26(3):186–191. doi: 10.1016/s1386-6346(03)00091-3. [DOI] [PubMed] [Google Scholar]
- 22.Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clinical Cancer Research. 2003;9(12):4415–4422. [PubMed] [Google Scholar]
- 23.Samuel MS, Suzuki H, Buchert M, et al. Elevated Dnmt3a activity promotes polyposis in ApcMin mice by relaxing extracellular restraints on Wnt signaling. Gastroenterology. 2009;137(3):902–913. doi: 10.1053/j.gastro.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 24.Shafiei F, Rahnama F, Pawella L, Mitchell MD, Gluckman PD, Lobie PE. DNMT3A and DNMT3B mediate autocrine hGH repression of plakoglobin gene transcription and consequent phenotypic conversion of mammary carcinoma cells. Oncogene. 2008;27(18):2602–2612. doi: 10.1038/sj.onc.1210917. [DOI] [PubMed] [Google Scholar]
- 25.Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. American Journal of Pathology. 2003;163(3):1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in Genetics. 2000;16(4):168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 27.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 29.Roncalli M, Bianchi P, Bruni B, et al. Methylation framework of cell cycle gene inhibitors in cirrhosis and associated hepatocellular carcinoma. Hepatology. 2002;36(2):427–432. doi: 10.1053/jhep.2002.34852. [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Ni M, Xu J, et al. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer. 2002;2(1):29–42. doi: 10.1186/1471-2407-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S, Lee HJ, Kim J-H, Lee H-S, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. American Journal of Pathology. 2003;163(4):1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh BK, Kim H, Park HJ, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. International Journal of Molecular Medicine. 2007;20(1):65–73. [PubMed] [Google Scholar]
- 33.Beaulieu N, Morin S, Chute IC, Robert M-F, Nguyen H, MacLeod AR. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. The Journal of Biological Chemistry. 2002;277(31):28176–28181. doi: 10.1074/jbc.M204734200. [DOI] [PubMed] [Google Scholar]
- 34.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 35.Kaneda M, Okano M, Hata K, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 36.Deng T, Kuang Y, Wang L, Li J, Wang Z, Fei J. An essential role for DNA methyltransferase 3a in melanoma tumorigenesis. Biochemical and Biophysical Research Communications. 2009;387(3):611–616. doi: 10.1016/j.bbrc.2009.07.093. [DOI] [PubMed] [Google Scholar]
- 37.Fan H, Zhao Z, Quan Y, Xu J, Zhang J, Xie W. DNA methyltransferase 1 knockdown induces silenced CDH1 gene reexpression by demethylation of methylated CpG in hepatocellular carcinoma cell line SMMC-7721. European Journal of Gastroenterology and Hepatology. 2007;19(11):952–961. doi: 10.1097/MEG.0b013e3282c3a89e. [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Fan H, Zhao Z-J, Zhang J-Q, Xie W. Identification of potential genes regulated by DNA methyltransferase 3B in a hepatocellular carcinoma cell line by RNA interference and microarray analysis. Yi Chuan Xue Bao. 2005;32(11):1115–1127. [PubMed] [Google Scholar]
- 39.Wang L, Wang W-L, Zhang Y, Guo S-P, Zhang J, Li Q-L. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatology Research. 2007;37(5):389–396. doi: 10.1111/j.1872-034X.2007.00042.x. [DOI] [PubMed] [Google Scholar]
- 40.Dahia PLM, Aguiar RCT, Alberta J, et al. PTEN is inversely correlated with the cell survival factor Akt/PKB and is inactivated via multiple mechanisms in haematological malignancies. Human Molecular Genetics. 1999;8(2):185–193. doi: 10.1093/hmg/8.2.185. [DOI] [PubMed] [Google Scholar]
- 41.Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132(1):127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Soria J-C, Lee H-Y, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clinical Cancer Research. 2002;8(5):1178–1184. [PubMed] [Google Scholar]
- 43.Salvesen HB, MacDonald N, Ryan A, et al. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. International Journal of Cancer. 2001;91(1):22–26. doi: 10.1002/1097-0215(20010101)91:1<22::aid-ijc1002>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 44.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133(3):403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D, Kim S, Koh H, et al. Akt/PKB promotes cancer cell invasion via increased motility and metalloproteinase production. The FASEB Journal. 2001;15(11):1953–1962. doi: 10.1096/fj.01-0198com. [DOI] [PubMed] [Google Scholar]
- 47.Shukla S, MacLennan GT, Hartman DJ, Fu P, Resnick MI, Gupta S. Activation of PI3K-Akt signaling pathway promotes prostate cancer cell invasion. International Journal of Cancer. 2007;121(7):1424–1432. doi: 10.1002/ijc.22862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials include two tables, one is about candidate DNMT3A siRNA sequences (19nt) for knocking down DNMT3A in HCC cell lines, the other is about 153 genes up-regulated in DNMT3A knockdown cell.
Supplementary materials also provided the depletion of DNMT3A in QSG-7701 cells to perform the same experiments, including RNA interference, cell proliferation, colony formation and PTEN expression.