To the Editor: Sheridan and colleagues recently reported that mutations in the tubulin gene TUBA8 result in polymicrogyria with optic nerve hypoplasia (PMGOH [MIM 613180]).1 This conclusion is based on the mapping of two consanguineous families of Pakistani origin to a 7.42 Mb region on chromosome 22q11.2 that contains ∼230 genes including TUBA8. Drawing on our previous finding that mutations in TUBA1A cause lissencephaly2 and that mutations in TUBB2B cause asymmetric polymicrogyria,3 Sheridan and colleagues sequenced TUBA8 and found a 14 bp deletion in intron 1 that affects splicing. They provide further evidence that TUBA8 is involved in the disease state by analyzing its expression in the developing mouse brain by in situ hybridization. They report that Tuba8 is widely expressed in developing neural structures, with strongest expression in the cortical plate at E15.5 and E18.5 and in the cortical plate, subplate, and hippocampus at P0.
A meaningful analysis of individual tubulin gene expression by in situ hybridization requires the use of probes that avoid cross-hybridization among the highly conserved coding regions, relying exclusively on either the variant 5′ or 3′ untranslated regions. The probe employed by Sheridan and colleagues was 443 nucleotides in length, of which 415 correspond to sequences contained within the conserved coding region. Consequently, this probe shares a very high sequence homology with other α-tubulins.4 An Ensembl BLAST search with the Sheridan probe against total mouse cDNA results in six other hits, each being at least 300 nucleotides in length with at least 80% sequence identity. Each of these hits corresponds to one of the six other members of the α-tubulin family and includes a 374 nucleotide stretch that shares 84.2% identity with the coding sequence of Tuba1a, a gene that is highly expressed in the developing CNS.5
To establish whether the results reported by Sheridan and colleagues are a consequence of cross-hybridization, we conducted in situ hybridization on the developing (E14.5, E16.5, and P0) and adult mouse brain employing their probe and two others that we designed. We first confirmed the sequence of Tuba8 mRNA by amplifying and cloning it from a C57BL/6 mouse adult olfactory bulb cDNA library (AK032157, GU591980) (Figure S1 available online). Next we designed two probes, a short probe (224 base pairs in length [nt 1569–1792]) and a long probe (545 base pairs in length [nt 1367–1911]) targeted to the highly variant 3′ UTR of Tuba8 (GU591980). These probes, along with the probe employed by Sheridan, were cloned into a pCRII-TOPO vector (Invitrogen, 45-0640), and the sequence was confirmed by Big Dye sequencing with a 3730 DNA Analyzer (Applied Biosystems). We checked the specificity of our probes by conducting an Ensembl BLAST search against total mouse cDNA. This revealed no other hits that could potentially form a stable hybrid with either our short or long UTR probes. After linearization of the probes, a T7 or SP6 promoter-driven in vitro transcription reaction was done in the presence of a DIG labeling mix (Roche, 11175025910). The denatured probes (sense or antisense) were then applied to pretreated coronal sections and hybridized overnight in a custom-built chamber.6 Staining was visualized by incubating sections with an alkaline phosphatase conjugated digoxygenin antibody (1:2000, 4°C, overnight) (Roche, 11093274910), followed by the application of BM-Purple AP (Roche, 1144207400).
When we used Sheridan's probe, we observed strong staining at E14.5, E16.5, and P0, particularly in the cortical plate, and to a lesser extent in the intermediate and ventricular zones (Figure 1). In contrast, we did not observe any signal at any of these time points with our Tuba8 3′UTR-specific probes, although we did observe positive staining in the Purkinje cell layer in the cerebellum and in the mitral and granule cell layers of the olfactory bulbs in the adult brain (Figure S2).
Figure 1.
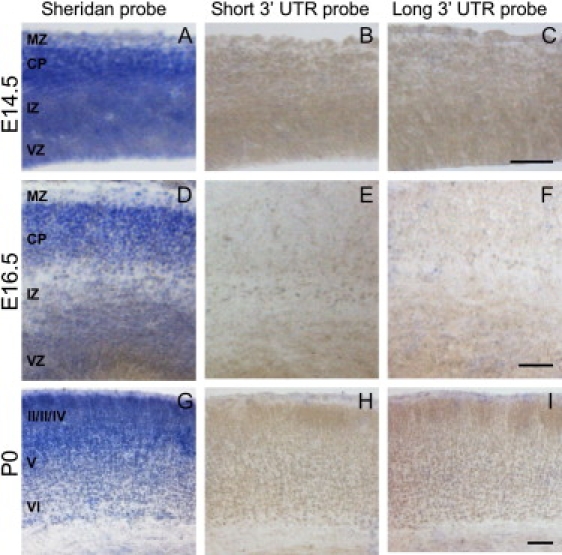
Tuba8 Expression in the Developing Mouse Brain
Coronal sections (14 μm) of the developing cortex (E14.5, E16.5, P0) showing our in situ hybridization results obtained with the probe employed by Sheridan and colleagues (A, D, G) and the short (B, E, H) and long (C, F, I) probe targeted to the unique 3′ UTR of Tuba8. High levels of staining are observed in the developing mouse brain with the Sheridan probe (particularly in the cortical plate [CP] and to a lesser extent in the intermediate zone [IZ] and ventricular zone [VZ]), but none is apparent with probes that exclusively target the 3′ UTR. We observed no staining when employing control sense probes (data not shown). Scale bars represent 50 μm. MZ indicates the marginal zone.
Because there is evidence for an alternatively spliced variant of Tuba8 with a shorter UTR, we confirmed our in situ hybridization results by undertaking real-time quantitative PCR (qPCR) with primers targeted to the coding region (Table S1).7 When designing intron-spanning primers for this task, we took care to ensure that each primer pair was specific, undertaking BLAST searches with multiple search engines (Ensemble, UCSC, and NCBI). In addition, we only used primer pairs with an efficiency between 95% and 105% that produced a melt curve indicative of a single product. To prepare templates for amplification, we dissected and flash froze embryonic brains (E10.5 [head], E12.5, E14.5, E16.5, E18.5), postnatal brains (P0, P6), various regions of the adult brain (8 weeks) and adult organs (C57BL/6), prior to RNA extraction (QIAGEN, 74804, 74704). Three independent RNA samples from each developmental time point, brain region, or organ were then quantitated and pooled for reverse transcription (Invitrogen, 45-0640). Quantitative PCR followed on a BioRad Cycler with SYBR green, alongside reverse transcriptase (RT) and cDNA negative controls. All reactions were performed in triplicate and normalized to the geometric mean of three reference genes (Pgk1, Tfrc, and Hprt).8,9
We compared the expression of Tuba8 to those tubulin genes (Tuba1a [MIM 611603], Tubb2b [MIM 610031], Tubb3 [MIM 600638]) that have been shown to cause distinct neurodevelopmental diseases.10 We found that Tuba 1a, Tubb2b, and Tubb3 are highly expressed in the developing mouse brain from E12.5 to P6, whereas Tuba8 is expressed at low levels (Figure 2A). For instance, at E14.5, a peak time for neuronal migration, Tuba1a is expressed at ∼94 times the control gene level, whereas Tuba8 is barely detectable, expressed at just 0.03 times the control gene level. We next analyzed expression levels of Tuba8 in different regions of the adult mouse brain by qPCR, again comparing it to Tuba1a, Tubb2b, and Tubb3 (Figure 2B). We observed regional differences in the expression levels of these genes, but generally Tuba1a was most highly expressed, followed by Tubb3 and Tubb2b. Tuba8 was again expressed at very low levels, with the exception of the olfactory bulbs and the cerebellum, where its modest level of expression was concordant with the results from our in situ hybridization experiments. We extended our expression analysis beyond the adult brain to the major adult organs in the mouse (Figure S3). We found that the level of Tuba8 expression is highest in testis (∼12 times control gene expression), followed by skeletal and heart muscle, confirming the results of Stanchi and colleagues who studied TUBA8 expression in adult human tissues.7
Figure 2.
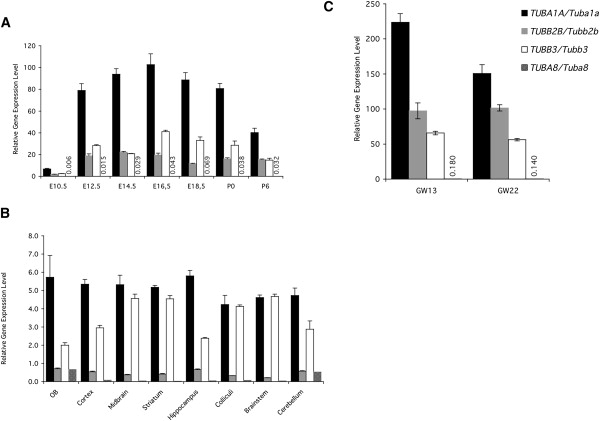
Real-Time qPCR Analysis of Tuba8 Expression Levels in Mouse and Human Brain
(A) Relative expression levels of Tuba1a, Tubb2b, Tubb3, and Tuba8 in the developing mouse brain (E10.5 [head], E12.5, E14.5, E16.5, E18.5, P0, and P6). Tuba8 is expressed at very low levels at all developmental stages.
(B) In the adult mouse brain, there are regional expression differences when comparing Tuba1a, Tubb2b, Tubb3, and Tuba8. The expression of Tuba8 is again low, with the exception of the olfactory bulbs and cerebellum, where it is expressed at modest levels.
(C) Expression of TUBA1A, TUBB2B, TUBB3, and TUBA8 in the developing human brain at GW13 (frontal lobe) and GW22 (total fetal brain). Again, TUBA8 is expressed at low levels. Error bars indicate the SEM.
Finally, we investigated the expression of TUBA1A, TUBB2B, TUBB3, and TUBA8 in human fetal brain at GW13 (Biochain, C1244051) and GW22 (Biochain, C1244035), again employing three reference genes for normalization (HPRT, PGK1, TBP). Consistent with our mouse data, we observed very high levels of expression of TUBA1A, TUBB2B, and TUBB3, but only low levels of TUBA8 (Figure 2C). For instance, in the frontal lobe at GW13, a peak time for neuronal migration in humans,11 expression of TUBA1A is ∼224 times the control level, whereas TUBA8 is expressed at just 0.18 times the control gene level. We replicated our qPCR results with alternative sets of primers (in both mouse and human), confirming that our qPCR results are not due to alternative splicing (data not shown).
The data presented here show that Tuba8 is expressed at a low level in the developing mouse and human brain. This raises a puzzling question: How is it that a gene that is preferentially expressed in testis, heart, and skeletal muscle presents as a rare brain malformation? This question is particularly pertinent given that those tubulin genes known to cause neurodevelopmental disorders are highly expressed in the developing CNS, whereas Tuba8 is not. We suggest three possible explanations. First, it is plausible that an unidentified mutation lies in one of the 230 unsequenced genes in the 7.42 Mb candidate interval that contributes to, or is responsible for, the observed phenotype. Second, it is conceivable that despite its low expression level, Tuba8 serves some unique function that is essential to the developing brain and that is not vital in other tissues. Third, one might imagine a scenario in which the deletion described by Sheridan results in the expression of a truncated tubulin polypeptide and that this might confer some deleterious effect. The generation of appropriate transgenic mouse models or the identification of additional genetically unrelated patients harboring mutations in TUBA8 would help to resolve these issues.
Supplemental Data
Supplemental Data include three figures and one table and can be found with this article online at http://www.cell.com/AJHG.
Supplemental Data
Web Resources
The URL for data presented herein is as follow:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Acknowledgments
This work was supported by the Oxford Partnership Comprehensive Biomedical Research Centre with funding from the Department of Health's NIHR Biomedical Research Centres funding scheme. N.J.C. acknowledges the receipt of a Grant (R01HD057028) from the NIH. D.A.K. is supported by an FWF project grant (P21092-B09).
References
- 1.Abdollahi M.R., Morrison E., Sirey T., Molnar Z., Hayward B.E., Carr I.M., Springell K., Woods C.G., Ahmed M., Hattingh L. Mutation of the variant alpha-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am. J. Hum. Genet. 2009;85:737–744. doi: 10.1016/j.ajhg.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keays D.A., Tian G., Poirier K., Huang G.J., Siebold C., Cleak J., Oliver P.L., Fray M., Harvey R.J., Molnar Z. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaglin X.H., Poirier K., Saillour Y., Buhler E., Tian G., Bahi-Buisson N., Fallet-Bianco C., Phan-Dinh-Tuy F., Kong X.P., Bomont P. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khodiyar V.K., Maltais L.J., Sneddon K.M., Smith J.R., Shimoyama M., Cabral F., Dumontet C., Dutcher S.K., Harvey R.J., Lafanechere L. A revised nomenclature for the human and rodent alpha-tubulin gene family. Genomics. 2007;90:285–289. doi: 10.1016/j.ygeno.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Gloster A., El-Bizri H., Bamji S.X., Rogers D., Miller F.D. Early induction of Talpha1 alpha-tubulin transcription in neurons of the developing nervous system. J. Comp. Neurol. 1999;405:45–60. doi: 10.1002/(sici)1096-9861(19990301)405:1<45::aid-cne4>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 6.Murtaugh L.C., Chyung J.H., Lassar A.B. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13:225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanchi F., Corso V., Scannapieco P., Ievolella C., Negrisolo E., Tiso N., Lanfranchi G., Valle G. TUBA8: A new tissue-specific isoform of alpha-tubulin that is highly conserved in human and mouse. Biochem. Biophys. Res. Commun. 2000;270:1111–1118. doi: 10.1006/bbrc.2000.2571. [DOI] [PubMed] [Google Scholar]
- 8.Boda E., Pini A., Hoxha E., Parolisi R., Tempia F. Selection of reference genes for quantitative real-time RT-PCR studies in mouse brain. J. Mol. Neurosci. 2009;37:238–253. doi: 10.1007/s12031-008-9128-9. [DOI] [PubMed] [Google Scholar]
- 9.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tischfield M.A., Baris H.N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W.M., Andrews C., Demer J.L., Robertson R.L. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaglin X.H., Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 2009;25:555–566. doi: 10.1016/j.tig.2009.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


