Abstract
In this work, we present a method for the integration of feature and intensity information for non rigid registration. Our method is based on a free-form deformation model, and uses a normalized mutual information intensity similarity metric to match intensities and the robust point matching framework to estimate feature (point) correspondences. The intensity and feature components of the registration are posed in a single energy functional with associated weights. We compare our method to both point-based and intensity-based registrations. In particular, we evaluate registration accuracy as measured by point landmark distances and image intensity similarity on a set of seventeen normal subjects. These results suggest that the integration of intensity and point-based registration is highly effective in yielding more accurate registrations.
1 Introduction
Non rigid image registration is a central task in medical image analysis. In the particular case of the brain, there are a number of important applications including comparing shape and function between individuals or groups, developing probabilistic models and atlases, measuring change within an individual and determining location with respect to a preacquired image during stereotactic surgery. The detailed comparison and nonrigid registration of brain images requires the determination of correspondence throughout the brain and the transformation of the image space according to this correspondence. In addition, a large number of other image analysis problems can in fact be posed as non rigid registration problems such as segmentation (via the use of an atlas), motion-tracking, etc.
There have been many approaches recently to nonrigid registration, with a particular emphasis on applications to brain imaging (see the collection [15]). Most commonly, non-linear registration methods use image intensities to compute the transformation (e.g. [2, 7, 14, 13, 8].) These techniques are potentially highly accurate but can be susceptible to local minima. In particular, the high anatomic variability of the cortex often results in intensity based methods yielding inaccurate results. Feature based and integrated feature-intensity methods have been developed to overcome such problems (e.g. [4, 6, 5, 11, 9, 1].) None of these methods, however, is able to handle outliers caused by large variations in sulcal anatomy, as well as irregular sulcal branching and discontinuity. We additionally note, that outliers can be present even in carefully manually segmented structures such as the ones used in this work. Brain structures are not always well defined and often part of their boundaries is set arbitrarily. For example, in the case of the amygdala typically manually traced on coronal slices, the selection of the posterior and the anterior extent is somewhat operator dependent. Hence a correspondence method that can explicitly handle outliers is important in this context.
In general, intensity-based methods are highly accurate in subcortical regions where the geometrical complexity is low. Feature-based registrations are often employed (typically with explicitly pre-segmented structures such as sulci) where accurate cortical registration is desired. In this paper we present a method which aim to integrate the strengths of both intensity and feature-based registration method. We test the results of these registrations one a set of 17 normal controls with manually extracted sulci and compare their performance to both point-based only and intensity-based only methods.
2 Methods
We first describe the components of our integrated method, namely the intensity-based registration module (section 2.1) and the robust point matching based-module (section 2.2). Then, the integrated method is presented in section 2.3.
2.1 Intensity-Based Module
We use a slightly modified form of the intensity-based non rigid registration method first described by Rueckert et al[13]. This method utilizes a free-form deformation transformation model based on tensor b-spline, and the normalized mutual information similarity metric. This metric can be expressed as:
| (1) |
where A and B are the two images, and H() is the image intensity entropy. This similarity metric is combined with a regularizing term to yield an optimization functional which is maximized in a multi-resolution framework. Our own implementation of this method which is used in the results section, first estimates a linear affine registration and then uses this as an input to estimate a full non-linear FFD transformation in a multiresolution manner. We modify the methodology in [13] in two ways: (a) we use a more efficient conjugate gradient optimization scheme, and (b) we implement an adaptive pre-processing scheme to better distribute the histogram bins over the intensity range in order to handle the often long tails at the upper end of the intensity of brain MRI images.
2.2 Robust-Point Based Matching Module
We present here a slightly modified form of the standard RPM methodology as can be found in Chui et al[3] and Papademetris et al[12]. The registration procedure consists of two alternative steps: (i) the correspondence estimation step and (ii) the transformation estimation step. In the following discussion we will label the reference point set as X and the transform point set as Y. The goal of the registration is to estimate the transformation G : X ↦ Y. We will label Gk the estimate of G at the end of iteration k. G0 is the starting transformation which can be the identity transformation.
Correspondence Estimation
Given the point sets X and Y we estimate the match matrix M, where Mij is the distance metric between points Gk (Xi) and Yj. The standard distance metric is defined as:
| (2) |
where |Xi − Yj| is the Euclidean distance between points Xi and Yj and T is the temperature that controls the fuzziness of the correspondence. If the correspondence problem is to be thought of as a linear assignment problem, the rows and columns of M must sum to 1. The framework is further extended to handle outlier points by introducing an outlier column C and an outlier row R. Ci is a measure of the degree of ‘outlierness’ of a point in the reference point set Xi and Rj is the same for a point in the transform point set Yj. C and R are initialized with constant values. The ability to model outliers allows this method to robustly match features of high variability such as cortical sulci. Once the normalization is completed we can compute the correspondence as follows. Let Vi be the corresponding point to Xi and wi the confidence in the match. Then Vi is defined as a normalized weighted sum of the points Yj where the weights are the elements of the match matrix M.
| (3) |
Note that a point that has a high value in the outlier column C will have low confidence and vice-versa. We note that in our integrated method (section 2.3) we simply use the correspondence piece of RPM.
Transformation Estimation
This is simply achieved by a regularized weighted least squares fit between Xi and Vi as follows:
| (4) |
where S(g) is a regularization functional (e.g. bending energy function) weighted by a function of the temperature f(T). This last weighting term is used to decrease the regularization as we approach convergence.
Deterministic Annealing Framework
The alternating estimation of M and G is performed in a deterministic annealing framework. Starting with a high value of T corresponding to a rough estimate of the maximum mis-alignment distance we first estimate M, and then G. Then T is decreased by multiplying it with an annealing factor and the process is repeated until T becomes sufficiently small. In our implementation, first an affine transformation is estimated using T = 15.0 ↦ 2.0mm and then a non rigid FFD transformation is estimated using T = 3.0 ↦ 1.0, with an annealing rate=0.93.
2.3 Integrated Method
Our method is closest in spirit to the work of Wang et al[16] and Harktens et al [9]. We first estimate an initial registration using the RPM method alone, to ensure that sulcal landmarks are correctly aligned. We then proceed to refine the estimate of the transformation by minimizing the following optimization functional which is a tradeoff between intensity similarity and adherence to point correspondence.
| (5) |
where the first term is the intensity similarity distance (eqn. 1), the second is a measure of adherence to the corresponding points as estimated by RPM (eqn. 4) weighted by the constant λ (N is the number of points in the reference point-set). We note that during the optimization of this functional the correspondences maybe re-evaluated at each iteration at a constant temperature T = 1.0mm which is equal to the minimum temperature used in the initial registration. In practice, however, keeping the correspondences fixed produced marginally better results. The transformation estimated has the exact same parameterization as the FFD transformation estimated by the RPM algorithm.
3 Results
In this section, we compare the performance of our new integrated algorithm to our previous point-based only method as well as a standard non-linear intensity-only based method. We evaluate the accuracy of the algorithms in matching cortical landmarks in a set of 17 normal controls.
All images used in this work were high resolution (1.2 × 1.2 × 1.2mm) 3D SPGR images acquired using a GE 1.5 T scanner (2 NEX, TR= 24msec, TE = 5msec, flip = 45°). The scans were of excellent quality and without movement artifacts. The brains were next stripped and the following four sulci were manually traced (a) left central sulcus, (b) right central suclus, (c) left superior frontal sulcus and (d) right superior frontal sulcus, as shown in figure 1. For the purpose of both the integrated and the point-only based methods, labeled point sets for each subject were constructed using approximately 5000 points for the outer cortical surface and 250 points for each sulcus resulting in a total point set of approximately 6000 points/subject. All subjects were registered to a normal control reference subject, an example registration is shown in figure 2
Fig. 1.
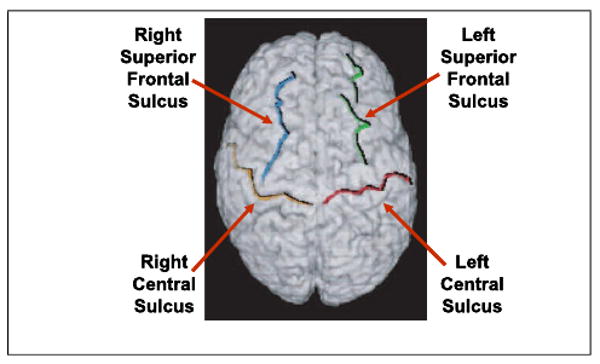
Sulci used in the evaluation of the registration methods. Note that these are major sulci, hence the matching error of the intensity based method is smaller that what is reported by Hellier et al. [10]
Fig. 2.
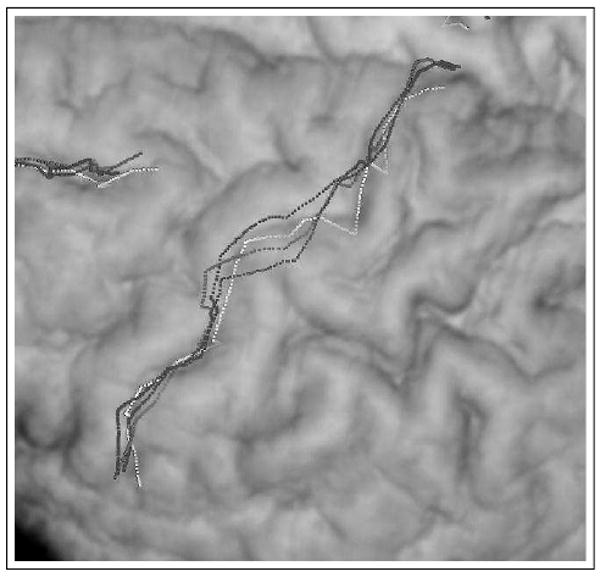
Example registration result. In this close-up of the central sulcus overlaid on a volume rendered stripped brain, the target surface is shown in white. The warped template is shown for three different registrations RPM − red, Integrated λ = 0.1) − blue and NMI − green.
For the purpose of comparison, the registrations were computed using (a) our point-based method RPM[12] and (b) our implementation of the non-linear intensity (NMI) based method of Rueckert et al [13]. To test the effect of the tradeoff parameter λ, the integrated algorithm was applied using seven different values of λ (INT-λ). We used the following measures to evaluate the quality of the registration:
1. Cross-Correlation Coefficient (CC)
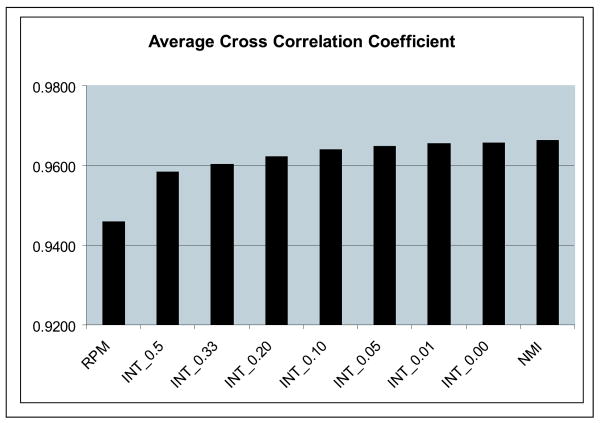
This measures the degree of intensity similarity. We use this measure rather than NMI as it was not explicitly optimized by methods (ii) and (iii). In practice, though, there was found to be a monotonic relationship between CC and NMI. The values of CC for all registration methods are shown in figure 3.
Fig. 3.
Average intensity similarity for the point-based registration method (RPM), the new integrated algorithm (INT-λ) with seven different values of the weight of adherence to the point correspondences λ and the intensity only similarity algorithm (NMI) as computed from N = 17 normal controls.
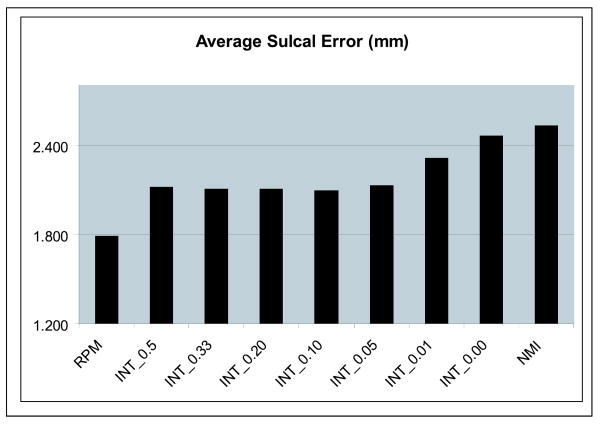
2. Average Sulcal matching error
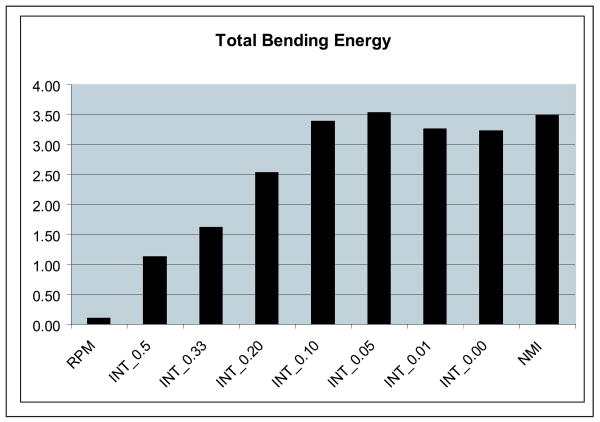
This was the mean distance of all points from the reference sulcus to the target sulcus using correspondences estimated by a nearest-neighbor matching method. The results for this are shown in figure 4. We also report the total bending energy of the calculated transformations to give a sense of the extent of the deformation in each case. Note that the bending energy in RPM is very low as there are no sub-cortical features and hence the registration is very smooth away from the cortical surface and the sulci. The integrated algorithms with high values of λ also have low bending energy; the bending energy gradually increased and approaches that of the NMI method a λ goes to zero. The average total bending energy for all the registration methods is shown in the chart in figure 5.
Fig. 4.
Average sulcal matching error for the point-based registration method (RPM), the new integrated algorithm (INT-λ) as above and the intensity only similarity algorithm (NMI) as computed from N = 17 normal controls.
Fig. 5.
Average bending energy for the registrations computed by the point-based registration method (RPM), the new integrated algorithm (INT-λ) and the intensity only similarity algorithm (NMI) as computed from N = 17 normal controls.
The intensity similarity metric results shown in figure 3 indicate that the integrated method approaches the intensity-based method as the value of λ goes to zero as expected, especially for λ < 0.05. As expected the RPM algorithm is superior in terms of sulcal distance error point matching Note that in particular for λ = 0.1 or 0.05, the performance of the integrated algorithm the integrated method performs almost as well as the individual components (i.e. RPM and NMI) in their area of respective strengths, i.e. it produces an optimal tradeoff between sulcal matching error and intensity similarity.
4 Conclusions
In this work, we present a new method for the integration of intensity based and feature based registrations with encouraging results. In general, while feature based methods are highly accurate in specific areas, their accuracy decreases in regions away from the pre-extracted points. Intensity based methods, on the other hand are highly accurate in subcortical regions but their accuracy decreases in the cortex. Combining the two approaches offers the opportunity of an optimal tradeoff between localized feature-driven registration accuracy in the cortex and accurate intensity registration in the sub-cortex. We believe that this improvement would be more obvious in the case of variable sulci, as opposed to the major sulci that were available for this work. (Further, the brains used for all registrations were accurately pre-stripped hence improving the surface registration for the intensity-based method.)
We believe that such an integration is necessary for the computation of both morphometric differences between subjects, as well as composite activation maps from fMRI in cortical areas where intensity only methods are relatively inaccurate [10]. We are currently investigating reformulating our method in terms of two interacting modules to yield an EM-like algorithm.
References
- 1.Cachier P, Bardinet E, Dormont D, Pennec X, Ayache N. Iconic feature based nonrigid registration: the PASHA algorithm. Computer Vision and Image Understanding. 2003;89:272–298. [Google Scholar]
- 2.Christensen GE, Miller MI, Vannier MW. Individualizing neuroanatomical atlases using a massively parallel computer. Computer [Google Scholar]
- 3.Chui H, Win L, Schultz RT, Duncan JS, Rangarajan A. A unified non-rigid feature registration method for brain mapping. Medical Image Analysis. 2003;7(2):113–130. doi: 10.1016/s1361-8415(02)00102-0. [DOI] [PubMed] [Google Scholar]
- 4.Collins DL, Le Goualher G, Evans AC. Medical Image Computing and Computer Assisted Intervention. Springer; Berlin: 1998. Non-linear cerebral registration with sulcal constraints; pp. 974–984. [Google Scholar]
- 5.Corouge I, Barillot C, Hellier P, Toulouse P, Gibaud B. Non-linear local registration of functional data. In: Viergever MA, Dohi T, Vannier M, editors. Medical Image Computing and Computer Aided Intervention (MICCAI) 2001. pp. 948–956. [Google Scholar]
- 6.Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comp Vision and Image Understanding. 1997;66(2):207–222. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- 7.Feldmar J, Malandain G, Declerck J, Ayache N. Extension of the ICP algorithm to non-rigid intensity-based registration of 3D volumes. Proc Workshop Math Meth Biomed Image Anal. 1996 June;:84–93. [Google Scholar]
- 8.Friston KJ, Ashburner J, Poline JB, Frith CD, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Human Brain Mapping. 1995;2:165–189. [Google Scholar]
- 9.Hartkens T, Hill DLG, Castellano-Smith AD, Hawkes DJ, Maurer CR, Jr, Martin AJ, Hall WA, Liu H, Truwit CL. Using points and surfaces to improve voxel-based non-rigid registration. Medical Image Computing and Computer Aided Intervention (MICCAI) 2002:565–572. [Google Scholar]
- 10.Hellier P, Barillot C, Corouge I, Gibaud B, Le Goualher G, Collins DL, Evans A, Malandain G, Ayache N, Christensen GE, Johnson HJ. Retrospective evaluation of inter-subject brain registration. IEEE Transactions on Medical Imaging. 2003;22(9):1120–1130. doi: 10.1109/TMI.2003.816961. [DOI] [PubMed] [Google Scholar]
- 11.Liu T, Shen D, Davatzikos C. Medical Image Computing and Computer Aided Intervention (MICCAI) Part II LLNCS 2879. Springer-Verlag; 2003. Deformable registration of cortical structures via hybrid volumetric and surface warping; pp. 780–787. [Google Scholar]
- 12.Papademetris X, Jackowski A, Schultz RT, Staib LH, Duncan JS. Medical Image Computing and Computer Aided Intervention (MICCAI) Part II LLNCS 2879. Springer-Verlag; 2003. Computing 3D non-rigid brain registration using extended robust point matching for composite multisubject fMRI analysis; pp. 788–795. [Google Scholar]
- 13.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Non-rigid registration using free-form deformations: Application to breast MR images. IEEE Transactions on Medical Imaging. 1999 doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 14.Thirion JP. Image matching as a diffusion process: An analogy with maxwell's demons. Med Image Anal. 1998;2(3):243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- 15.Toga AW. Brain Warping. Academic Press; San Diego: 1999. [Google Scholar]
- 16.Wang Y, Staib LH. Physical model based non-rigid registration incorporating statistical shape information. Med Image Anal. 2000;4:7–20. doi: 10.1016/s1361-8415(00)00004-9. [DOI] [PubMed] [Google Scholar]