Abstract
More than 270 million people worldwide have hearing loss that affects normal communication. Although astonishing progress has been made in the identification of more than 50 genes for deafness during the past decade, the majority of deafness genes are yet to be identified. In this study, we mapped a previously unknown autosomal-recessive nonsyndromic sensorineural hearing loss locus (DFNB91) to chromosome 6p25 in a consanguineous Turkish family. The degree of hearing loss was moderate to severe in affected individuals. We subsequently identified a nonsense mutation (p.E245X) in SERPINB6, which is located within the linkage interval for DFNB91 and encodes for an intracellular protease inhibitor. The p.E245X mutation cosegregated in the family as a completely penetrant autosomal-recessive trait and was absent in 300 Turkish controls. The mRNA expression of SERPINB6 was reduced and production of protein was absent in the peripheral leukocytes of homozygotes, suggesting that the hearing loss is due to loss of function of SERPINB6. We also demonstrated that SERPINB6 was expressed primarily in the inner ear hair cells. We propose that SERPINB6 plays an important role in the inner ear in the protection against leakage of lysosomal content during stress and that loss of this protection results in cell death and sensorineural hearing loss.
Main Text
Genetic causes of hearing loss are estimated to account for 68% of cases in newborns and 55% of cases by the age of four.1 Autosomal-recessive, dominant, and X-linked inheritance accounts for 77%, 22%, and 1% of the genetic deafness, respectively.2 Most cases of genetic deafness fall into the category of sensorineural hearing loss and are caused by pathologies of the inner ear or auditory nerves; these can be identified with audiological investigations. Hearing loss can be classified into syndromic (20%–30%) and nonsyndromic (70%–80%) forms based on the presence or absence of distinctive clinical or laboratory features. More than 50 dominant and/or recessive genes for nonsyndromic sensorineural hearing loss have been identified and most of the 35 genes for the autosomal-recessive form were initially mapped in consanguineous families or population isolates.
During our studies on hereditary deafness, which were approved by Ankara University Ethics Committee (Turkey), by the IRB at the University of Miami (USA), and by the Ethics Committee of the Institute of Child Health, Athens (Greece), we ascertained a Turkish family, family 728, in which four children with sensorineural hearing loss were born to consanguineous parents. The father, whose parents were also first cousins, had sensorineural hearing loss as well (Figure 1A). Diagnosis of sensorineural hearing loss was established via standard audiometry. Audiograms of family 728 showed that the hearing loss was moderate to severe with some residual hearing in all affected individuals (Figure 1B). All affected members of the family had oral communication with partial help of lip reading. Individual 201, at 54 years old the eldest affected in the family, had the most severe hearing loss with more severe involvement of high frequencies. The youngest affected family member, 106, was 23 years old with hearing loss involving all frequencies. He self-reported that he started having more difficulties in hearing after age 20. A progressive nature of hearing loss was reported by affected individuals but has not been verified with audiograms. The age at the onset of hearing loss could not be precisely determined because previous audiograms were not available. The remainder of the examination was completely normal including normal anterior chamber and fundus of the eyes. Affected individuals did not have delays in gross motor development. Neither did they have balance problems, vertigo, dizziness, or spontaneous and positional nystagmus. Tandem walking was normal and Romberg test was negative. CT scans of the temporal bone in two affected family members were normal as well. EKGs, liver and kidney function tests, serum electrolytes, urinalysis, CBC, and leukocyte subgroups were all within normal limits in affected subjects.
Figure 1.

The DFNB91 Locus, Audiograms, and Molecular Studies in Family 728
(A) Haplotypes created with SNP markers show complete cosegregation of a locus at 6p25 (DFNB91) in family 728.
(B) Audiograms of seven members of family 728. 301, 102, and 103 have heterozygous, 201, 101, 105, and 106 have homozygous p.E245X mutation in SERPINB6.
(C) Electropherograms showing the p.E245X (c.733G>T) mutation in SERPINB6.
(D) Relative quantity values of SERPINB6 cDNA obtained from peripheral blood leukocytes of homozygotes and heterozygotes in family 728. Difference between homozygotes and heterozygotes is statistically significant (p = 0.034).
(E) The western blotting results of SERPINB6 in peripheral blood leukocytes in homozygous and heterozygous members of family 728. The 42 kDa protein is clearly visible in NIH/3T3, HeLa cells, and a healthy person with wild-type SERPINB6. A reduced SERPINB6 band is visible in a heterozygote. The SERPINB6 protein band is completely absent in homozygotes.
DNA was extracted from peripheral leukocytes of each member of family 728 via phenol chloroform method. Two affected individuals (728-101 and 728-201) were prescreened and found to be negative for mutations in GJB2 (MIM 121011) via direct sequencing of both exons and for the m.1555A>G mutation in MTRNR1 (MIM 561000) via an RFLP method via previously reported protocols.3,4 Genome-wide SNP genotyping was done in eight members of family 728 (201, 301, 101, 102, 103, 104, 105, and 106) via Illumina 1M duo beadchips and assays (Illumina, CA). The DNA samples were processed according to Illumina procedures for processing of the Infinium II assay, the BeadChips were scanned on the Illumina BeadArray Reader, and data were extracted by the Illumina Beadstudio software (Illumina). Before analysis, overall sample call rate, gender consistency checks, relationship inference (via the Graphical Representation of Relationships program),5 and the Mendelian inconsistency rates were used for quality assessment. Genotypes were transferred into Excel files and sorted according to genomic positions along with all 35 previously identified autosomal-recessive nonsyndromic deafness genes. The cosegregation of the flanking genotypes for each gene was visually evaluated. None of the known deafness loci cosegregated with the phenotype, thereby excluding these as possible causes and suggesting that a mutation in a previously unknown deafness gene was responsible in this family. Copy number variants (CNVs) were assessed by determining the relative loss or gain of fluorescent signal intensity from SNP or CNV probes on the array via PennCNV program6 and no segregating CNVs were detected.
Regions of autozygosity from the SNP screen were first sought with PLINK.7 All five affected members of family 728 were concordant for homozygous SNP marker blocks that were larger than one megabase in five chromosomal locations (Table S1 available online). Haplotypes were constructed for homozygous genomic segments and their cosegregation with deafness was assessed visually. Only the homozygous block between 289,878 bp (rs7762811) and 3,908,468 bp (rs13205752) (total length was 3,618,590 bp) at 6p25 cosegregated with hearing loss in the family (Figure 1A) (NCBI Build 36.1; hg18). The other four large shared homozygous streches in affected family members did not cosegregate with the phenotype and were not further analyzed. A genome-wide two-point linkage analysis was performed with SuperLink.8 Sixteen SNP markers on chromosomes 6, 8, 16, and 21 were detected with a LOD score of more than 3 (Table S2). Visual evaluation of haplotypes in these regions confirmed that the largest autozygous segment cosegregating with the phenotype was the same 3,618,590 bp region at 6p25. There were five other cosegregating segments on chromosomes 8, 16, and 21 with much smaller sizes ranging from 8,791 bp to 273,430 bp (Table S2). Before concentrating on the largest autozygous region on chromosome 6, the other five cosegregating regions were evaluated for their gene content with the UCSC genome browser. There were no known genes within the regions at chromosomes 16 and 21. The cosegregating segment on chromosome 8 was 14,852 bp long and partially included MSRA (MIM 601250). Sequencing of all six exons as well as intron-exon boundaries of this gene in two affected members of family 728 did not detect a mutation.
The only remaining autozygous segment that could contain a previously unrecognized deafness gene in family 728 was at 6p25. Multipoint linkage analysis of this segment was conducted with Fastlink9 assuming a fully penetrant autosomal-recessive disease, with a disease allele frequency of 0.0001. Both inbreeding loops were retained in the analysis. SNPs spanning the homozygous region were chosen for linkage analysis based on tagging and heterozygosity in the parents. Allele frequencies were obtained from the CEU HapMap. Linkage analysis detected a maximum lod score of 5.0 between recombinant markers rs7762811 and rs13205752 at 6p25 (Figure 1A). Examination of haplotypes between these two SNPs not included in the multipoint analysis did not identify any recombinations. This locus was designated as DFNB91 by the HUGO Nomenclature Committee.
The linkage interval at 6p25 included 24 known protein coding genes listed on the UCSC database (Figure S1). None of these genes have been previously reported to cause any forms of hearing loss in humans or in animals. The coding exons and intron-exon boundaries of all 24 genes were sequenced in family 728 (Figure S1 and Table S3). A homozygous nonsense mutation, p.E245X (c.733G>T) (according to GenBank accession number NM_004568.4), was identified in SERPINB6 (MIM 173321) in all affected members of family 728 (Figure 1C). No other DNA sequence change in this gene was detected. This mutation removes 131 amino acids in the carboxy terminal of the protein including the reactive center loop. It completely cosegregated with deafness in family 728. The identified mutation was not found in 300 Turkish controls via an amplification refractory mutation detection system (ARMS) protocol that was shown to be sensitive and specific with DNA sequence analysis-proven homozygous, heterozygous, and wild-type samples (Table S3). One previously unreported intronic change and 21 previously reported SNPs were detected in other sequenced genes. None of these changes were predicted to have an effect on function (Table S4).
A premature stop codon was likely to activate nonsense-mediated mRNA decay response, thus leading to a decreased SERPINB6 mRNA expression. SERPINB6 was known to be expressed in white blood cells. Total RNA was isolated from peripheral blood samples of seven members of family 728 and cDNA was synthesized. The sequencing of SERPINB6 in cDNA containing the point mutation with exonic primers detected the p.E245X (c.733G>T) mutation in both homozygotes and heterozygotes, showing that the identified mutation was present at the mRNA level. The quantitative expression analysis of SERPINB6 was performed with TaqMan PCR assays in quadriplicate for each individual on cDNA samples via ABIPrism 7900 HT Sequence Detection system 2.3 (Applied Biosystems Inc., CA) (for methods, see Table S3). The mRNA expression was significantly reduced in homozygotes compared to those in heterozygotes (Figure 1D) (p = 0.034; Mann-Whitney U test), indicating that the premature stop codon destabilizes mRNA, leading to mRNA decay.
The protein expression of the mutant SERPINB6 was evaluated in peripheral leukocytes of homozygotes and heterozygotes (for methods, see Table S3). The expected SERPINB6 band was not detected in homozygotes, whereas the 42 kDa protein was visible in a normal control and a heterozygote (Figure 1E). The trunctated protein product of SERPINB6 with the p.E245X mutation was estimated to be 27 kDa via Current Protocols database dna-rna-protein molecular weight calculator software. A truncated protein was not observed in heterozygotes or homozygotes, suggesting that the truncated protein is either not produced or rapidly degraded.
For identification of other families with SERPINB6 mutations, the probands of a total of 256 unrelated multiplex families with autosomal-recessive nonsyndromic sensorineural hearing loss were screened. These families were from Turkey (202), Greece (26), and the USA (28). The vast majority of affected members of these families had severe to profound congenital or prelingual onset sensorineural hearing loss. Sequencing of all seven exons revealed seven additional previously unreported single-nucleotide changes (Table S5). However, these changes are unlikely to be the cause of phenotype because (1) they did not cosegregate with deafness in the families and (2) in silico analyses with PolyPhen and SIFT predicted that they do not affect protein function and are therefore likely to be benign. An additional 286 Palestinian probands from multiplex autosomal-recessive nonsyndromic deafness families were screened for the p.E245X mutation with a restriction fragment length polymorphism (RFLP) method because the mutation removes the recognition site of the BslI restriction enzyme (Table S3). None of the samples were found to be positive for the screened mutation.
Given the observed hearing loss in humans, immunostaining and in situ hybridization were used to identify the site for Serpinb6 expression in developing mouse inner ear. Immunohistochemistry was performed with developing mouse inner ear tissues according to the protocol as described before.10 In brief, frozen sections of the inner ear tissues were dried for 15 min at 37°C and rehydrated in 1×PBS for 5 min. Then, the sections were subjected to an antigen unmasking treatment via the Antigen Unmasking Solution (Vector Laboratories, UK) according to the manufacturer's protocol. The blocking, primary antibody and second primary antibody incubation followed the standard protocol. Antibodies used in the study are goat anti-SERPINB6 (human) (sc-21143, Santa Cruz Biotechnology, CA); rabbit anti-Myo7a (Proteus BioSciences Inc., CA), and rabbit anti-Ptprq (a gift from Dr. Guy Richardson, University of Sussex, UK). The secondary antibodies were anti-goat Alexa 594 and anti-rabbit Alexa 488 (Invitrogen Corporation, CA) for fluorescent labeling. Data visualization and acquisition were performed with a Zeiss Axioscope 2 fluorescent microscope. A weak Serpinb6 signal was detected in E13.5 utricle sensory epithelium but not in hair cells (Figures 2A–2D). At E16.5, more prominent Serpinb6 was detected in crista hair cells (Figures 2E–2H). Hair cell expression of Serpinb6 was sustained in postnatal mice (data not shown). In cochlea, Serpinb6 was detected in cochlear hair cells in embryo (Figures 2I–2L) and in hair cells and the greater epithelial ridge (GER) region in early postnatal age (Figures 2M–2P). In both cochlear and utricular hair cells, Serpinb6 was found in cytoplasm (Figures 2E and 2M). There are five mouse Serpinb6 orthologs including Serpinb6a, b, c, d, and e, and the human SERPINB6 antibody recognizes mouse Serpinb6a and Serpinb6c. Mouse Serpinb6a has the highest homology to human SERPINB6 with 75% identity, whereas Serpinb6c is 68% identical to SERPINB6. To further confirm immunohistochemistry results from the human SERPINB6 antibody study, we performed in situ hybridization with the mouse Serpinb6a antisense probe. The mouse Serpinb6a cDNA was purchased from Thermo Scientific Open Biosystems (clone ID: 5342439), and the preparation of riboprobes and in situ hybridization followed the protocol described.11 The Serpinb6a in situ results matched that from immunostaining with weak expression in E18.5 and significantly increased expression in P6 GER and cochlear hair cells (Figures 2Q and 2S).
Figure 2.
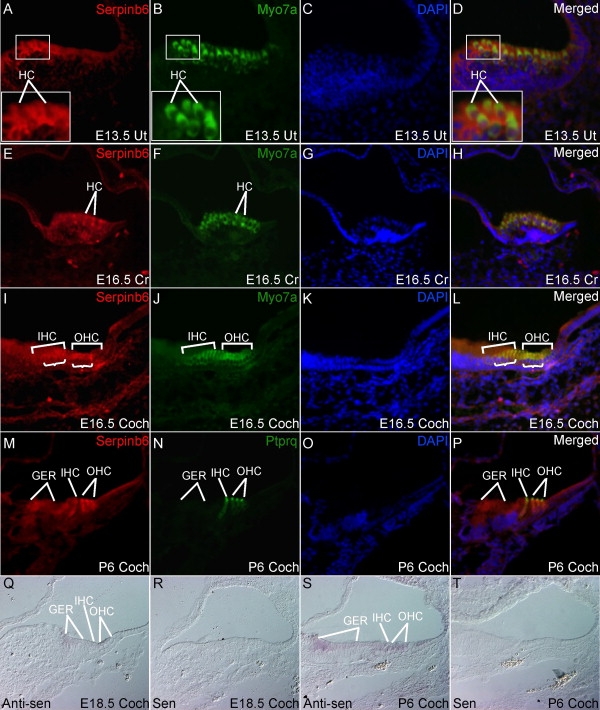
Detection of Serpinb6 in the Developing Mouse Inner Ear by Immunohistochemistry and In Situ Hybridization
(A–D) Weak Serpinb6 was detected in E13.5 utricular sensory epithelium. However, Serpinb6 was not in utricular hair cells, which were labeled with myo7a (B). Lines show examples of hair cells that are devoid of Serpinb6 (A).
(E–H) Distinct Serpinb6 was detected in E16.5 crista hair cells (lines).
(I–L) Weak Serpinb6 was detected in E16.5 cochlear hair cells. Multiple inner and outer hair cells were shown due to a section angle (J).
(M–P) Serpinb6 was upregulated in P6 inner and outer hair cells (IHC, OHC), with mainly cytoplasm distribution. Ptprq labeled hair bundles (N)40. In addition, Serpinb6 was detected in the GER at this stage.
(Q–T) In situ hybridization showed very weak Serpinb6a expression in E18.5 cochlear hair cells and GER (Q), whereas the expression was significantly increased in P6 cochlear hair cells and GER (S). Control sense probe did not produce any signal (R, T).
Abbreviations: Coch, cochlea; Cr, crista; Ut, utricle; anti-sen, antisense probe; sen, sense probe. Magnification: 40×.
Proteases are important components of a number of physiological processes, including coagulation, cell migration, phagocytosis, fibrinolysis, cell-mediated cytotoxicity, complement fixation, and apoptosis. To avoid nonspecific tissue damage, protease activity is regulated by inhibitors, such as members of the serpin superfamily. Inhibitory serpins bind their target proteases irreversibly, via a conformational change that deforms the protease and stabilizes the serpin-protease complex.12 Serpin dysfunction or deficiency is the underlying factor in a variety of human diseases, including emphysema, angioedema, thrombosis, and dementia13,14 because of uncontrolled tissue damage by different proteases.
In humans, the largest serpin clade (clade B) contains 13 intracellular proteins.13 Although their physiological roles remain largely unknown, there is evidence that they are involved in the regulation of cell growth, differentiation, and cytoprotection. A distinct subset of clade B serpins comprises SERPINB1 (MIM 130135), SERPINB6, and SERPINB9 (MIM 601799), which are encoded by a gene cluster on chromosome 6.15 SERPINB6 (also named placental thrombin inhibitor, PTI, cytoplasmic antiproteinase, and protease inhibitor-6) was discovered in 1993 in human placental tissue.16 Its reactive center is located at Arg-341 and Cys-342, and it lacks a classical N-terminal signal sequence, showing that it is not an extracellular protein.17 Intracellular serpins, Serpins b6, b9, and b1 in particular, have been implicated in cytoprotective roles. For example, Serpinb9 inhibits the cytotoxic lymphocyte protease granzyme B.18,19 Serpinb6 is a potent inhibitor of the monocyte/granulocyte protease cathepsin G, which is stored in azurophilic granules and then released into phagolysosomes or secreted during inflammation,20 and kallikreins,21 which are stored in cytoplasmic storage vesicles analogous to leukocyte granules. Any proteases released into the interior of the cell during cellular stress would be rapidly inactivated by intracellular serpins. Although cells can tolerate and repair lysosomal ruptures to a certain degree, as the amount of damage increases, cells may undergo apoptosis or necrosis. A dramatic support for this model has recently been reported where a C. elegans intracellular serpin, srp-6, exhibited a prosurvival function by blocking necrosis. Minutes after hypotonic shock, srp-6 null animals underwent a catastrophic series of events culminating in lysosomal disruption, cytoplasmic proteolysis, and death. This “hypo-osmotic stress lethal” phenotype was dependent upon calpains and lysosomal cysteine peptidases, two in vitro targets of srp-6. By protecting against both the induction of and the lethal effects from lysosomal injury, srp-6 also blocked death induced by heat shock, oxidative stress, hypoxia, and cation channel hyperactivity.22
To assess the effects of overexpression of wild-type and mutant SERPINB6 under osmotic stress, we studied transiently transfected human cell lines and evaluated the integrity of the lysosomal content. For the evaluation of the lysosomal integrity, we used a pH-dependent marker of lysosomes, LysoTracker-red, in cultured cells as reported previously.23 The lysates of HeLa cells were probed for SERPINB6 with monoclonal antibody (SA-622, Enzo LifeSciences, PA), which showed that HeLa cells expressed SERPINB6 endogenously. Constructs encoding Serpinb6-GFP fusion protein were subcloned from an untagged Human cDNA clone (SC321547, OriGene Technologies, MD) via PCR amplification and TOPO cloning via the pcDNA3.1/NT-GFP-TOPO cloning kit (Invitrogen Corporation, CA). The primer sequences used for generating the constructs are available in Table S3. In brief, the SERPINB6-GFP-F primer was used in conjunction with the SERPINB6-WT-R or the SERPINB6-MUT-GFP-R to generate a PCR amplicon for either the wild-type or mutant constructs, respectively. The proper sized amplicon was selected for gel extraction and then cloned into the pcDNA3.1/NT-GFP-TOPO. Colonies were selected and screened via Sanger sequencing with Applied Biosystems BigDye Terminator Cycle Sequencing Kit (Applied Biosystems Inc., CA).
HeLa cells were transfected with the SERPINB6-WT-GFP, the SERPINB6-MUT-GFP, and the pcDNA3.1-Control-GFP expression vectors in 96-well BD Falcon plates (353948, BD Bioscience, CA) with Lipofectamine 2000 and incubated at 37°C with 5% CO2. Twenty-four hours after transfection, cells were incubated in a 75 nM working concentration of LysoTracker Red 99-DND (Invitrogen Corporation) and then either continued to be cultured in PBS or introduced to osmotic shock by replacing the culture media with molecular biology grade water. After 5 min, plates were measured on Cyntellect's adherent cell cytometer Celigo (Celigo Inc., CA). Three-channel, 8-bit stitched images were generated covering whole wells to identify the surface area and number of cells along with fluorescent intensities. We measured three channels: bright field, to count and locate cells; green, to measure number and intensity of the expressed recombinant protein; and red, to measure intensity of LysoTracker. Images were analyzed with the Celigo software (Celigo Inc.). The intensity of the red channel signal was averaged per each replicate in PBS and water. A Student's two-tailed t test was performed by comparing two histograms of intensity distributed over 35 bins from 0 to 255, representing the dynamic range at which intensity can be measured across an 8-bit image.
HeLa cells transfected with wild-type SERPINB6 showed less reduction in red florescence (lysosomal content assessed through the maintenance of their lysosomal pH gradient and acidic environment) than that of the truncated mutant protein or the control GFP expression vector. Differences for the average intensities of LysoTracker between PBS and water incubation were 43.23%, 65.48%, and 60.47% for SERPINB6-WT-GFP, SERPINB6-Mut-GFP, and control-GFP, respectively (Figure 3), with all shifts being found statistically significant with p values of 0.011, 0.00005, and 0.0001, respectively. The SERPINB6-Mut-GFP and control-GFP showed 44.4% and 39.0% more intensity decrease, respectively, in LysoTracker signal as compared to the SERPINB6-WT-GFP protein when incubated in water; p values are 0.003 and 0.024, respectively.
Figure 3.
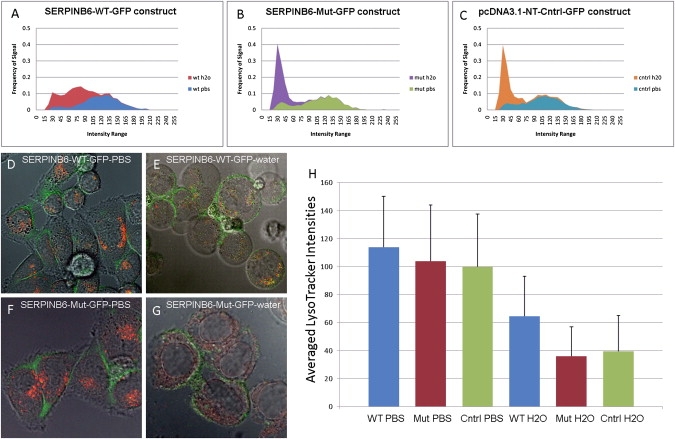
The Effects of Transient Overexpression of SERPINB6 on Lysosomes in HeLa Cells
(A–C) Distribution of LysoTracker Red 99-DND Florescence in HeLa cells transfected with different GFP constructs. Distribution of the intensity range and frequency of LysoTracker-red was different in SERPIN-B-6-WT cells from both mutant and control cells after water treatment. Histograms show that most lysosomes lost their integrity in mutant and control cells, while some lysosomes remain less disrupted in SERPINB6-WT cells.
(D–G) Representative examples of images of wild-type and mutant SERPINB6-transfected HeLa cells. Green signal shows the overexpressed wild-type or mutant SERPINB6, which is localized in the cytoplasm. Red signal shows intact lysosomes. After osmotic stress, cells became round and lysosomes became indistinguishable with a general pinkish hue in the cytosol. However, this effect was less pronounced in cells where wild-type SERPINB6 is overexpressed.
(H) Averaged LysoTracker Intensities with standard deviation with PBS and water. Averaged LysoTracker intensity difference in WT SERPINB6-overexpressed cells is less than those in mutant SERPINB6-overexpressed and control cell lines (p = 0.003 and 0.024, respectively, for the mutant and control cells).
Further, we observed that cells incubated in PBS show discernable red structures representing lysosomes. When PBS was replaced with water, LysoTracker morphology changed from bright punctuate spots to red spots with a general pinkish hue in the cytosol (Figure 3). This shift can also be seen in the histogram distributions (Figures 3A–3C), which indicates that lysosomes cannot maintain their acidic environment, leading to leakage and disruption.23 Observed changes in red fluoresecence was roughly equivalent between the Control-GFP and the SERPINB6 Mutant while disintegrity of lysosomes in cells from the overexpressed SERPINB6-WT constructs was significantly less (Figures 3A–3G). These results demonstrate that wild-type SERPINB6 has a protective effect for lysosomal integrity in osmotic stress in a human cell line, similar to the results of a previous study for srp-6 shown in C. elegans.22 Loss of this protective effect, as shown in our experiments with the truncated protein, is associated with increased lysosomal disintegrity. We also showed that the truncated protein is localized in the cytoplasm, similar to the wild-type SERPINB6, and did not have an additional adverse effect on lysosomal integrity. These results are consistent with the theory that the phenotype associated with the homozygous truncated SERPINB6 is due to loss of function of the protein.
Given its increased expression in hair cells in the aging inner ear, it is highly plausible to envision a role for serpins in the inner ear cells, and particularly in hair cells, in the protection against various noxious stimuli including noise, ototoxic drugs, or trauma during aging process. These stimuli as well as normal physiological hearing processes may induce cellular stress, such as oxidative stress, that results in death of hair cells as individuals get older.24 Leakage of lysosomal content resulting from various stress conditions is likely to occur in hair cells. To reduce hair cell damage and remain functional, SERPINB6 may be required to counter the potentially cytotoxic components of leaking lysosomal proteases.
In conclusion, several lines of evidence supported the hypothesis that the identified SERPINB6 mutation was responsible for hearing loss: (1) it completely cosegregated with hearing loss in a large family; (2) the identified mutation removed the functional domain of the SERPINB6 protein, SERPINB6 mRNA expression was reduced, and protein expression was absent in homozygotes; (3) the identified mutation was absent in 300 ethnically matched controls; and (4) SERPINB6 was expressed in the inner ear, in particular in hair cells. The phenotype associated with a SERPINB6 mutation was moderate to severe, likely to be progressive sensorineural hearing loss. Absence of SERPINB6 mutations in a large number of probands with congenital severe to profound hearing loss suggests that the typical phenotype caused by SERPINB6 mutations is not congenital profound deafness. Mutations in recessive deafness genes usually cause severe to profound congenital or prelingual onset sensorineural hearing loss. Nevertheless, biallelic mutations in LOXHD1 (MIM 613072), MYO3A (MIM 606808), and PJVK (MIM 610219) have been reported to cause progressive sensorineural hearing loss, suggesting that age-dependent hair cell failure is a common mechanism for progressive autosomal-recessive sensorineural hearing loss.25–27 The loss of function of an intracellular protease inhibitor, SERPINB6, resulting from a homozygous truncating mutation provides a highly plausible model for age-dependent hair cell failure.
Acknowledgments
We are thankful to all participating families. We thank Professor Phillip I. Bird from Monash University, Australia, for helpful suggestions and critical reading of the manuscript. This study was partially supported by The Scientific & Technological Research Council of Turkey Grants (105S464 and 108S045), funds from the University of Miami to M.T., NIH R01 DC006908 to Z.-Y.C., National Natural Science Foundation of China to Z.-Y.C. (30728030), the Federick and Ines Yeatts inner ear hair cell regeneration fellowship to M.H., NIH R01 DC005575 to X.Z.L., and a grant from Oticon Fonden, Denmark, to M.B.P.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Current Protocols, http://www.currentprotocols.com/tools/dnarnaprotein-molecular-weight-calculator
Hereditary Hearing Loss Homepage, http://webh01.ua.ac.be/hhh/
National Institutes of Health (NIH) HapMap-CEU, http://www.ncbi.nih.gov/SNP/snp_viewTable.cgi?pop=1409
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
PolyPhen, http://genetics.bwh.harvard.edu/pph/
SIFT, http://sift.jcvi.org/
UCSC Genome Browser, http://genome.ucsc.edu
References
- 1.Morton C.C., Nance W.E. Newborn hearing screening—A silent revolution. N. Engl. J. Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Morton N.E. Genetic epidemiology of hearing impairment. Ann. N Y Acad. Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 3.Tekin M., Duman T., Bogoclu G., Incesulu A., Comak E., Fitoz S., Yilmaz E., Ilhan I., Akar N. Frequency of mtDNA A1555G and A7445G mutations among children with prelingual deafness in Turkey. Eur. J. Pediatr. 2003;162:154–158. doi: 10.1007/s00431-002-1129-z. [DOI] [PubMed] [Google Scholar]
- 4.Tekin M., Arici Z.S. Genetic epidemiological studies of congenital/prelingual deafness in turkey: Population structure and mating type are major determinants of mutation identification. Am. J. Med. Genet. A. 2007;143A:1583–1591. doi: 10.1002/ajmg.a.31702. [DOI] [PubMed] [Google Scholar]
- 5.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. GRR: Graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 6.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: An integrated hidden markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishelson M., Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18(Suppl 1):S189–S198. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 9.Schäffer A.A. Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum. Hered. 1996;46:226–235. doi: 10.1159/000154358. [DOI] [PubMed] [Google Scholar]
- 10.Sage C., Huang M., Vollrath M.A., Brown M.C., Hinds P.W., Corey D.P., Vetter D.E., Chen Z.Y. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc. Natl. Acad. Sci. USA. 2006;103:7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M., Sage C., Li H., Xiang M., Heller S., Chen Z.Y. Diverse expression patterns of LIM-homeodomain transcription factors (LIM-HDs) in mammalian inner ear development. Dev. Dyn. 2008;237:3305–3312. doi: 10.1002/dvdy.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huntington J.A., Read R.A., Carrell R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 13.Silverman G.A., Bird P.I., Carrell R.W., Church F.C., Coughlin P.B., Gettins P.G.W., Irving J.A., Lomas D.A., Luke C.J., Moyer R.W. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 14.Stein P.E., Carrell R.W. What do dysfunctional serpins tell us about molecular mobility and disease? Nat. Struct. Biol. 1995;2:96–113. doi: 10.1038/nsb0295-96. [DOI] [PubMed] [Google Scholar]
- 15.Scott F.L., Eyre H.J., Lioumi M., Ragoussis J., Irving J.A., Sutherland G.A., Bird P.I. Human ovalbumin serpin evolution: phylogenic analysis, gene organization, and identification of new PI-8-related genes suggest that two interchromosomal and several intrachromosomal duplications generated the gene clusters at 18q21-q23 and 6p25. Genomics. 1999;62:490–499. doi: 10.1006/geno.1999.6021. [DOI] [PubMed] [Google Scholar]
- 16.Coughlin P.B., Tetaz T., Salem H.H. Identification and purification of a novel serine proteinase inhibitor. J. Biol. Chem. 1993;268:9541–9547. [PubMed] [Google Scholar]
- 17.Scott F.L., Coughlin P.B., Bird C., Cerruti L., Hayman J.A., Bird P. Proteinase inhibitor 6 cannot be secreted, which suggests it is a new type of cellular serpin. J. Biol. Chem. 1996;271:1605–1612. doi: 10.1074/jbc.271.3.1605. [DOI] [PubMed] [Google Scholar]
- 18.Bird C.H., Sutton V.R., Sun J., Hirst C.E., Novak A., Trapani J.A., Bird P.I. Selective regulation of apoptosis: The cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 1998;18:6387–6398. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirst C.E., Buzza M.S., Bird C.H., Warren H.S., Cameron P.U., Zhang M., Ashton-Rickhardt P.G., Bird P.I. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J. Immunol. 2003;170:805–815. doi: 10.4049/jimmunol.170.2.805. [DOI] [PubMed] [Google Scholar]
- 20.Scott F.L., Hirst C.E., Sun J., Bottomley S.P., Bird C.H., Bird P.I. The intracellular serpin proteinase inhibitor 6 (PI-6) is expressed in monocytes and granulocytes and is a potent inhibitor of the azurophilic granule proteinase, cathepsin G. Blood. 1999;93:2089–2097. [PubMed] [Google Scholar]
- 21.Scott F.L., Sun J., Whisstock J.C., Kato K., Bird P.I. SerpinB6 is an inhibitor of kallikrein-8 in keratinocytes. J. Biochem. 2007;142:435–442. doi: 10.1093/jb/mvm156. [DOI] [PubMed] [Google Scholar]
- 22.Luke C.J., Pak S.C., Askew Y.S., Naviglia T.L., Askew D.J., Nobar S.M., Vetica A.C., Long O.S., Watkins S.C., Stolz D.B. An intracellular serpin regulates necrosis by inhibiting the induction and sequelae of lysosomal injury. Cell. 2007;130:1108–1119. doi: 10.1016/j.cell.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng J., Yan T., Feng Y., Zhai Q. Involvement of lysosomes in the early stages of axon degeneration. Neurochem. Int. 2010;56:516–521. doi: 10.1016/j.neuint.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Van De Water T.R., Lallemend F., Eshraghi A.A., Ahsan S., He J., Guzman J., Polak M., Malgrange B., Lefebvre P.P., Staecker H. Caspases, the enemy within, and their role in oxidative stress-induced apoptosis of inner ear sensory cells. Otol. Neurotol. 2004;25:627–632. doi: 10.1097/00129492-200407000-00035. [DOI] [PubMed] [Google Scholar]
- 25.Grillet N., Schwander M., Hildebrand M.S., Sczaniecka A., Kolatkar A., Velasco J., Webster J.A., Kahrizi K., Najmabadi H., Kimberling W.J. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am. J. Hum. Genet. 2009;85:328–337. doi: 10.1016/j.ajhg.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh T., Walsh V., Vreugde S., Hertzano R., Shahin H., Haika S., Lee M.K., Kanaan M., King M.C., Avraham K.B. From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Natl. Acad. Sci. USA. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delmaghani S., del Castillo F.J., Michel V., Leibovici M., Aghaie A., Ron U., Van Laer L., Ben-Tal N., Van Camp G., Weil D. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat. Genet. 2006;38:770–778. doi: 10.1038/ng1829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


