Abstract
The accumulation of unfolded or misfolded proteins in the endoplasmic reticulum (ER) results in the condition called “ER stress,” which induces the unfolded protein response (UPR), a complex cellular process that includes changes in expression of many genes. Failure to restore homeostasis in the ER is associated with human diseases. To identify the underlying changes in gene expression in response to ER stress, we induced ER stress in human B cells and then measured gene expression at ten time points. We followed up those results by studying cells from 60 unrelated people. We rediscovered genes that were known to play a role in the ER-stress response and uncovered several thousand genes that are not known to be involved. Two of these are VLDLR and INHBE, which showed significant increase in expression after ER stress in B cells and in primary fibroblasts. To study the links between UPR and disease susceptibility, we identified ER-stress-responsive genes that are associated with human diseases and assessed individual differences in the ER-stress response. Many of the UPR genes are associated with Mendelian disorders, such as Wolfram syndrome, and complex diseases, including amyotrophic lateral sclerosis and diabetes. Data from two independent samples showed extensive individual variability in ER-stress response. Additional analyses with monozygotic twins revealed significant correlations within twin pairs in their responses to ER stress, thus showing evidence for heritable variation among individuals. These results have implications for basic understanding of ER function and its role in disease susceptibility.
Introduction
The endoplasmic reticulum (ER) is the organelle where proteins and lipids are processed1 and intracellular calcium is regulated. When cells such as human B-lymphocytes have to cope with increased protein loads in response to cellular demands that require synthesis and secretion of proteins such as immunoglobulins, they expand and cause the differentiation of the ER.2 However, if unfolded or misfolded proteins accumulate in the ER, the overall result is termed “ER stress.” The cell responds by the unfolded protein response (UPR), a coordinated series of cellular events that increase the capacity of the ER to process the unfolded proteins and reduce the protein loads.3,4 These processes, many of which involve changes in gene expression, are initiated when the ER chaperone protein BiP5 dissociates from the transmembrane ER-stress-sensor molecules, ATF6,6,7 IRE1,8 and PERK.9 ER stress occurs in a wide variety of tissues and species and is associated with numerous human diseases.10–14 Although ER stress arises in normal cellular functions as well as in diseases, many human genes and pathways involved in the process remain to be identified. Characterization of these genes will lead to a better understanding of ER functions and of diseases caused by abnormalities in the UPR.
In this study, we characterized the temporal response and individual differences in response to ER stress by exposing B cells from unrelated individuals to tunicamycin or thapsigargin. First, we carried out a comprehensive search for genes involved in UPR by studying the gene expression patterns at ten time points in these cells. Second, we focused on two time points and studied the responses in cells from 60 individuals and a replicate sample with 14 individuals in order to assess individual variability in gene expression response to ER stress. Third, we studied the genetic contribution to the ER-stress response in genetically identical monozygotic (MZ) twins. The results allowed us to identify a large number of human genes involved in UPR; among them are genes such as inhibin beta E (INHBE [MIM 612031]) and very low density lipoprotein receptor (VLDLR [MIM 192977]), which have not been implicated in UPR and yet are strongly induced in B cells undergoing ER stress. We also showed that there is extensive individual variability in gene expression response to ER stress, and we demonstrated that there is likely a genetic component to this variation. Many of these variable ER-stress-responsive genes play a role in Mendelian disorders and complex diseases, suggesting the importance of proper ER function in human health.
Materials and Methods
Samples and Induction of ER Stress
Immortalized B cells from 60 unrelated individuals (grandparents in the HapMap CEPH-Utah pedigrees) and 26 MZ twin pairs (14 of European ancestry, 12 African American) were obtained from Coriell Cell Repositories (Camden, NJ, USA). All twin pairs were normal and apparently healthy. Zygosity testing was done at the Coriell Institute (11 twin pairs) or in our lab (15 twin pairs) by genotyping of 28 microsatellite markers.
Cells were grown at 37°C in 5% CO2 in RPMI medium 1640 containing 15% fetal bovine serum (FBS), 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin. As a way to minimize possible batch effects, cells from the 60 unrelated individuals were grown in four batches, and for the twin samples, all but three pairs were grown in only two batches, of 12 and 11 twin pairs (24 and 22 cultures), respectively. In order to monitor effects resulting from culturing and hybridizing cells in batches, some cells from batch 1 were grown again with each successive batch.
To induce ER stress, we transferred cells to fresh RPMI 1640, supplemented as above, at a concentration of 106 cells/ml, grew them for 18 hr, and then treated them at the indicated time points with either 500 nM thapsigargin (Sigma-Aldrich) dissolved in DMSO (Sigma-Aldrich) or 4 μg/mL tunicamycin (Sigma-Aldrich) in DMSO. These standard doses of thapsigargin and tunicamycin are used in many studies for the induction of ER stress.15,16 These drug dosages did not lead to excessive cytoxicity. Untreated control cultures were grown in RPMI 1640 containing 0.5% DMSO.
To monitor for ER stress, we assayed for x box-binding protein 1 (XBP1 [MIM 194355]) splicing by performing PCR with the use of cDNA from cells as template and the following primers: forward, 5′-GCTGAAGAGGAGGCGGAAG-3′; reverse, 5′- GTCCAGAATGCCCAACAGG-3′. Amplicons were resolved on 2.5% agarose gels.
Affymetrix Expression Arrays and Analysis
RNA was prepared from the treated and untreated cells with the use of the RNeasy kit (QIAGEN), labeled with biotin with the use of the GeneChip Expression 3′-Amplification One-Cycle cDNA Synthesis Kit (Affymetrix), and hybridized to Affymetrix U133 Plus 2.0 GeneChip arrays in accordance with manufacturers' protocols.
The microarray data were normalized with MAS5.0 and log2 transformed with Expression Console v 1.1.1 software (Affymetrix). Analyses were carried out on (1) genes called “present” in 25% or more of the arrays (of the ∼55,000 transcripts on the microarray, ∼26,000 fit this criterion and are therefore considered to be expressed in our B cells) or, (2) for the time-course experiments, genes that were called “present” in two or more time points. We used a different selection criterion in the time-course study to include genes that were induced at some but not all parts of the UPR. However, we found that there are few such genes; > 80% of the genes are either expressed or not expressed at all time points.
DMSO was used as a solvent for thapsigargin and tunicamycin. To account for possible effects of the DMSO, we treated cells with DMSO alone and with thapsigargin or tunicamycin in DMSO. For each gene, we subtracted the expression value (log2) in the DMSO-treated samples from that (log2) in the thapsigargin- or tunicamycin-treated cells. In order to present the response to ER stress as a fold change (FC), we took the anti-log2 of the expression responses.
Samples used in various analyses include the following: first, for the time-course study, we used ten unrelated individuals from the collection of twins of European ancestry. The samples were pooled at each time point and treatment and were hybridized onto duplicate arrays. To identify those genes that change expression levels over time, we carried out analysis of variance (ANOVA; p < 10−5). Second, to identify individual variability in gene expression response to ER stress, we analyzed data from 60 (HapMap CEPH-Utah collection) unrelated individuals and then replicated the findings in 14 (European twin collection) unrelated individuals. For each “expressed” gene, we carried out a paired t test to compare gene expression in DMSO treatment versus tunicamycin or thapsigargin treatment. Genes with a corrected p < 10−5 (Bonferroni correction) were considered “ER-stress responsive.” To determine individual variability, we calculated variance of their expression response across 60 and 14 individuals separately.
Fibroblasts and Keratinocytes
Primary fibroblasts from the foreskin of healthy newborns were cultured in MEM medium supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin. Keratinocytes (ATCC) were cultured in Dermal Cell Basal medium supplemented with Keratinocyte Growth Kit (ATCC). As with the immortalized B cells, the fibroblasts and keratinocytes were treated with 4 μg/ml tunicamycin in DMSO or with DMSO alone. RNA was extracted at 2, 4, and 8 hr after treatment and used for expression analysis of VLDLR and INHBE by quantitative RT-PCR per the manufacturer's protocol (Applied Biosystems). Expression of GAPDH and ACTB were used as controls for normalization, and changes in expression were calculated relative to cells treated with DMSO alone. Primers were as follows: INHBE forward, 5′-ACTACAGCCAGGGAGTGTGG-3′; INHBE reverse, 5′-AGTGAGCAGGGAGCTGTAGG-3′; VLDLR forward, 5′-TCTGTTGGACACACGTACCC-3′; VLDLR reverse, 5′-CCTCAAAGGTCAACATTTGTCA-3′.
Intraclass Correlation Coefficient Analysis
For each ER-stress-responsive gene, ANOVA was performed as an assessment of differences between twin pairs, relative to differences within pairs. From the ANOVA, we computed the intraclass correlation coefficient (ICC).17,18
Cis-Acting Elements
For each ER-stress-responsive gene, we identified the RefSeq sequence (if there were multiple RefSeq sequences for a gene, we used the one that represented the longest transcript). Within those sequences and 2 kb up- and downstream, we looked for exact matches to the ER stress response element (ERSE) (CCAAT-N9-CCACG) and the unfolded protein response element (UPRE) (TGACGTGG/A) sequences. The positions of the ERSE or UPRE in Table S1 (available online) correspond to those in build hg18 of the UCSC reference genome.
Disease-Susceptibility Genes
We searched Online Mendelian Inheritance in Man (OMIM) and the GWAS catalog19 for 1752 ER-stress-responsive genes (corrected p < 0.05, t test; FC ≥ 1.5) to identify those that have been implicated in Mendelian and complex diseases, respectively.
Results
ER Stress Induced by Thapsigargin or Tunicamycin
We induced ER stress in immortalized B cells from healthy individuals with two standard drugs, thapsigargin and tunicamycin, that inhibit protein folding and modification in the ER. Thapsigargin is an inhibitor of the sarcoplasmic or endoplasmic reticulum Ca2+ ATPase (SERCA); it depletes the ER calcium stores and therefore prevents proper functions of molecular chaperones, such as calreticulin, that process nascent polypeptides.20 Tunicamycin inhibits N-glycosylation, which leads to an accumulation of underglycosylated proteins that cannot exit the ER.21 In the cells that we treated with thapsigargin or tunicamycin, ER stress was induced as shown by splicing of XBP1, a hallmark of UPR22 (Figure 1).
Figure 1.
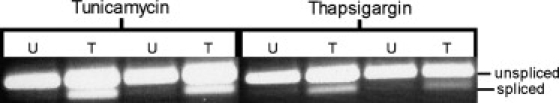
XBP1 Splicing in Cells Treated with Thapsigargin and Tunicamycin
Cells from two individuals with (T) and without (U) treatment with thapsigargin and tunicamycin. In treated cells, XBP1 splicing, a hallmark of ER stress, was observed.
Temporal Gene Expression Response to Thapsigargin and Tunicamycin
To study the temporal response of B cells to ER stress, we treated cells from ten healthy individuals with thapsigargin or tunicamycin and used microarrays to profile gene expression at ten time points (baseline and 15 min, 30 min, 1 hr, 2 hr, 4 hr, 8 hr, 12 hr, 24 hr, and 48 hr after treatment). For each time point and treatment, we pooled RNA samples from ten individuals and hybridized them onto Affymetrix microarrays. For each gene, we calculated changes in expression levels by comparing the signal intensity of the treated samples with that of the baseline and also to treatment with DMSO alone. To identify genes that changed significantly at one or more time points after treatment, we carried out an ANOVA.
In the tunicamycin-treated samples, 26,631 genes were expressed at two or more time points. We focused on these genes for further analysis. Among the expressed genes, 1301 genes (5%) changed significantly (p < 10−5, ANOVA) at one or more time points after exposure to tunicamycin. We expect to find less than one gene that showed this level of significance by chance alone; thus, we conclude that many of these genes indeed play a role in response to tunicamycin-induced ER stress. The genes that showed significant changes in gene expression include those in the classic UPR pathways, such as signaling factors (ERN1 [MIM 604033]), chaperones (CALM1 [MIM 114180], CANX [MIM 114217]), ER-associated degradation factors (EDEM1 [MIM 607673], EDEM2 [MIM 610302]) and cell-death regulators (TRIB3 [MIM 607898], DDIT3 [MIM 126337]). Figure 2 shows the temporal expression patterns of genes in the classic UPR pathways.
Figure 2.
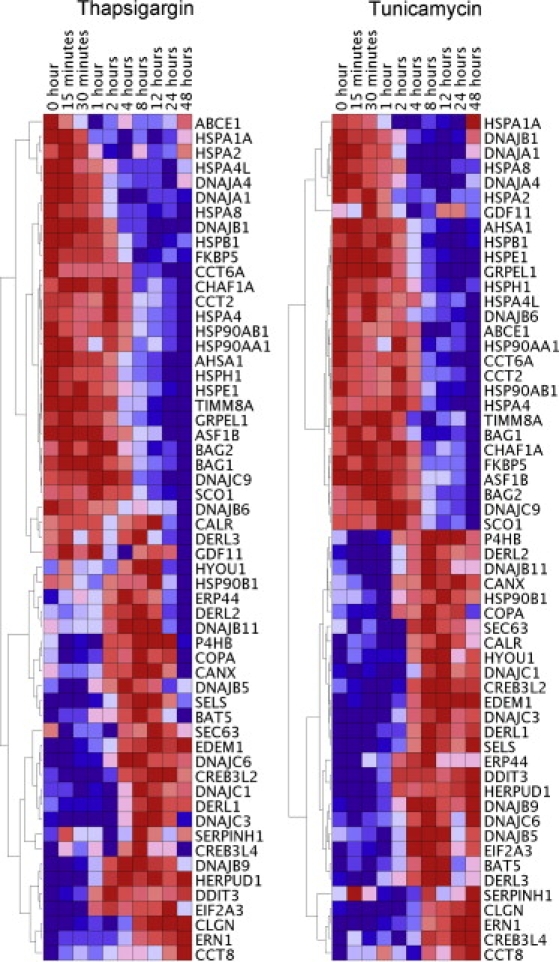
Temporal Gene Expression Pattern of Classic UPR Genes in Cells at Baseline and Nine Time Points after Exposure to Tunicamycin and Thapsigargin
Hierarchical clustering based on pairwise correlations for 57 classic UPR genes. Analyses were carried out with the use of GenePattern software.
To study the temporal response patterns, we clustered the genes by their patterns of expression using correlation with hierarchical clustering and carried out a post hoc t test. Only a small number of genes showed significant changes in expression at the 15 and 30 min time points (14 genes and 24 genes at p < 0.0001 [t test], respectively). Among these early-response genes are signaling factors (PIK3CA [MIM 171834], LCK [MIM 153390]), transcription factors (ZNF294), and cell-cycle regulators (MCM2 [MIM 116945], CDC42 [MIM 116952]). In contrast, the late-response genes are enriched for those involved in ER-Golgi transport (PRKD2 [MIM 607074], COPB2 [MIM 606990]) and ubiquitin degradation (UBE2B [MIM 179095], RWDD4A). The majority of genes that showed significant expression changes were induced or repressed within 8 hr after tunicamycin treatment.
Thapsigargin and Tunicamycin Induce Similar Changes in Gene Expression
Thapsigargin evoked a similar pattern of gene expression changes as tunicamycin. Forty-six percent (or 596) of 1301 genes whose expression levels changed significantly (p < 10−5, ANOVA) after tunicamycin treatment also showed significant (p < 10−5) changes after thapsigargin treatment. Many of the remaining genes showed changes in expression after thapsigargin treatment, although they did not reach the significance threshold of p < 10−5. Highly similar temporal patterns of expression were seen after tunicamycin and thapsigargin treatments (Figure 2 and Figure 3). For each gene, we compared the gene expression patterns after treatments with the two drugs by calculating correlation coefficients. The median correlation was 0.87 (average, 0.80, range, −0.88 to 1.0). Figure 3 shows gene expression patterns of four genes after treatment with the two drugs. These results suggest that most of the expression changes are due to ER stress and are not a specific response to tunicamycin or thapsigargin.
Figure 3.
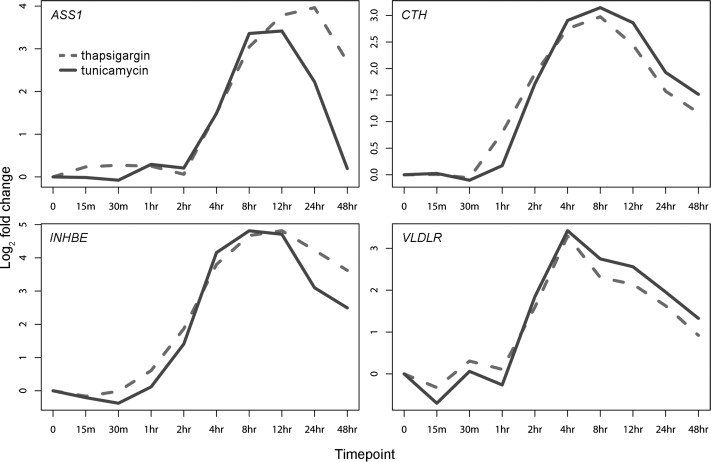
Examples of ER-Stress-Responsive Genes that Showed Similar Expression Patterns in Response to Tunicamycin and Thapsigargin
Cis-Acting Elements in ER-Stress-Responsive Genes
Transcription factors such as XBP1 and ATF6 coordinate many of the gene expression changes in the UPR. There are two well-characterized cis-acting ER-stress-responsive elements for binding of these transcription factors. One is the ERSE, CCAAT-N9-CCACG, where NF-Y binds to the CCAAT part and pATF6α and pATF6β bind to the CCACG portion.23,24 The second is the UPRE, TGACTGG/A, a binding site for XBP1.25,26 We looked for ERSE and UPRE sequences in the genes that showed significant changes (p < 10−5) in gene expression after tunicamycin and thapsigargin treatments. We found that 25% of the genes contain one or more UPREs and that < 1% (seven genes) contain an ERSE (see Table S1) within or 2 kb upstream and downstream of the genic regions. Among the genes with UPRE are those that are known to play a role in UPR, including DNA chaperones (DNAJC3 [MIM 601184], DNAJB6 [MIM 611332]), genes in the apoptotic pathways (BID [MIM 601997], BCL11A [MIM 606557]), and genes that thus far are not known to play a role in UPR, such as VLDLR and INHBE. The seven genes with ERSE include two genes in the classic UPR pathway (HSP90B1 [MIM 191175], CANX), three solute carriers (SLC5A3 [MIM 600444], SLC7A5 [MIM 600182], SLC33A1 [MIM 603690]), a mitochondrial ribosomal protein (MRPS6 [MIM 611973]), and an uncharacterized gene (FAM107B). These results allow further characterization of these genes by placing them in either the ATF6 or the XBP1 pathway.
ER-Stress-Responsive Genes
The time-course results provide us with a temporal pattern of gene expression responses to ER stress. However, the number of time points limits how many samples we can study. For the analysis, we used a pool of samples (ten individuals in the pool). Thus, genes with subtle effects would not be detected. We also did not obtain information on individual differences in response. To address these individual differences, we picked two time points (baseline and 8 hr after exposure) and studied changes in gene expression 8 hr after treatment with tunicamycin in 60 individuals.
Among the 23,196 array probes that are expressed in our B cells, 10,729 (46%, corresponding to 7031 genes) showed significant changes in gene expression with tunicamycin treatment (p < 10−5, t test). The sample size allows us to identify genes that show modest but significant response to ER stress. Almost half of the expressed genes showed significant changes in gene expression in response to ER stress, thus illustrating the complexity of UPR. There were 2533 array probes (11%, corresponding to 1752 genes; Table S2) that showed ≥ 1.5-fold increase or decrease in expression. Some genes showed large changes in expression; for example, seven that showed > 10-fold change in expression are: inhibin-beta E (INHBE), cation-transport regulator homolog 1 (CHAC1), cell division cycle 14 (CDC14 [MIM 603504]), an expressed sequence tag (237127_at), aldehyde dehydrogenase family 1 (ALDH1L2), solute carrier family 7 member 11 (SLC7A11 [MIM 607933]), and cystathionase (CTH). Among them, only CHAC1 and SLC7A11 are known to participate in UPR. However, genes with large fold changes in expression are not necessarily the most biologically important. Here, by studying a large sample, we can detect subtle changes in gene expression. Many genes, such as presenilin-associated rhomboid-like protein (PARL [MIM 607858]) and LRRC47, showed only modest changes in expression (< 1.5-fold), but the changes were observed in nearly all 60 individuals (Figure 4). The consistency of the data provides a strong suggestion that they play a role in UPR.
Figure 4.
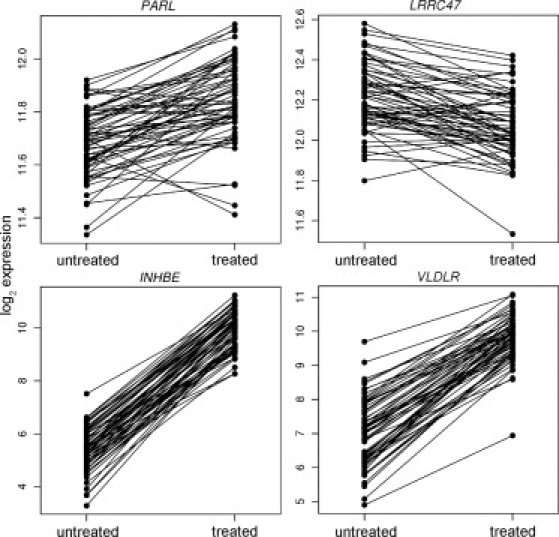
Examples of Genes that Showed Significant Changes in Response to ER Stress among 60 Unrelated Individuals
Shown are two genes (PARL and LRRC47) that showed significant but modest fold change after ER stress. In addition, two genes (INHBE and VLDLR) that were not known to play a role in ER stress showed significant and large fold change after exposure to tunicamycin.
To follow up these findings, we studied an additional sample. We treated cells from an independent set of 14 unrelated individuals with thapsigargin (rather than tunicamycin) for 4 hr and measured changes in gene expression. Despite a smaller sample size, among the 2533 array probes that showed significant and ≥ 1.5-fold changes in expression levels in the tunicamycin-treated cells, we found that 1500 probes showed similar changes (Table S2) after thapsigargin treatment. This further supports the idea that these genes are indeed responding to ER stress and not just to tunicamycin. To study these “ER-stress-responsive genes,” we first looked for genes that belong to canonical ER stress pathways. Known UPR genes that showed significant changes in gene expression include CHOP/DDIT3 (fold difference = 8.2), IRE1/ERN1 (2.5), activating transcription factor 3 (ATF3 [MIM 603148]) (2.3), homocysteine- and endoplasmic reticulum stress-inducible protein, ubiquitin-like domain-containing, 1 (HERPUD1 [MIM 608070]) (2.8), protein phosphatase 1, regulatory subunit 15a (GADD34/PPP1R15A [MIM 611048]) (2.3), SIL1 (BIP/HSPA5 [MIM 608005]) (4.3), growth arrest- and dna damage-inducible gene (GADD45A [MIM 126335]) (1.7), eukaryotic translation initiation factor 2-alpha kinase 3 (PERK/EIF2AK3 [MIM 604032]) (1.7), and activating transcription factor 4 (ATF4 [MIM 604064]) (2.5). Other “classic UPR” genes in which we detected significant changes in expression are listed in Table 1.
Table 1.
Changes in Expression Levels of Genes in the Classic UPR Pathway
| Gene Symbol | p Value (t Test) | Fold Change |
|---|---|---|
| ABCE1 | 1.34 × 10−43 | −1.6 |
| AHSA1 | 3.53 × 10−51 | −1.9 |
| ASF1B | 1.55 × 10−39 | −1.6 |
| BAG1 | 1.06 × 10−35 | 1.5 |
| BAG2 | 6.35 × 10−39 | −1.7 |
| BAT5 | 5.68 × 10−32 | 1.5 |
| CALR | 1.82 × 10−38 | 1.7 |
| CANX | 8.20 × 10−29 | 1.6 |
| CCT2 | 1.10 × 10−38 | −1.9 |
| CCT6A | 3.42 × 10−38 | −1.6 |
| CCT8 | 3.98 × 10−11 | 1.5 |
| CHAF1A | 1.18 × 10−35 | −1.9 |
| CLGN | 2.49 × 10−12 | 2.3 |
| COPA | 5.77 × 10−27 | 1.6 |
| CREB3L2 | 2.91 × 10−41 | 2.0 |
| CREB3L4 | 5.12 × 10−33 | 1.8 |
| DDIT3 | 5.29 × 10−55 | 8.2 |
| DERL1 | 7.26 × 10−35 | 1.7 |
| DERL2 | 8.45 × 10−40 | 2.4 |
| DERL3 | 4.44 × 10−34 | 3.9 |
| DNAJA1 | 4.43 × 10−40 | −1.8 |
| DNAJA4 | 1.48 × 10−10 | −1.7 |
| DNAJB1 | 4.82 × 10−41 | −2.5 |
| DNAJB11 | 4.07 × 10−42 | 2.8 |
| DNAJB5 | 6.02 × 10−29 | 2.1 |
| DNAJB6 | 4.49 × 10−29 | −1.9 |
| DNAJB9 | 8.45 × 10−37 | 6.0 |
| DNAJC1 | 5.22 × 10−38 | 2.2 |
| DNAJC3 | 1.47 × 10−37 | 2.5 |
| DNAJC6 | 1.41 × 10−14 | 1.8 |
| DNAJC9 | 4.59 × 10−35 | −1.5 |
| EDEM1 | 1.10 × 10−42 | 2.5 |
| EIF2A3 | 2.18 × 10−25 | 1.7 |
| ERN1 | 2.27 × 10−36 | 2.5 |
| ERP44 | 1.52 × 10−28 | 1.7 |
| FKBP5 | 9.27 × 10−42 | −1.7 |
| GDF11 | 2.21 × 10−15 | −1.7 |
| GRPEL1 | 1.76 × 10−34 | −1.7 |
| HERPUD1 | 4.14 × 10−37 | 2.8 |
| HSP90AA1 | 1.03 × 10−37 | −1.7 |
| HSP90AB1 | 2.05 × 10−42 | −1.5 |
| HSP90B1 | 6.04 × 10−33 | 2.0 |
| HSPA1A | 3.27 × 10−33 | −3.0 |
| HSPA2 | 6.62 × 10−21 | −2.1 |
| HSPA4 | 2.14 × 10−37 | −1.7 |
| HSPA4L | 1.56 × 10−35 | −1.5 |
| HSPA8 | 5.31 × 10−37 | −2.3 |
| HSPB1 | 3.95 × 10−29 | −1.5 |
| HSPE1 | 2.42 × 10−47 | −2.0 |
| HSPH1 | 6.27 × 10−43 | −2.5 |
| HYOU1 | 2.49 × 10−45 | 3.1 |
| P4HB | 3.87 × 10−34 | 1.8 |
| SCO1 | 1.03 × 10−28 | −1.5 |
| SEC63 | 6.82 × 10−25 | 1.7 |
| SELS | 3.53 × 10−45 | 2.8 |
| SERPINH1 | 3.58 × 10−11 | 1.5 |
| TIMM8A | 1.66 × 10−34 | −2.4 |
However, there are also many genes that have not been reported to play a role in UPR. Among the 100 genes that showed the most significant changes and largest fold change in gene expression, only 38 have been reported in the literature (PubMed) as participating in UPR. Others, such as INHBE and VLDLR, showed significant (p = 10−51 and 10−37, respectively) and large fold changes (23- and 7-fold changes, respectively) and yet have not been implicated in UPR. Table 2 shows some of these newly identified ER-stress-responsive genes and their functional categories according to the Gene Ontology database.27
Table 2.
Examples of Genes that Have Not been Reported to Participate in the ER-Stress Response
| Functional Annotation | Gene Symbola | p Value (t Test) | Fold Change |
|---|---|---|---|
| Organic acid metabolism | ALDH1L2 | 3.13 × 10−21 | 6.8 |
| ALOX5AP | 6.72 × 10−34 | 3.4 | |
| ASS1 | 6.00 × 10−37 | 5.9 | |
| HPDL | 5.03 × 10−30 | −0.3 | |
| PCK2 | 4.13 × 10−57 | 3.4 | |
| PHGDH | 5.65 × 10−45 | 4.9 | |
| PSAT1 | 3.36 × 10−42 | 5.1 | |
| SLC7A5 | 8.90 × 10−46 | 3.9 | |
| Transporter | CDRT4 | 5.33 × 10−16 | 3.5 |
| PHGDH | 5.65 × 10−45 | 4.9 | |
| SLC1A5 | 7.20 × 10−51 | 3.8 | |
| SLC3A2 | 4.88 × 10−56 | 4.8 | |
| SLC7A5 | 8.90 × 10−46 | 3.9 | |
| VLDLR | 4.83 × 10−38 | 6.8 | |
| Response to stress | ALOX5AP | 6.72 × 10−34 | 3.4 |
| NFE2L1 | 1.25 × 10−53 | 3.3 | |
| Kinase | ALPK2 | 1.39 × 10−31 | 3.7 |
| RPS6KA2 | 8.26 × 10−32 | 4.5 | |
| Glycoprotein | FKBP14 | 3.27 × 10−40 | 3.9 |
| GPR84 | 8.84 × 10−29 | 4.7 | |
| INHBE | 2.52 × 10−51 | 23.6 | |
| KISS1R | 1.50 × 10−24 | −0.3 | |
| SLC1A5 | 7.20 × 10−51 | 3.8 | |
| SLC3A2 | 4.88 × 10−56 | 4.8 | |
| SLC7A5 | 8.90 × 10−46 | 3.9 | |
| TSLP | 2.60 × 10−22 | 4.3 | |
| VLDLR | 4.83 × 10−38 | 6.8 | |
| Calcium ion binding | EML2 | 9.66 × 10−41 | 3.9 |
| FKBP14 | 3.27 × 10−40 | 3.9 | |
| SNTB1 | 1.13 × 10−35 | 3.3 | |
| VLDLR | 4.83 × 10−38 | 6.8 | |
Data are from 60 individuals of European descent treated with tunicamycin.
From 100 genes that showed the most significant and largest fold change in response to ER stress.
Very Low Density Lipoprotein Receptor and Inhibin Beta-E
The expression levels of INHBE and VLDLR increased significantly after ER stress. Neither gene has been implicated in UPR. The temporal patterns of expression of INHBE and VLDLR after treatment with thapsigargin or tunicamycin are shown in Figure 3, and their expression levels in 60 individuals at baseline and 8 hr after thapsigargin treatment are shown in Figure 4. These results showed consistent induction of these two genes across time points and individuals; thus confirming their roles in UPR. To determine whether the expression responses of INHBE and VLDLR are specific to cultured B cells or are also seen in other cell types, we induced ER stress in primary human fibroblasts and keratinocytes and measured expression levels of these genes at 2, 4, and 8 hr after tunicamycin treatment. In primary fibroblasts, significant (p < 0.01) changes were observed for INHBE and VLDLR. Maximum inductions of INHBE (4.2-fold) and VLDLR (2.9-fold) were seen 4 and 8 hr after ER stress, respectively. In keratinocytes, INHBE was not expressed, but VLDLR was expressed and showed 2.8-fold induction after ER stress. As reported above, the roles of INHBE and VLDLR in the ER-stress response are further supported by the presence of unfolded protein response element (UPRE), TGACTGG/A, a binding site for XBP125,26 in these genes (Table S1).
Annotations of UPR-Responsive Genes
To characterize the collection of ER-stress-responsive genes, we annotated the genes by using Gene Ontology Annotations and the KEGG pathway tool. We found enrichments of genes that are involved in RNA processing, response to protein stimulus, and the cell cycle (see Table 3 for complete list). Similarly, the KEGG pathway tool showed enrichment for genes that participate in pyrimidine metabolism, cell cycle, folate biosynthesis, and N-glycan biosynthesis (Bejamini-corrected p < 0.05).
Table 3.
Functional Categories that Are Enriched in ER-Stress-Responsive Genes
| GO Annotation | p Value (Benjamini) |
|---|---|
| RNA processing | 3.50 × 10−12 |
| Ribosome biogenesis and assembly | 7.19 × 10−12 |
| Metabolic process | 1.62 × 10−10 |
| Response to protein stimulus | 1.74 × 10−8 |
| Nucleotide binding | 3.26 × 10−8 |
| Protein folding | 7.67 × 10−8 |
| Response to DNA damage | 1.59 × 10−6 |
| Helicase activity | 2.04 × 10−6 |
| tRNA metabolic process | 3.20 × 10−5 |
| Splicing | 4.85 × 10−4 |
| Cell cycle | 7.00 × 10−3 |
UPR-Responsive Genes and Disease Susceptibility
ER stress is implicated in many human diseases, from Mendelian disorders, such as cystic fibrosis, to complex diseases, including diabetes and neurodegenerative disorders. The large number of ER-stress-responsive genes identified in this study provides us an opportunity to study the contribution of ER stress to human diseases more comprehensively. We queried OMIM and the catalog of published results from GWAS19 to identify the ER-stress-responsive genes that have been implicated in Mendelian and complex diseases. We found that 191 and 357 of the ER-stress-responsive genes are disease susceptibility genes in the OMIM and GWAS catalog, respectively.
Examples of ER-stress-responsive genes associated with Mendelian disorders include WFS1 (MIM 606201, associated with Wolfram syndrome 1 [MIM 222300]); CDGSH iron sulfur domain protein 2 (CISD2 [MIM 611507], associated with Wolfram syndrome 2 [MIM 604928]); 3-prime repair exonuclease 1 (TREX1 [MIM 606609], associated with Aicardi-Goutières syndrome [MIM 225750]); ATP-binding cassette, subfamily a, member 12 (ABCA12 [MIM 607800], associated with Harlequin ichthyosis [MIM 242500]); SIL1 (MIM 608005, associated with Marinesco-Sjogren syndrome [MIM 248800]); DNA cross-link repair protein 1C (DCLRE1C [MIM 605988], associated with Omenn syndrome [MIM 603554]); and EIF2AK3 (MIM 604032, associated with Wolcott-Rallison syndrome [MIM 226980]). ER-stress-responsive genes that have been implicated in complex human diseases include (1) UDP-GAL:beta-GlcNAc beta-1,4-galactosyltransferase, polypeptide 6 (B4GALT6 [MIM 604017]); casein kinase I, gamma-3 (CSNK1G3 [MIM 604253]); inositol 1,4,5-triphosphate receptor, type 2 (ITPR2 [MIM 600144]); RNA-binding motif protein, single strand-interacting, 1 (RBMS1 [MIM 602310]); and ZNF746, all of which are associated with amyotrophic lateral sclerosis, (2) tyrosine kinase, B-lymphocyte specific (BLK [MIM 191305]); V-ETS avian erythroblastosis virus E26 oncogene homolog 1 (ETS1 [MIM 164720]); interferon regulatory factor 5 (IRF5 [MIM 607218]); and ubiquitin-conjugating enzyme E2l 3 (UBE2L3 [MIM 603721]), all of which are associated with systemic lupus erythematosus (MIM 152700), and (3) cyclin-dependent kinase 2 (CDK2 [MIM 116953]); interferon induced with helicase C domain protein 1 (IFIH1 [MIM 606951]); protein-tyrosine phosphatase, nonreceptor-type, 2 (PTPN2 [MIM 176887]); and RAS-associated protein RAB5b (RAB5B [MIM 179514]), all of which are associated with type 1 diabetes (MIM 222100) (see Table 4 for additional examples). These results provide further mechanistic suggestions of ER stress in the pathophysiology of human diseases.
Table 4.
ER-Stress-Responsive Genes Associated with Disease Susceptibility
| Disease | Susceptibility Genes Identified in GWAS that Respond to ER Stress |
|---|---|
| Acute lymphoblastic leukemia | ARID5B, ZNF230 |
| AIDS | TGFBRAP1, LTB, MICB, TNF |
| Alzheimer disease | CLU, CR1, FAM113B, PICALM, RFC3, TOMM40, SASH1 |
| Amyotrophic lateral sclerosis | B4GALT6, C9orf72, CSNK1G3, ITPR2, RBMS1, SELL, SLC39A11, ZNF746 |
| Bipolar disorder | CTNNA2, DCTN5, PALB2, MCTP1 |
| HDL cholesterol | CTCF, FADS1, NR1H3, MMAB, MVK, TTC39B |
| Multiple sclerosis | ASAP1, C1GALT1, CD40, CD58, CENPC1, HLA-B, IL12A, IRF8, JAR1D2, KIF1B, METTL1, MPHOSPH9, PTGER4, RAB38, RFK, RPL5, SLC25A36, TNFRSF1A, WDR7, ZMIZ1 |
| Parkinson disease | AAK1, ATF6, PRRG4, QSER1 |
| Rheumatoid arthritis | REL, TRAF1, TNFRSF14 |
| Systemic lupus erythematosus | BLK, ETS1, HIC2, IKZF1, IRF5, PXK, RASGRP3, TNIP1, UBE2L3 |
| Type 1 diabetes | C12orf30, C6orf173, CD69, CDK2, CTSH, HLA-E, IFIH1, IKZF4, IL10, PGM1, PHTF1, PTPN2, RAB5B, SUOX |
| Type 2 diabetes | CAMK1D, CDC123, NOTCH2, THADA, VEGFA, IRS1, WFS1 |
Individual Variation in Expression Response to ER Stress
Using gene expression responses of the 60 individuals, we assessed individual variability in response to ER stress. For each gene, we calculated variance of fold change in response to tunicamycin-induced ER stress, and we ranked the genes by the variances. We found that many genes showed extensive individual variation but that others showed less variability. The fold-change values for ten genes were plotted in Figure 5. Eight genes were highly variable, and two (EIF3B [MIM 603917] and SOS2 [MIM 608674]) showed little individual variation in their expression responses to ER stress.
Figure 5.
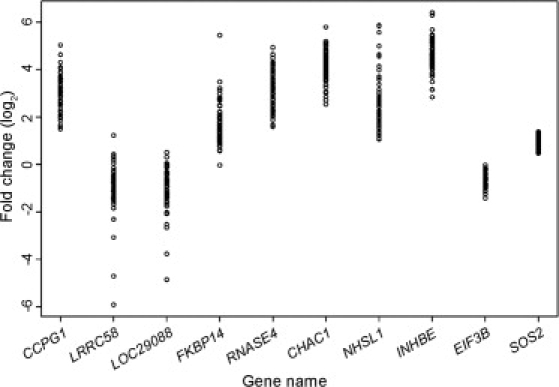
Extensive Individual Variation in Gene Expression Response to ER Stress
Fold changes for ten genes in B cells from 60 unrelated individuals at 8 hr after exposure to tunicamycin are shown. Eight of the genes are highly variable, and two (EIF3B and SOS2) showed less variability.
To study the variable genes, we annotated them by using Gene Ontology. However, the variable genes did not show enrichment of any functional groupings. Then, we looked for known functions of the variable and less variable genes in the ER by searching for the names of the genes and the words “endoplasmic reticulum” in PubMed. We found that significantly more of the less variable genes were known to have a role in the ER. Thirty of the 78 least variable genes are found in PubMed abstracts that have the words “endoplasmic reticulum,” whereas only 13 of the 78 most variable genes have such association (p = 0.0001, chi-square). This result is not too surprising, given that individual variability complicates molecular validations. Genes that are highly variable are induced in some individuals but not in others. Thus, these variable genes are less likely to have been shown to play a role in the ER; experiments with small samples would have yielded inconsistencies. These variable responsive genes can be detected only when a large number of samples are studied. However, the individual variation is important because these genes are more likely to be disease-susceptibility genes than are genes that are less polymorphic.
Genetic Component to ER-Stress Response
To assess whether there is a genetic component to the individual variation in gene expression response to ER stress, we examined the phenotypes in genetically identical MZ twins. We induced ER stress in immortalized B cells from 14 twin pairs of European descent and 12 African American twin pairs, then we measured gene expression by using the same microarrays as in the above experiments. To evaluate twin similarity in response to ER stress, we carried out the ANOVA separately with data from the twins of European and African American descent. For each gene, the ANOVA provides VA and VW, estimates of the variance among and variance “within” MZ twin pairs. The fraction VA / (VA + VW) is known as the ICC (intraclass correlation coefficient) and is a standard measure of within-pair similarity.17,18 An estimate of the ICC close to its maximum value of 1 indicates high similarity of MZ twins for that measurement; an estimate close to 0 indicates that the twins are no more similar for that measurement than expected by chance. Furthermore, the ANOVA allows a test of the significance of the ICC; that is, a test for significant correlation within MZ twin pairs.
We used the ICC to evaluate MZ twin similarity in response to ER stress. The ICC is not a direct indicator of this heritable contribution, but it is the natural measure to use with data from a sample of MZ twins.18 We considered whether to pool data from the two ethnic groups before carrying out the ANOVA, in order to increase sample size. However, we were concerned that there might be underlying differences between the groups that could result in misleading findings. For this reason, we carried out the ICC analysis separately for the two sets of twins.
The results show that for many genes, twins are more similar in their response to ER stress than are nontwins. Figure 6 is an example of our findings. It shows individual twin-pair results for gene phosphoserine phosphatase-like (PSPHL [MIM 604239]). Twins are significantly more similar than unrelated individuals in their ER-stress response (ICC = 0.81 and 0.69 for pairs of European descent and African American pairs, respectively). Overall, there are 289 ER-stress responsive genes that show an ICC > 0.5. Examples of some of these genes are listed in Table 5. These results are based on a small sample size (14 and 12 pairs of twins), but they are suggestive that individual variation in expression response to ER stress is genetically regulated.
Figure 6.
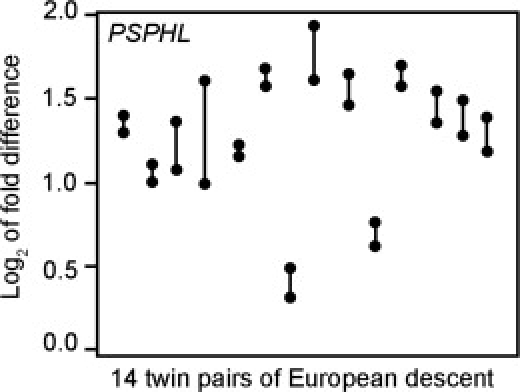
Gene Expression Response of PSPHL to ER Stress of Members of MZ Twin Pairs
ER stress was induced in B cells from 14 pairs of twins of European descent. A vertical line connects the members of a single MZ pair. Similar results were found for African American twins (European descent ICC = 0.8, p = 9 x10−5; African American ICC = 0.69, p = 3.3 × 10−3).
Table 5.
Examples of UPR Genes that Showed Significant ICC
| Gene Symbol |
European-Descent Twins (n = 14) |
African American Twins (n = 12) |
||
|---|---|---|---|---|
| Fold Changea | ICCb | Fold Changea | ICCb | |
| PSPHL/CO9 | 2.46 | 0.88 | 1.37 | 0.79 |
| PHF13 | −1.41 | 0.75 | −1.37 | 0.72 |
| ANXA2 | 1.25 | 0.79 | 1.25 | 0.67 |
| SFRS1 | −1.57 | 0.75 | −1.35 | 0.66 |
| CD24 | 2.27 | 0.73 | 2.30 | 0.66 |
| BID | −1.13 | 0.77 | −1.24 | 0.60 |
| CLIC4 | 1.91 | 0.82 | 1.71 | 0.53 |
| IRF2BP2 | 1.40 | 0.76 | 1.18 | 0.57 |
| CFLAR | 1.43 | 0.73 | 1.12 | 0.57 |
| ARHGDIB | −1.30 | 0.76 | −1.28 | 0.53 |
| IFNGR1 | 1.50 | 0.78 | 1.52 | 0.51 |
| MS4A1 | −1.79 | 0.71 | −2.05 | 0.51 |
| FASN | −1.54 | 0.71 | −1.24 | 0.49 |
| SLC29A1 | −1.84 | 0.72 | −1.94 | 0.47 |
| TMEM48 | −1.19 | 0.71 | −1.35 | 0.47 |
4 hr after ER stress.
p < 0.005.
Discussion
Individual differences in the efficiency of the ER to carry out its cellular functions underlie human diseases such as autoimmune and neurodegenerative disorders. The ER is the organelle where key cellular functions, including protein synthesis, protein modification, calcium maintenance, and lipid synthesis, occur. To respond to cellular needs, the ER synthesizes transmembrane proteins and lipids that constitute most of the cell's organelles from the ER itself to mitochondria, Golgi, and the plasma membrane. In cells, such as B-lymphocytes, which secrete proteins in the form of immunoglobulins, or hepatocytes, where lipoproteins are produced, the ER is abundant. The efficiency in which cells can produce antibodies, insulin, and lipoproteins affects the organism's ability to fight infections, its ability to deal with sugar and fat loads, and ultimately its susceptibility to diseases. Accumulation of proteins in cells triggers UPR, which is a complex response that includes changes in transcription of genes involved in protein degradation, transport, and synthesis. In this study, we carried out gene expression studies in cells from a large number of individuals treated with two ER-stress-inducing drugs, tunicamycin and thapsigargin, to assemble a comprehensive list of genes that participate directly and indirectly in the UPR. We “rediscovered” the known genes, but we also found many genes that were not known to play a role in UPR. Among the top 100 genes that showed the most significant and highest fold induction or repression in response to ER stress, only 40% are known “UPR genes.” The validity of many of the findings is supported by “rediscovering” known UPR genes, consistent results from two independent samples and from two drugs that induce ER stress by different mechanisms. Some of the “new” genes participate in known UPR pathways such as protein transport and degradation, but the roles of others, including VLDLR and INHBE, whose expression levels were induced greatly (∼7-fold and 23-fold, respectively) in both B cells and fibroblasts, remain to be determined. VLDLR was also induced in response to ER stress in primary keratinocytes. Hence, these results provide a basis for refining our knowledge of known UPR pathways and for identifying new ones.
To examine the connection between ER function and disease susceptibility, we assessed individual variation in ER function. We found extensive variation in gene expression response to ER stress. Previously, we showed that there is extensive variation in gene expression at baseline28 and in response to radiation.29 Results from this study suggest that individuals differ not only in response to external stimuli, such as radiation, but also to cellular stress, such as protein load. Among the genes that show variable response to ER stress are chemokines, such as lymphotactin (XCL1 [MIM 600250]), which play a key role in immune response and are associated with disorders such as rheumatoid arthritis [MIM 180300]30 and IgA nephropathy [MIM 161950].31 There are also genes that are associated with type 1 diabetes, including PTPN2, major histocompatibility complex, class II, DR alpha (HLA-DR1 [MIM 142860]), and interleukin 1-alpha (IL1A [MIM 147760]),32,33 and WFS134 and CISD235 are associated with Wolfram syndromes 1 and 2, syndromic forms of diabetes. Understanding the functions of these variable UPR genes will provide a mechanistic link between ER function and human diseases.
We also examined the ER-stress response in MZ twins in order to assess genetic contributions to individual variability in response to ER stress. Results from 26 MZ twin pairs, 14 and 12 of European and African American descent, respectively, showed significant MZ twin similarity; the expression response for many genes is highly correlated within the twin pairs. These results suggest a heritable contribution to the expression response of some genes to ER stress and the existence of germline genetic variation that influences response to ER stress. In genetics of gene expression studies,36,37 we and others have shown that DNA variants influence gene expression at baseline and in response to radiation.38–43 The results from this study suggest that similar genetic approaches can be used to identify DNA sequence variants that influence the ER-stress response. Those results will provide a basic understanding of regulation of the function of the ER and provide candidate susceptibility genes for diseases that are result of ER stress, such as diabetes.
Acknowledgments
We thank Haig Kazazian and Morris Birnbaum for comments and discussions, Alan Bruzel for molecular analyses, Michael Morley and Kris Halasa for data analyses, and Colleen McGarry for manuscript preparation. This work was supported by grants from the National Institutes of Health and by a pilot grant from the University of Pennsylvania Diabetes and Endocrine Research Center.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Catalog of Published GWAS, http://www.genome.gov/gwastudies/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/
Accession Numbers
Microarray data from this study are available at NCBI's Gene Expression Omnibus (GEO), under accession no. GSE19519.
References
- 1.Hultin T. On the functions of the endoplasmic reticulum. Biochem. Pharmacol. 1961;5:359–368. doi: 10.1016/0006-2952(61)90029-6. [DOI] [PubMed] [Google Scholar]
- 2.Shands J.W., Jr., Peavy D.L., Smith R.T. Differential morphology of mouse spleen cells stimulated in vitro by endotoxin, phytohemagglutinin, pokeweed mitogen and staphylococcal enterotoxin B. Am. J. Pathol. 1973;70:1–24. [PMC free article] [PubMed] [Google Scholar]
- 3.Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 4.Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S., Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haas I.G., Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida H., Haze K., Yanagi H., Yura T., Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 7.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J.S., Shamu C.E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 9.Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 10.Kim I., Xu W., Reed J.C. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 11.Austin R.C. The unfolded protein response in health and disease. Antioxid Redox Signal. 2009;11:2279–2287. doi: 10.1089/ars.2009.2686. [DOI] [PubMed] [Google Scholar]
- 12.Todd D.J., Lee A.H., Glimcher L.H. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat. Rev. Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman R.J. Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schröder M., Kaufman R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 15.Morris J.A., Dorner A.J., Edwards C.A., Hendershot L.M., Kaufman R.J. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J. Biol. Chem. 1997;272:4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- 16.Yan W., Frank C.L., Korth M.J., Sopher B.L., Novoa I., Ron D., Katze M.G. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. USA. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokal R.R., Rohlf F.J. 3rd edition. W. H. Freeman and Co.; New York: 1995. Biometry: The principles and practice of statistics in biological research. 213–215. [Google Scholar]
- 18.McRae A.F., Matigian N.A., Vadlamudi L., Mulley J.C., Mowry B., Martin N.G., Berkovic S.F., Hayward N.K., Visscher P.M. Replicated effects of sex and genotype on gene expression in human lymphoblastoid cell lines. Hum. Mol. Genet. 2007;16:364–373. doi: 10.1093/hmg/ddl456. [DOI] [PubMed] [Google Scholar]
- 19.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booth C., Koch G.L.E. Perturbation of cellular calcium induces secretion of luminal ER proteins. Cell. 1989;59:729–737. doi: 10.1016/0092-8674(89)90019-6. [DOI] [PubMed] [Google Scholar]
- 21.Tkacz J.S., Lampen O. Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem. Biophys. Res. Commun. 1975;65:248–257. doi: 10.1016/s0006-291x(75)80086-6. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol. Cell. Biol. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R.J., Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H., Matsui T., Hosokawa N., Kaufman R.J., Nagata K., Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.-Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 29.Smirnov D.A., Morley M., Shin E., Spielman R.S., Cheung V.G. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaschke S., Middel P., Dorner B.G., Blaschke V., Hummel K.M., Kroczek R.A., Reich K., Benoehr P., Koziolek M., Müller G.A. Expression of activation-induced, T cell-derived, and chemokine-related cytokine/lymphotactin and its functional role in rheumatoid arthritis. Arthritis Rheum. 2003;48:1858–1872. doi: 10.1002/art.11171. [DOI] [PubMed] [Google Scholar]
- 31.Ou Z.L., Hotta O., Natori Y., Sugai H., Taguma Y., Natori Y. Enhanced expression of C chemokine lymphotactin in IgA nephropathy. Nephron. 2002;91:262–269. doi: 10.1159/000058402. [DOI] [PubMed] [Google Scholar]
- 32.Moore F., Colli M.L., Cnop M., Esteve M.I., Cardozo A.K., Cunha D.A., Bugliani M., Marchetti P., Eizirik D.L. PTPN2, a candidate gene for type 1 diabetes, modulates interferon-gamma-induced pancreatic beta-cell apoptosis. Diabetes. 2009;58:1283–1291. doi: 10.2337/db08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ounissi-Benkalha H., Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol. Med. 2008;14:268–275. doi: 10.1016/j.molmed.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Inoue H., Tanizawa Y., Wasson J., Behn P., Kalidas K., Bernal-Mizrachi E., Mueckler M., Marshall H., Donis-Keller H., Crock P. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome) Nat. Genet. 1998;20:143–148. doi: 10.1038/2441. [DOI] [PubMed] [Google Scholar]
- 35.Amr S., Heisey C., Zhang M., Xia X.J., Shows K.H., Ajlouni K., Pandya A., Satin L.S., El-Shanti H., Shiang R. A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am. J. Hum. Genet. 2007;81:673–683. doi: 10.1086/520961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheung V.G., Spielman R.S. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet. 2009;10:595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 41.Göring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 42.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 43.Stranger B.E., Dermitzakis E.T. The genetics of regulatory variation in the human genome. Hum. Genomics. 2005;2:126–131. doi: 10.1186/1479-7364-2-2-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


