Abstract
Structural variation includes many different types of chromosomal rearrangement and encompasses millions of bases in every human genome. Over the past three years the extent and complexity of structural variation has become better appreciated. Diverse approaches have been adopted to explore the functional impact of this class of variation. As disparate indications of the important biological consequences of genome dynamism are accumulating rapidly, we review the evidence that structural variation has an appreciable impact on cellular phenotypes, disease and human evolution.
Introduction to genomic structural variation
Structural variation (SV) is a broad term for genetic variants that alter chromosomal structure; it encompasses both balanced changes (inversions and some translocations) and those that alter DNA copy number [Copy Number Variation (CNV)]. The genome manifests a size continuum of genomic variants from single base deletions to whole-chromosomal aneuploidies. Structural variation is generally used to refer to larger changes, typically larger than 1kb 1,2, although this is an arbitrary threshold.
The phenotypic relevance of SV in genomes was first appreciated more than 70 years ago with the observation that the bar eye phenotype in Drosophila melanogaster is caused by a tandem chromosomal duplication 3. Over the next six decades, knowledge of SV in humans accrued slowly, largely through observations by cytogeneticists of the role that large chromosomal rearrangements play in sporadic severe developmental disorders (such as Down Syndrome 4), and by researchers who studied specific disease-related regions of the genome in painstaking detail (e.g. 5). These early studies revealed that SV contributes to all classes of disease with a genetic aetiology: sporadic development syndromes, Mendelian diseases, complex disorders and infectious diseases, as well as health-related metabolic phenotypes (Table 1).
Table 1. Examples of diseases and pharmacogenetic phenotypes influenced by structural variation.
| Type of Disease phenotype | Disease | Structural Variant | Reference |
|---|---|---|---|
| Rare (sporadic) disease | Williams-Beuren Syndrome | Deletion of ELN + others | 65 |
| Velo-Cardio-Facial Syndrome | Deletion of TBX1 + others | 66 | |
| Autism | Deletion in 16p11.2 | 48-50 | |
| Rare (Mendelian) disease | Haemophilia A | Inversion disrupting F8 | 67 |
| Charcot-Marie-Tooth type 1A | Duplication of PMP22 | 36 | |
| Juvenile Nephronophthisis | Deletion of NPHP1 | 37 | |
| Common Disease | Psoriasis | Multiallelic CNV of Beta-defensins | 43 |
| Systemic Lupus Erythematosus | Multiallelic CNV of C4 | 44 | |
| Malaria susceptibility | Deletion of alpha-globin | 40 | |
| HIV susceptibility | Multi-allelic CNV of CCL3L1 | 41 | |
| Pharmacogenetic | Codeine metabolism | Multi-allelic CNV of CYP2D6 | 68 |
| Carcinogen metabolism | Deletion of GSTM1 | 69 |
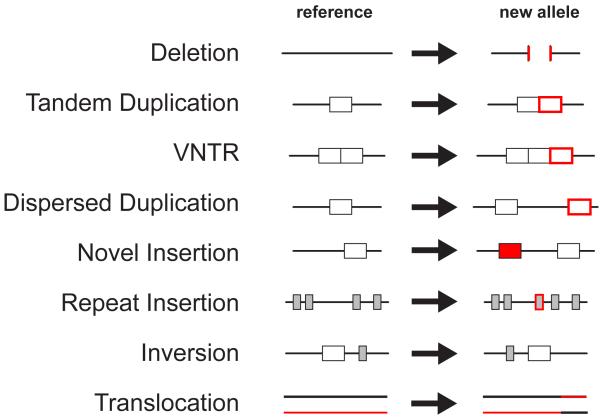
Unlike other forms of genetic variation, for example, single nucleotide polymorphisms (SNPs), SV cannot be easily studied from single sequence reads, and as a result characterisation of this form of variation lagged behind other forms of variation. It was the advent of microarray technologies to measure DNA copy number in 1998 6 and publication of the draft human sequence in 2001 that enabled genome-wide surveys for structural variation 7. Over the past 3 years, studies applying both microarray and sequencing technologies have revealed that the structural variation in the human genome is extensive and complex, with many different types of variation contributing to structural diversity 8-12 (Figure 1).
Figure 1. Types of structural variant.
Eight different types of structural variant are depicted, defined relative to the reference genome sequence
As databases of structural variation remain far from saturation, our understanding of the functional importance of SV is clearly in its infancy. Nevertheless, the biological impact of this form of variation has become apparent through a variety of complementary approaches. In this review we consider the current picture of the functional impact of SV from three different biological perspectives: the cell, the organism and the population. Specifically, we examine its impact on levels of gene expression (a cellular trait), disease (an organismal phenotype) and evolutionary change (a property of populations).
Discovery of structural variation
Historically, structural variation was primarily assayed cytogenetically in diseased genomes, and until recently these variants were not collated in any systematic fashion. More recently, genomic technologies (such as microarrays and sequencing technologies, see Table 2) have been applied to genome-wide surveys of SV in apparently healthy individuals. The relative merits and challenges of these technologies are reviewed elsewhere 13,14. These technologies can identify SVs much smaller than those identified cytogenetically, which, as a result, are often termed ‘submicroscopic’ variants. These surveys have been catalogued in genomic databases, most notably the Database of Genomic Variants (http://projects.tcag.ca/variation). This database currently (March 2008) contains entries for 11,966 individual variants >1kb in size, the vast majority of which are CNVs (n=11,784), rather than inversions (n=182). This is due to both the likely lower prevalence of inversions in the genome 10 and the difficulty of identifying inversions. Many CNVs have been identified independently, and so these 11,784 CNVs probably represent ~ 5,000 non-redundant variants. Most of the current technologies for discovering SV only provide the approximate size and location of the variant, rather than single nucleotide resolution, and are better able to identify larger variants. As a result, we have a much less complete catalogue of smaller SVs. Moreover, modelling of the variants that have been discovered strongly suggests that smaller SVs are much more frequent in the genome than longer SVs 15. Thus it seems likely that the majority of SVs remain to be identified.
Table 2. Genomic technologies for identifying structural variation.
| Technology | Genome-wide | High-throughput | Detects balanced variants | Detects breakpoint sequences | Ref. |
|---|---|---|---|---|---|
| Quantitative amplification (e.g. MAPHa, MLPAb, qPCRc) | − | ++ | − | − | 70,71 |
| Microarray (e.g. SNP genotyping, CGH) | + | + | − | − | 6,72 |
| Read-pair sequencing | + | − | + | + | 10,12 |
| Assembly comparisons | + | − | + | ++ | 73,74 |
Multiplex Amplified Probe Hybridisation
Multiplex Ligation Probe Amplification
quantitative-Polymerase Chain Reaction
The number of SVs identified in any individual genome has increased dramatically as the resolution of the technologies has improved. The first genome-wide studies identified of the order of 10 variants in an individual 8,9; more recently hundreds of variants have been identified in individuals 10,12 and once the limited (albeit improving) resolution of these technologies have been taken into account it is likely that each diploid genome harbours >1,000 SVs (MEH unpublished observations).
Structural variation is generated by a variety of different mutational mechanisms, including non-allelic homologous recombination 16, non-homologous end joining, retrotransposition and replication-based mechanisms 17. It is likely that there are other mechanisms in operation but elucidation of these will require more systematic characterisation of the precise sequence content of SVs. These mutational processes do not operate homogenously across the genome, and so this leads to the observation that some genomic regions are much more structurally dynamic than others: for example, regions rich in duplicated sequences tend to be highly variable in structure, both in terms of the population frequency and the size of structural variants 11,18. Duplicated sequences are enriched in sub-telomeric and pericentromeric regions, and both are enriched for large structural variants.
Impact of structural variation on gene expression
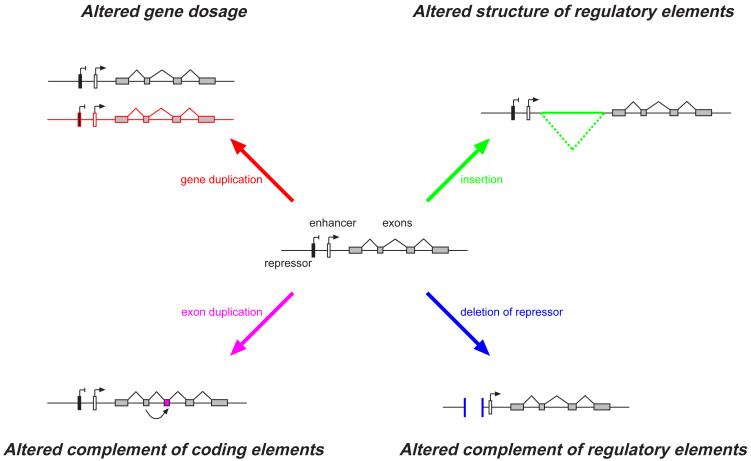
One of the key features of structural variation is that it encompasses large numbers of nucleotides. Many SVs affect large functional units such as genes, and these functional units can be fully contained in the SV. Substantial diversity in size allows many models of functional variation to be driven by SVs. All forms of SV are capable of major reorganization of the landscape of functional elements in the regions affected 19 (Figure 2). As described above, CNVs have been the predominant type of structural variation that has been ascertained and studied to investigate the functional consequences of SV, mainly due to the availability of CNV data in large population samples, such as the HapMap collections.
Figure 2. Influence of structural variation on gene regulation.
A gene is represented by a set of exons (grey boxes), an enhancer (white box) and repressor (black box). Four general mechanisms by which a Structural Variant can impact upon gene expression are depicted. For each mechanism, an exemplar structural variant (in colour) is shown relative to the central reference gene structure.
Levels of gene expression are one of few cellular traits that are both highly heritable 20 and amenable to accurate high-throughput genome-wide quantitation. Early small-scale or gene-specific studies identified examples of CNVs that had an impact on the levels of gene expression either from a population perspective 21 or in a disease context 22-24. One of the simplest models for the functional impact of CNVs is the change in levels of expression of genes within or surrounding the affected genomic region. An intuitive model suggests that an increase in the copy number of a specific gene will, on average, lead to corresponding increase in the expression level of that gene, and vice versa. But in addition, it is likely that deletions or insertions of additional DNA might lead to a variety of effects that are not as simple as the “more gene copies - more expression” expectation.
A recent study 25 looked at the relative impact of CNVs and SNPs on genome-wide gene expression in the 270 lymphoblastoid cell-lines of the HapMap collection. Although in most cases (~80%) the copy number was positively correlated with gene expression, the remaining 20% were negatively correlated. In addition, more than half of the CNVs that were associated with gene expression did not encompass coding sequences of their associated genes, suggesting that the mechanisms by which CNVs and in general SVs mediate their functional effect are quite diverse. One of the key conclusions from this study was that ~20% of the measurable genetic impact on gene expression is driven by CNVs and the rest by SNPs. This is likely an underestimate of the importance of CNVs because the current CNV maps are hugely biased toward large CNVs. Smaller CNVs are more abundant and are expected to have more specific effects than the large ones since they will likely affect individual functional units (e.g. a single enhancer). The field has not yet touched the effect of other SVs such as inversions or translocations, where the change in location of a genomic region can lead to the abolishment of functionality of some genomic elements. An even more interesting scenario is the creation of new combinations of genomic elements (e.g. bringing a gene close to a heterologous enhancer) that may be neutral, detrimental, or even advantageous for the organism, raising intriguing possibilities for evolution.
Impact of structural variation on disease
In principle, a disease with a genetic aetiology can be caused by any type of genetic lesion; some of these lesions will be SNPs, some will be SVs. Both of these variants can alter gene regulation (see above) or generate novel coding variation (e.g. nonsynonymous SNPs and fusion genes).
Largely because of the ongoing contribution of classical cytogenetics to the molecular ascertainment of SV, it is the diseases that have traditionally been referred to cytogenetics clinics for which we have most evidence for the involvement of SV, namely sporadic disorders where causal chromosomal rearrangements are suspected. Many such disorders have been identified where a single variant (de novo or inherited) is sufficient to cause the disease, ranging from the chromosomal 21 trisomies causing Down Syndrome that are recurrent within the population to non-recurrent interstitial deletions observed in a single individual. Screening duplicated regions of the genome that might be prone to chromosomal rearrangement, in patient cohorts has proven to be to fruitful in identifying new genetic syndromes (known as ‘Genomic Disorders’ 26) that result from recurrent rearrangements 27-29.
Individual pathogenic SVs are often rare, which makes proving their causal role challenging. Identifying individuals with overlapping SVs and similar phenotypic features is often critical for confirming a causal role. In this regard, the rapid growth in internet databases such as Decipher (http://www.sanger.ac.uk/PostGenomics/decipher/), which allow easy integration of clinical phenotypes and associated SV data across a global consortium, is catalysing the identification of novel disorders 30,31. In the absence of clear genotype-phenotype correlations across individuals, the size of a structural variant and its absence from either parent are regarded as the primary criteria for assessing the causal role of a novel variant. Thus long de novo deletions are typically regarded as being more likely to be causal than short, inherited duplications 32. As the resolution of SV detection increases, many variants may be identified in each patient, and consequently maps of SV from cohorts of apparently healthy individuals are increasingly being used to discriminate between benign and pathogenic variants (e.g. 33-35). Excluding structural variants from further consideration on the basis of their presence in these ‘control’ maps allows the prioritisation of potentially clinically-relevant variants, but, in not taking into account allele frequency, makes implicit assumptions about the inheritance pattern of the underlying causal variant(s) and is not without potential dangers (see below).
For Mendelian diseases, the mapping of the disrupted genes (typically by linkage) has sometimes revealed that a structural variant is the predominant disease allele (e.g. Charcot-Marie-Tooth disease type I A 36 and Juvenile nephronophthisis 37), but even when single nucleotide changes are the predominant causal variant, screening of patients without the common causative alleles often reveals a minor contribution of structural variants in the same gene, for example, rare single exon deletions and duplications, which disrupt the reading frame (e.g. 38).
Currently, association studies are the most popular method for identifying genetic variants underlying common or infectious diseases such as Type 2 Diabetes or Malaria. Genome-wide association studies are currently only practicable using SNPs, and allow the detection of associated haplotypes containing the causal variant. For most known common disease-associated haplotypes, the actual causal variant has not been fine-mapped. Thus the role of structural variants in common disease has hitherto been largely limited to the testing of candidate variants, such as CNVs encompassing the genes: alpha-globin, Fcγ receptor 3B (FCGR3B), Chemokine (C-C motif) Ligand 3-like-1 (CCL3L1), Beta-defensins and Complement component 4 (C4). This approach has yielded several reports of CNV-disease associations in recent years, most notably for infectious disease susceptibility and immune-related diseases 39-44, and we can expect that these will grow rapidly as the existing maps of common structural variants are mined for plausible candidate variants.
There is some evidence that power to detect the causal role of SV in a disease through SNP-based association studies is attenuated by recurrent SV mutation and the technical difficulty of genotyping SNPs in the duplicated regions that are enriched near SVs 11,25. This motivates the testing for SV-associations by assaying the structural variants directly. The ability to mine CNV from intensity data underlying genome-wide SNP genotyping platforms has led to the prospect that CNV-disease associations might be tested directly from these data, as well as through linkage disequilibrium with genotyped SNPs 21,45. Mining of CNVs from genome-wide SNP genotyping data in Autism cohorts has strengthened the evidence that some CNVs are sufficient to cause the disease, as well as identifying novel variants enriched in autistic individuals that warrant further testing 46,47. Some of these variants, such as de novo microdeletions of 16p11.2, can reach a frequency of 1% in Autistic individuals 48-50. This observation that a proportion of Autism can be explained by de novo chromosomal rearrangements provides an intriguing link between sporadic and common genetic disease 51; it remains to be seen whether Autism is a special case in this regard.
Impact of structural variation on evolutionary change
Because SVs can have significant effects on phenotypic traits, we would expect them to underlie some of the changes that have been important in the evolution and differentiation of modern humans. From an evolutionary point of view, most genetic variants found in the population are neutral, a few are disadvantageous and subject to negative selection, and even fewer are advantageous and experience positive selection, or in some circumstances balancing selection. Genome-wide surveys support the idea that most SVs are neutral: for example, they collectively reveal the same patterns of population clustering and population differentiation as the predominantly neutral SNPs 10. But the exceptions, particularly those that have been positively selected, are of great interest. Each form of selection affects the frequency of the SV and over evolutionary time and leaves an imprint in the surrounding pattern of variation. The simplest way to assess the evolutionary impact of an individual SV would be by counting the number of offspring from parents with different forms of the SV, but this is rarely possible. Alternatively, studies of SV frequency in different populations and variation of the surrounding region can be used (Box 1). In addition, genomewide analyses can address more subtle questions about classes of SVs: for example, have the SVs we now find in the population been filtered by negative selection?
Box 1: Detecting positive selection on an individual SV from genetic information.
Consider a new mutation that is positively selected in a population. It will increase rapidly in frequency, carrying with it its surrounding haplotype, thus lying in an unusually long haplotype and showing large frequency differences between populations. As it approaches fixation, it will show an unusual allele frequency spectrum (e.g. with an excess of high-frequency derived alleles). After fixation, functional changes (e.g. to amino acids) can continue to accumulate unusually fast. For each of these characteristics - long haplotypes, population differentiation, skewed allele frequency spectrum, excess functional change - an appropriate statistical test can be used to assess the strength of the evidence for selection (compared with a neutral model, or an empirical distribution), and detect selection over different timescales, up to 25,000, 75,000, 250,000 and millions of years, respectively 64.
Although these characteristics of positive selection are as relevant for beneficial SVs as they are for advantageous SNPs, it is often not simple to adapt methods developed for SNPs to detect positive selection on SVs, largely as a result of current limitations in interrogating SVs. Many SVs cannot be genotyped: few methods can distinguish between seven and eight copies, for example. And even if they can be genotyped, it may not be possible to assign haplotypes: e.g. a diploid genotype of 10 copies could correspond to haplotypes of 1+9, 2+8 etc., which cannot be distinguished. One way around these problems has been to develop methods that use the experimental information, such as CGH intensity, directly rather than via inferred genotypes or haplotypes 11.
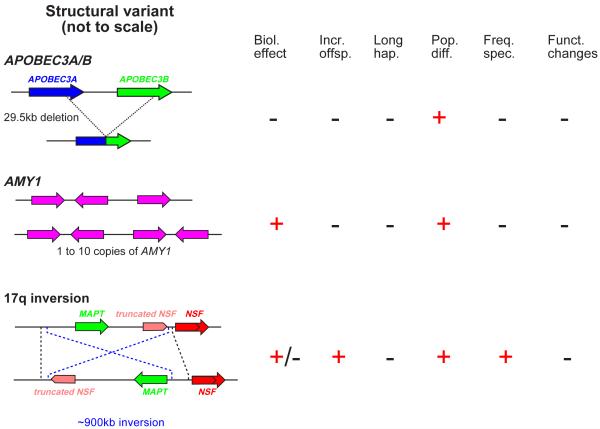
We will consider three examples of individual SVs of particular interest that illustrate a range of evolutionary changes and some of the complexities in the interpretation of signals of selection around SVs. First, APOBEC proteins help to defend us against retroviruses by deaminating C residues to U, and are encoded by a small gene family. Some individuals have APOBEC3A and APOBEC3B genes, whereas others carry a 30-kb deletion that produces a fusion gene which has the same amino acid sequence as APOBEC3A, so the effect is to delete APOBEC3B and potentially alter APOBEC3A regulation (Figure 3)52. Population differentiation for this SV was unusually high (FST=0.28, in the empirical top 2%), and simple haplotype analysis indicated a significantly extended haplotype surrounding the deletion in East Asian populations; but when the size of the deletion itself was taken into account, the haplotype structure was no longer unusual 52, showing the additional care needed for assessing SVs. The phenotype under selection is unclear.
Figure 3. Examples of evidence for selection on structural variants.
(a) Selection might have favoured an APOBEC3 deletion, (b) an increase in AMY1 copy number in populations with a high starch diet (c) and an inversion polymorphism encompassing a number of genes, including MAPT and a truncated copy of NSF. The presence or absence of evidence for positive selection acting on each variant is summarised on the right hand side: an understanding of the relevant biological effect (Biol. effect), an increased number of offspring (Incr. offsp.), an unusually long haplotype (Long hap.), elevated population differentiation (Pop. Diff.), a skewed allele frequency spectrum (Freq. spec.) or an excess of functional changes (Funct. changes). A ‘-‘ indicates either that there was no evidence or that evidence was not sought.
Second, salivary amylase (AMY1) begins the digestion of starch in food. Individual chromosomes can carry from one to ten AMY1 genes, leading to a corresponding level of protein in the saliva 53. Populations depending on high-starch diets have a higher diploid copy number than those with low-starch diets (6.7±2.4 v 5.4±2.0), and the degree of population differentiation between the population pair Japanese-Yakut was again unusual when compared empirically with other CNVs 53. Here, it is worth noting that AMY1 copy numbers for individual chromosomes could not be determined, so standard methods for detecting selection (Box 1) could not be applied (Figure 3).
Third, several genes, including microtubule-associated protein Tau (MAPT), lie in a ~900-kb inversion/indel polymorphism on chromosome 17q 54. The two haplotypes H1 and H2 show several remarkable features. They differ substantially in sequence, corresponding to a divergence time of ~3 million years ago, and also in diversity. H1 has a typical level of diversity, but H2 has very low diversity, and is common only in European and Middle Eastern populations, suggesting recent positive selection in these populations. It was possible to test this hypothesis directly in deep Icelandic pedigrees and female H2 carriers were found to have ~3.5% more children than non-carriers. The long-term presence of both H1 and H2 lineages in the population could be explained by ancient balancing selection or population substructure such as the introduction of H2 from an archaic population like Neanderthals 54,55. Increased H2 fertility was associated with a higher female recombination rate, but many aspects of the selection acting at this locus remain unclear.
From a genomic perspective, the distribution of SVs is biased away from genes and other functional elements 11 and some studies show that deletions might be rarer than SNPs 45, suggesting the general action of negative selection. Deletions tend to be shorter than duplications (a recent study identified average sizes of 43kb vs 120kb respectively 11) and so might be subject to stronger purifying selection. Interestingly, there is evidence of over-representation of some gene types in CNVs, including those related to immunity and sensory perception, and this has been interpreted as suggestive of positive selection 56.
Our understanding of the impact of SVs on human evolution is thus developing. A few candidates for relevant adaptations are known, but much remains poorly understood even for these. This area is likely to develop rapidly in the near future.
Concluding remarks and future perspectives
Here we have documented the functional impact of structural variation on cells, organisms and populations. These studies have applied a broad range of genotyping and phenotyping methodologies, but have been limited by a far from complete catalogue of SV in the human genome.
Our ability to identify novel structural variants at ever greater resolution is developing rapidly. We envisage having a comprehensive catalogue of common SV within the next few years through the efforts of several large-scale projects, such as the Human Structural Variation Project 57 and the Genome Structural Variation Consortium 11 as well as genome resequencing studies such as the 1000 genomes project (www.1000genomes.org). This catalogue will enable the development of genome-wide SV genotyping platforms for application to studies of both cellular and organismal phenotypes, and population genetics. Although microarray-based methods are potentially well-suited to performing large-scale CNV genotyping, the path to developing genome-wide genotyping of balanced structural variants remains extremely challenging 58. Currently, it is not even possible to genotype CNVs genome-wide in a robust manner, and this degree of uncertainty in genotype inference reduces the power of association studies, and potentially increases the risk of false-positive associations. The availability of genome-wide platforms for SV genotyping will catalyse genetic studies of other metabolomic and proteomic traits, which hitherto have had little investigation with respect to SV (but see 59,60). Irrespective of the phenotype in question, new statistical methods are going to be required to analyse the data produced by these SV assays, which, while superficially similar to SNP data, present novel challenges for statistical inference, for example, in handling diploid information and multi-allelic loci.
In addition, we need to develop biological and genetic models of greater complexity, for while it is true that we will discover large functional effects of SVs relatively simply, the mere fact that SVs affect large regions of DNA, many of which will also be variable at the sequence level, suggests that the range of models required to understand complex phenotypes will be much broader and the details of the models quite complex. Irrespective of the exact number of SVs in a human genome, they are likely to increase the statistical space for genetic association studies of human disease dis-proportionally to the amount of sequence they affect.
Demonstrating the functional impact of rarer structural variants is likely to remain a long-term challenge. Proving a causal role for any variant seen only in a single patient is impossible on genetic grounds alone. Moreover, clinical geneticists are increasingly using published CNV maps in apparently healthy individuals as control datasets (for example 33-35), thus blurring the boundaries between association studies and clinical genetics. There are several potential concerns here. First, the individuals in these studies are rarely phenotyped as comprehensively as one would wish for a properly-controlled association study. Second, as these CNV maps dig deeper into rarer variants, it can be expected than many recessive and variably expressive dominant alleles will also be represented in ‘control’ datasets. For example, one of the HapMap individuals used in many of the current CNV maps is heterozygous for the deletion causing autosomal recessive Juvenile Nephronophthisis 11. Simply excluding structural variants seen in a control population could lead to causal variants being overlooked. Filtering the set of structural variants identified in a patient to exclude those found in a control cohort presupposes that the underlying causal variant is a fully penetrant dominant mutation. Although this filtering might represent a useful first pass approach, it will certainly bias against the discovery of other modes of inheritance underlying these phenotypes.
The difficulties of ascribing a biological impact to a novel structural variant could be mitigated if the consequence of that variant could be predicted with confidence from existing functional data. However, despite the increasing richness of the functional annotation of the genome, it remains difficult to predict the functional impact of a novel structural variant 61. It should become possible to integrate functional datasets from a broad range of sources, including interaction networks 62 and gene perturbation phenotypes in model organisms, to develop a much more predictive understanding of dosage-sensitivity in the human genome.
Recent work 63 examining the influence of rare SNPs on common disease through gene re-sequencing in case and control cohorts has shown that summing rare alleles in an informed manner (for example, nonsynonymous SNPs in a candidate gene) can establish the influence of rarer variants on a complex phenotype. It is worth noting that CNV-mining from genome-wide SNP genotyping platforms is similar to gene resequencing studies in that there is no ascertainment bias with respect to the frequency of the variant; both rare and common CNVs can be detected, and so (unlike an assay targeted to known CNVs) it lends itself to these analyses of rare alleles.
As the evidence for the functional impact of SVs accrues, we can expect to see more functional studies of specific variants using model organisms and cellular assays. These functional studies require substantial investment of time and effort and it is only natural that they tend to be pursued only once there is robust evidence for the causal role of any given variant. The number of SVs that meet this critical evidential threshold can be expected to increase rapidly over the coming years.
The research summarised in this review covers a broad range of disciplines, including genome biology, population genetics and medical genetics. It is readily apparent that only through adopting such complementary approaches can the full functional impact of any form of variation be fully appreciated. We foresee that the integration of data from these, and other, disciplines is critical to fully reap the benefits of the incipient revolution of whole-genome resequencing.
Acknowledgements
Our work is supported by The Wellcome Trust.
References
- 1.Feuk L, et al. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Freeman JL, et al. Copy Number Variation: New Insights in Genome Diversity. Genome Research. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 3.Bridges CB. The Bar ‘gene’: a duplication. Science. 1936;83:210–211. doi: 10.1126/science.83.2148.210. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs PA, et al. The somatic chromosomes in mongolism. Lancet. 1959;1(7075):710. doi: 10.1016/s0140-6736(59)91892-6. [DOI] [PubMed] [Google Scholar]
- 5.Higgs DR, et al. A review of the molecular genetics of the human alpha-globin gene cluster. Blood. 1989;73(5):1081–1104. [PubMed] [Google Scholar]
- 6.Pinkel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20(2):207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 7.IHGSC Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 9.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 10.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 11.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherer SW, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39(7 Suppl):S7–15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39(7 Suppl):S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad DF, et al. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38(1):75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 16.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18(2):74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 17.Lee JA, et al. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 18.Sharp AJ, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77(1):78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1(6):e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33(3):422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 21.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nature Genetics. 2006;38(1):86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 22.Merla G, et al. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet. 2006;79(2):332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JA, et al. Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol. 2006;59(2):398–403. doi: 10.1002/ana.20732. [DOI] [PubMed] [Google Scholar]
- 24.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupski J. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 27.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38(9):1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 28.Sharp AJ, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16(5):567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 29.Mefford HC, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81(5):1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redon R, et al. Interstitial 9q22.3 microdeletion: clinical and molecular characterisation of a newly recognised overgrowth syndrome. Eur J Hum Genet. 2006;14(6):759–767. doi: 10.1038/sj.ejhg.5201613. [DOI] [PubMed] [Google Scholar]
- 31.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38(9):1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 32.Lee C, et al. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39(7 Suppl):S48–54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 33.Jacquemont ML, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43(11):843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Froyen G, et al. Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: important role for increased gene dosage of XLMR genes. Hum Mutat. 2007;28(10):1034–1042. doi: 10.1002/humu.20564. [DOI] [PubMed] [Google Scholar]
- 35.Ullmann R, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28(7):674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 36.Lupski JR, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66(2):219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 37.Konrad M, et al. Large homozygous deletions of the 2q13 region are a major cause of juvenile nephronophthisis. Hum Mol Genet. 1996;5(3):367–371. doi: 10.1093/hmg/5.3.367. [DOI] [PubMed] [Google Scholar]
- 38.White SJ, et al. Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat. 2004;24(1):86–92. doi: 10.1002/humu.20054. [DOI] [PubMed] [Google Scholar]
- 39.Aitman TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439(7078):851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 40.Flint J, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321(6072):744–750. doi: 10.1038/321744a0. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307(5714):1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 42.Fellermann K, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79(3):439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollox EJ, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40(1):23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80(6):1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinds DA, et al. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38(1):82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 46.Szatmari P, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17(4):628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 50.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 51.Zhao X, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104(31):12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kidd JM, et al. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3(4):e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39(10):1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37(2):129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 55.Hardy J, et al. Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens. Biochem Soc Trans. 2005;33(Pt 4):582–585. doi: 10.1042/BST0330582. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen DQ, et al. Bias of Selection on Human Copy-Number Variants. PLoS Genet. 2006;2(2):e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eichler EE, et al. Completing the map of human genetic variation. Nature. 2007;447(7141):161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner DJ, et al. Assaying chromosomal inversions by single-molecule haplotyping. Nat Methods. 2006;3(6):439–445. doi: 10.1038/nmeth881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazarus P, et al. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics. 2005;15(11):769–778. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 60.Dumas ME, et al. Direct quantitative trait locus mapping of mammalian metabolic phenotypes in diabetic and normoglycemic rat models. Nat Genet. 2007;39(5):666–672. doi: 10.1038/ng2026. [DOI] [PubMed] [Google Scholar]
- 61.Van Vooren S, et al. Array comparative genomic hybridization and computational genome annotation in constitutional cytogenetics: suggesting candidate genes for novel submicroscopic chromosomal imbalance syndromes. Genet Med. 2007;9(9):642–649. doi: 10.1097/gim.0b013e318145b27b. [DOI] [PubMed] [Google Scholar]
- 62.Kim PM, et al. Positive selection at the protein network periphery: Evaluation in terms of structural constraints and cellular context. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0710183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romeo S, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39(4):513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312(5780):1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 65.Ewart AK, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5(1):11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 66.Scambler PJ, et al. Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet. 1992;339(8802):1138–1139. doi: 10.1016/0140-6736(92)90734-k. [DOI] [PubMed] [Google Scholar]
- 67.Lakich D, et al. Inversions Disrupting the Factor-Viii Gene Are a Common-Cause of Severe Hemophilia-A. Nature Genetics. 1993;5(3):236–241. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- 68.Gasche Y, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351(27):2827–2831. doi: 10.1056/NEJMoa041888. [DOI] [PubMed] [Google Scholar]
- 69.Bell DA, et al. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J Natl Cancer Inst. 1993;85(14):1159–1164. doi: 10.1093/jnci/85.14.1159. [DOI] [PubMed] [Google Scholar]
- 70.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Armour JA, et al. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28(2):605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bignell GR, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14(2):287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khaja R, et al. Genome assembly comparison identifies structural variants in the human genome. Nat Genet. 2006;38(12):1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]