Abstract
Fecal indicator bacteria (FIB), commonly used to regulate sanitary water quality, cannot discriminate among sources of contamination. The use of alternative quantitative PCR (qPCR) methods for monitoring fecal contamination or microbial source tracking requires an understanding of relationships with cultivated FIB, as contamination ages under various conditions in the environment. In this study, the decay rates of three Bacteroidales 16S rRNA gene markers (AllBac for general contamination and qHF183 and BacHum for human-associated contamination) were compared with the decay rate of cultivated Escherichia coli in river water microcosms spiked with human wastewater. The following five sets of microcosms were monitored over 11 days: control, artificial sunlight, sediment exposure, reduced temperature, and no autochthonous predation. Decay was characterized by estimation of the time needed to produce a 2-log reduction (t99). No treatment-associated differences in the decay of the 4 targets were evident except with reduced predation, where E. coli, qHF183, and BacHum markers had lower levels of decay by day 3. However, there were substantial target-associated differences. Decay curves for the AllBac marker indicated a larger persistent population than those of the other targets. Exposure to sunlight, sediment, and reduced predation resulted in more rapid decay of the human-associated markers relative to cultivable E. coli, but there were no differences in t99 values among the 4 targets under control conditions or at reduced temperatures. Further evaluation of epidemiological relationships will be needed in order to relate the markers directly to health risk. These findings suggest that the tested human-associated markers can complement E. coli as indicators of the human impact on sanitary water quality under the constrained conditions described in this paper.
Recreational water quality standards for freshwater are commonly based on the cultivable concentration of the fecal indicator bacterium (FIB) Escherichia coli (42). Epidemiological studies demonstrated a correlation between E. coli concentration and rates of gastrointestinal illness among swimmers (19, 32, 46), and water quality criteria based on E. coli have been established by the U.S. Environmental Protection Agency for the protection of human health (41). However, E. coli are found in many hosts, both human and nonhuman, that carry different cohorts of human-pathogenic microorganisms (13), and E. coli types have limited host specificity (4, 16, 17). E. coli also have been shown to reproduce in the environment under some conditions (7, 12, 23, 37, 49). These limitations of E. coli as an indicator of public health risk are well recognized (34) and may result in revision of U.S. recreational water quality criteria (44).
In the pursuit of alternate assessment strategies, researchers in the field of microbial source tracking (MST) have developed and applied host-associated molecular markers of fecal contamination. Prominent among candidate MST protocols are those that detect and quantify the 16S rRNA gene of the order Bacteroidales fecal anaerobes. In particular, the human feces-associated (HF) Bacteroidales marker HF183, developed by Bernhard and Field (6) for detection by the PCR, is commonly used to detect human-source fecal contamination (14, 18, 20, 38). Several HF quantitative PCR (qPCR) assays (24, 33, 35) that target the same 16S rRNA gene cluster in the genus Bacteroides as that used by Bernhard and Field (6) have been designed.
Whereas current standards are based on cultivable E. coli, PCR-based MST protocols inherently measure both living and dead cells. Because of this, further validation of these molecular markers for MST in the current regulatory context requires an understanding of marker persistence compared with the persistence of cultivated E. coli over time and under various environmental conditions. Although research into live-dead discrimination of Bacteroidales shows some promise (2, 3, 48), measurement of total (DNA) marker remains the common method because it requires considerably less time and manipulation and is more easily applied in most laboratories.
Comparison of cultivation- and PCR-based parameters is complicated by potentially different decay characteristics of the targets. The decay of cultivated bacterial targets, such as E. coli, is driven by a number of environmental factors. Several studies have shown that E. coli decay is more rapid with exposure to sunlight, predation, and salinity and less rapid at colder temperatures (1, 10, 21, 28, 29). Turbidity and association with sediments also have been shown to enhance the survival of E. coli in water (10, 15, 27, 31).
Less is known about the persistence of Bacteroidales DNA under various environmental conditions. In a laboratory microcosm study, Kreader (25) detected Bacteroides distasonis in river water spiked with human feces for up to 2 weeks at 4°C by conventional PCR but for only 1 to 2 days at 24°C. In addition to temperature, predation was shown to be associated with more rapid marker decay. Seurinck et al. (35) used qPCR to quantify the HF marker (qHF183) in freshwater over time, and a 2-log reduction in the copy number was reached after 6 days at 28°C, a 1-log reduction was seen after 10 days at 12°C, and no significant reduction was seen in 28 days at 4°C. Okabe and Shimazu (30) investigated the decay of various Bacteroidales markers in microcosms spiked with feces by qPCR and found longer persistence with lower temperature and higher salinity. Bell et al. (5) spiked horse feces into stream water microcosms and used the AllBac general Bacteroidales qPCR assay (26) to compare the effects of temperature, fecal aggregate size, and concentration on decay rates in unfiltered (normal community) and filtered (reduced biological activity, including predation) water. As in the report by Kreader (25), temperature and predation were shown to be the controlling factors, while aggregate size and initial concentration had no significant effects on decay rates.
Reports on the effects of sunlight on Bacteroidales decay have been variable. Bae and Wuertz (3) found no difference in the times needed to produce a 2-log reduction (t99) for either the general or HF Bacteroidales-based DNA markers BacUni and BacHum (24) in sunlight-exposed or dark seawater microcosms incubated at 14°C (t99 was approximately 7 days for both markers). This was in contrast to a seawater microcosm study by Walters et al. (48), in which the HF DNA marker BacHum exhibited significantly faster decay in natural sunlight (t90 = 1.77 days) than in the dark (t90 = 8.72 days) at 17°C.
In a freshwater application, Walters and Field (47) used qPCR to estimate HF marker concentrations in microcosms spiked with feces, incubated them at 13°C, and compared them with cultivated FIB. They found that exposure to natural sunlight did not significantly affect the persistence of either total (from both live and dead cells) HF Bacteroidales DNA markers or cultivable E. coli. However, measurement of rRNA by reverse transcriptase qPCR, rather than measurement of DNA by qPCR, indicated that exposure to sunlight did result in the more rapid decay of HF markers associated with living cells.
The objective of this study was to measure the relative decay of cultivated E. coli and general and HF Bacteroidales 16S rRNA gene copy numbers in wastewater-spiked river water as it ages under different conditions. Microcosms were constructed to simulate environmental stresses, spiked with sewage, and monitored over time. One potential application of these results is the use of an HF marker-to-AllBac ratio to help interpret the contribution of human sources to overall fecal load or of an HF marker-to-E. coli ratio to interpret the extent to which human sources contribute specifically to E. coli density. If decay rates among the general marker, HF markers, and E. coli are similar, these ratios could be considered stable over time and take the science one step closer to this eventual use.
MATERIALS AND METHODS
Microcosms and treatments.
Microcosm experiments were conducted using constant-temperature walk-in incubators at the Soil Microbial Ecology Laboratory in the School of Environment and Natural Resources at The Ohio State University. Fresh river water from the Olentangy River near the Ohio State University was collected into autoclaved, 20-liter polypropylene carboys (Nalgene Nunc, Rochester, NY), using a sump pump after flushing with river water. Sediment was collected from the river bottom using a presterilized petite Ponar sediment sampler. Large organic debris (leaves and sticks) was picked from the sediment manually before addition to selected microcosms.
Each of the 15 microcosms consisted of 15 liters of river water spiked with 1% (vol/vol) settled, untreated wastewater from the Olentangy Environmental Control Center (Delaware, OH). The series of microcosms was divided in sets of 3 among 4 test treatments and 1 set of controls. The four test treatments were the following: (i) artificial sunlight, with typical distribution of UV (UVA and UVB) and photosynthetically active radiation (PAR) (12 h on/12 h off); (ii) reduced temperature (15°C); (iii) removal of autochthonous predation (autoclaved river water); and (iv) exposure to sediment (10 g/liter added and settled). All microcosms except those in the reduced temperature treatment were incubated at 25°C, and the experiment was carried out for 11 days. The initial turbidity of source river water was 102 nephelometric turbidity units (NTU); by the first day of the experiment, turbidities fell to between 45 and 69 NTU (the sediment microcosms had higher values than the others), and by the end of the experiment, turbidities dropped to between 22 and 47 NTU. The potential effect of the settling observed through this reduction in turbidity was addressed by shaking each microcosm to resuspend, and resampling after the 11-day sampling was collected and processed.
For all treatments except artificial sunlight, carboy lids were fitted with aquarium tubing and air release valves through rubber stoppers, and incubations took place in the dark under constant mixing using an air stone and aquarium air pump. Serological pipets (25 ml) were inserted through the rubber stoppers to allow for sampling. For the artificial sunlight experiments, 5-gallon buckets were bleached (6% hypochlorite), rinsed with distilled water, and covered with 0.05-mm-thick FEP-Teflon film, which is 96% transmissive to UVA and UVB light (Dupont Corporation, Wilmington, DE). Submersible, recirculating pumps were placed in the buckets to minimize condensation on the UV-transparent film, which was observed with the aeration method of mixing. The microcosms were acclimated in the incubators overnight at treatment temperatures prior to being spiked with wastewater. Submersible temperature loggers (Hobo Pendant; Onset Computer Corporation, Bourne, MA) were placed in 2 of the 3 light microcosms, the darkened control that was held in the same room, an additional control that was held in a 25°C room, and one test microcosm that was held in a 15°C room.
Artificial sunlight.
A 400-W Philips MSR 400 HR hydrargyrum medium-arc iodide (HMI) lamp (Koninklijke Philips Electronics, Netherlands) was used to approximate the levels of natural UVA, UVB, and PAR, based on the American Society for Testing and Materials (ASTM) standard terrestrial solar spectral irradiance distributions for U.S. latitude 37°N (1a). The lamp's spectral power distribution is similar to that of sunlight (continuous spectrum from 250 to 800 nm; power plateau from 400 to 700 nm). It was tested prior to and during the experiment using radiometers to measure output in the UVA (320- to 400-nm) and UVB (280- to 320-nm) ranges (Solarmeter 5.0 and 6.2; Solartech, Inc., Harrison Township, MI) and the PAR (400- to 700-nm) range (Field Scout quantum light meter; Spectrum Technologies, Inc., Plainfield, IL).
Target intensities for UVA, UVB, and PAR (from ASTM typical summertime values) were 8.9 mW/cm2 (UVA+UVB), 0.3 mW/cm2 (UVB), and 1,950 μmol/m2/s (PAR). The intensities of irradiation that the experimental apparatus delivered to the microcosms were 0.68 mW/cm2 (UVA+UVB), 0.06 mW/cm2 (UVB), and 240 μmol/m2/s (PAR). These intensities, approximately 1/10 of the target intensities, delivered 17 to 69% (mean, 26%) of the daily total PAR (mmol/m2) measured in a separate study at Huntington Beach on Ohio's Lake Erie shore during the summer of 2008 (our unpublished data).
Sampling.
Sampling was carried out 1 h after inoculation, with 10 ml/liter wastewater, and again at the same times on days 1, 3, 5, and 11. For each sample, volumes of 40 ml were removed to 50-ml polypropylene Oak Ridge centrifuge tubes (Nalgene Nunc, Rochester, NY) for marker DNA analysis. Increasing volumes (10 ml to 100 ml) were removed for E. coli analysis over time, as different volumes were required to remain within assay limits of detection. A 10-ml sample was collected separately from each microcosm for turbidity analyses.
Enumeration of E. coli.
The Colilert assay using the Quanti-Tray/2000 was used to measure cultivable E. coli in each microcosm sample, according to the manufacturer's protocol (IDEXX Laboratories, Inc., Westbrook, ME). Dilutions were made as needed, with 10−1 and 10−2 dilutions on day 0 and with direct analysis of 100-ml volumes by day 5.
Preparation of internal DNA processing control.
A pure culture of Pantoea stewartii was prepared as described previously (39). Estimates of concentrations were made by plate counting, and 1-ml culture aliquots were stored at −80°C. This Gram-negative-control species, with morphological characteristics similar to those of the Bacteroidales, was spiked into each of the microcosm samples prior to processing and DNA extraction to monitor recovery efficiency. Percent recovery of the spike was used to adjust marker concentrations. The genetic target for detection of P. stewartii was the exopolysaccharide synthesis gene (cpsD), carried as a single copy on the chromosome (9). Previous work (39) demonstrated that adjustment of observed qPCR data according to the level of spike recovery reduced variability in replicate analyses, suggesting that it could be used to enhance precision in studies involving trends analysis and treatment comparisons. Each of the 40-ml samples from the microcosms was spiked with approximately 1.5 × 106 CFU of P. stewartii at the time of collection.
Sample processing and DNA extraction.
Immediately after spike addition, samples were vortexed and then concentrated at 2,880 × g for 10 min in a Sorvall RC2-B centrifuge (Thermo Electron Corp., Asheville, NC), and supernatant was decanted. After the addition of 500 μl of MoBio PowerSoil DNA extraction kit lysis buffer (MoBio, Inc., Carlsbad, CA), cells were vortexed and stored at −80°C prior to DNA extraction. The MoBio kit protocol was modified to use all the supernatant, as described by Stoeckel et al. (39), and extraction controls using PCR-grade water were included each day. The DNA was eluted in 250 μl of C6 reagent, and purified DNA was stored at −20°C.
qPCR assays.
Genomic (P. stewartii) and plasmid (all other assays) DNA for qPCR standards was prepared according to the work of Stoeckel et al. (39). All assays were performed according to published protocols (24, 26, 35, 40). Sequences for primers and probes used in the study are listed in Table 1. Reactions were run on the Bio-Rad iQ5 instrument (Bio-Rad Laboratories, Inc., Hercules, CA) in 25-μl volumes, using 5 μl of the 250-μl extracts as a template. No-template controls containing PCR-grade water, along with 6-point standard curves, were run in triplicate with each set of reactions. Test reactions were carried out in duplicate. Unamended river water and sewage samples were processed as negative controls for the internal spike and as positive controls for the background occurrence of the markers in the sample matrices. Limits of detection (LOD) for each assay were defined as the fifth percentile among observed threshold cycles (CT) across all blanks and negative-control reactions. Inhibition of the qPCR was monitored by making 10−1 and 10−2 extract dilutions and was considered significant when the difference in CT between dilutions was ≤3. Inhibition was not detected in any sample.
TABLE 1.
Primer and probe identities, target genes, sequences, and amplicon lengths for general fecal, HF and internal control qPCR assays used in this studya
| Primer/probe | Target gene | Sequence (5′-3′) | Size (bp) | Reference |
|---|---|---|---|---|
| AllBac296F | General Bacteroidales 16S rRNA | GAGAGGAAGGTCCCCCAC | 106 | 26 |
| AllBac412R | CGCTACTTGGCTGGTTCAG | |||
| AllBac-Bhq | CCATTGACCAATATTCCTCACTGCTGCCT | |||
| BacHum160F | HF Bacteroidales 16S rRNA | TGAGTTCACATGTCCGCATGA | 81 | 24 |
| BacHum241R | CGTTACCCCGCCTACTATCTAATG | |||
| BacHum193FBhq | TAGGGGTTCTGAGAGGAAGGTCCCCC | |||
| HF183F | HF Bacteroidales 16S rRNA | ATCATGAGTTCACATGTCCG | 82 | 35 |
| Bac242R | TACCCCGCCTACTATCTAATG | |||
| cpsRT74F | P. stewartii cpsD | TGCTGATTTTAAGTTTTGCTA | 82 | 40 |
| cpsRT177R | AAGATGAGCGAGGTCAGGATA | |||
| cps133-Bhq | TCGGGTTCACGTCTGTCCAACT |
Internal control, P. stewartii.
Normalization of standard curves.
Variability among qPCR standards was minimized by the generation of composite standard curves, in which acceptance parameters were established based on efficiency between 0.80 and 1.10 and R2 values of >0.99 (Sigma qPCR technical guide). The CT for every observation was adjusted according to the average difference in CT between the run standard curve and the composite standard curve (ΔCT). When an additional standard curve was added to the composite, 100 iterations were performed to reposition the composite standard curve and recalculate ΔCT values for each constituent qPCR run.
Data analyses.
Analytical replicates were expressed as the geometric mean marker copy number per 100 ml or E. coli most probable number (MPN) per 100 ml, and experimental replicates were expressed as the mean log10 reductions and t99 ± standard deviations. Decay was characterized by the log10 reduction for each experimental parameter at each time point. Using the following equations, decay rates were calculated based on a first-order decay model, Nt/N0 = 10−kt (11), and t99 values were calculated using the decay constant (k) in the following equation, t99 = −2/k.
In many cases, the full time scale of data collected did not fit the first-order decay model; hence, those data that were approximately linear, plus all data relevant to 99% reduction whether linear or not, were used in the calculations (as described further in Results). Differences among the mean values were evaluated at a 95% confidence interval using Tukey's honestly significant difference (HSD) post-hoc multiple-comparisons test in conjunction with analysis of variance (ANOVA) in SPSS 17.0 (SPSS, Inc., Chicago, IL).
RESULTS
In this study, 15 river water microcosms were sampled on days 0, 1, 3, 5, and 11 after being spiked with human wastewater to compare the decay of Bacteroidales general and HF markers against cultivated E. coli under the effects of artificial sunlight, exposure to sediment, reduced temperature, and reduced predation. Decay (log10 reduction) at each sampling time was compared for each of the four targets within (Table 2) and across (Table 3) treatments, and t99 were compared across treatments (Table 4). Temperature, monitored continuously over the duration of the study of the five representative microcosms, remained stable. Within the three constant-temperature rooms used for this experiment, means and standard deviations were 24.2 ± 0.1°C, 25.2 ± 0.6°C, and 15.4 ± 0.0°C at reduced temperature (data not shown). Irradiation by the lamp used to create artificial sunlight caused an increase in temperature for treated microcosms compared to that of the control in the same room (28.3 ± 1.3°C for the treated microcosm compared to 25.2 ± 0.6°C for the control that was shielded from light).
TABLE 2.
Level of significance for log10 reduction values among microbial targets for each treatment and daya
| Target | Treatment day | Level of significanceb |
||||
|---|---|---|---|---|---|---|
| Control | Light | Sediment | Reduced temp | Reduced predation | ||
| E. coli | 1 | a | a | a | a | a |
| qHF183 | a | b | c | a | bc | |
| BacHum | a | b | c | a | ab | |
| AllBac | a | b | b | a | c | |
| E. coli | 3 | b | a | b | a | a |
| qHF183 | ab | <LOD | c | a | b | |
| BacHum | b | <LOD | c | a | ab | |
| AllBac | a | a | a | a | ab | |
| E. coli | 5 | b | <LOD | b | a | a |
| qHF183 | <LOD | <LOD | c | <LOD | c | |
| BacHum | <LOD | <LOD | <LOD | <LOD | bc | |
| AllBac | a | Detected | a | a | b | |
Log10 reduction values were plotted in Fig. 1.
<LOD, one or more observations below the LOD and statistical comparisons not done; detected, observation was above the LOD but below the lowest standard curve value. Lowercase letters denote statistically significant differences in the remaining concentration in each treatment on each day, as evaluated by Tukey's HSD test (P < 0.05).
TABLE 3.
Level of significance for log10 reduction values among treatments within each microbial target and daya
| Treatment | Target day | Level of significanceb |
|||
|---|---|---|---|---|---|
| E. coli | qHF183 | BacHum | AllBac | ||
| Control | 1 | a | ab | a | a |
| Light | ab | abc | ab | a | |
| Sediment | a | a | ab | a | |
| Red temp | a | bc | bc | a | |
| Red predation | b | c | c | a | |
| Control | 3 | a | a | a | a |
| Light | a | <LOD | <LOD | a | |
| Sediment | a | a | a | a | |
| Red temp | a | a | a | a | |
| Red predation | b | b | b | a | |
| Control | 5 | a | <LOD | <LOD | a |
| Light | <LOD | <LOD | <LOD | a | |
| Sediment | a | <LOD | <LOD | a | |
| Red temp | a | <LOD | <LOD | a | |
| Red predation | b | Detected | Detected | a | |
Log10 reduction values were plotted in Fig. 1.
<LOD, one or more observations below the LOD and statistical comparisons not done; detected, observation was above the LOD but below the lowest standard curve value. Lowercase letters denote statistically significant differences in the remaining concentration of each target on each day, as evaluated by Tukey's HSD test (P < 0.05).
TABLE 4.
Mean t99 for all targets and time needed to achieve 235 E. coli MPN/100 ml under different treatments, as estimated from log10 reductions
| Treatment (temp [°C]) | Mean t99 (SD) (days)a |
Mean time (days) needed to achieve 235 E. coli MPN/100 ml (SD) | |||
|---|---|---|---|---|---|
| E. coli | qHF183 | BacHum | AllBac | ||
| Control (25) | 2.02 (0.28) a | 2.16 (0.47) a | 1.74 (0.16) a | 3.28 (1.26) a | 1.98 (0.28) |
| Light (25) | 2.18 (0.18) b | 1.71 (0.23) a | 1.54 (0.42) b | 2.55 (0.23) b | 2.13 (0.18) |
| Sediment (25) | 2.80 (0.42) b | 1.88 (0.47) a | 2.08 (0.10) a | 4.44 (0.34) c | 2.74 (0.41) |
| Reduced temp (15) | 3.01 (1.07) a | 2.53 (0.36) a | 2.35 (0.53) a | 2.73 (0.84) a | 2.94 (1.04) |
| Reduced predation (25) | 7.14 (0.34) b | 2.78 (0.03) a | 3.03 (0.23) a | 3.75 (0.86) a | 6.98 (0.33) |
Letters denote statistically significant differences among the t99 of microbial targets within treatments, as evaluated by Tukey's HSD test (P < 0.05).
Recovery of internal processing control.
The internal control spike, P. stewartii, was monitored to measure recovery efficiency for each sample through the steps of sample processing and DNA extraction (39). Recovery of the internal control ranged from 2.5% to 73% of the estimated 1.5 × 106 CFU spike concentration. Concentrations of the Bacteroidales targets were estimated by dividing the observed concentration by the proportional spike recovery. Coefficients of variation (CV) for marker concentrations in triplicate microcosms ranged from 1.2% to 870% (mean overall CV = 57%) before adjustment for spike recovery and from 0.53% to 58% (mean overall CV = 16%) after adjustment. The reduced CV in adjusted measurements indicated that use of the internal processing control enhanced measurement precision and validated its use in this application.
Starting concentrations and assay LOD.
Following equilibration and spiking of microcosms, mean concentrations of the four targets were 2.12 × 104 E. coli MPN/100 ml, 8.38 × 108 AllBac copies/100 ml, and 3.76 × 107 and 1.02 × 107 copies/100 ml for qHF183 and BacHum, respectively. The LOD for BacHum and qHF183 qPCR assays were 4 and 6 copies/5-μl extract. The LOD for AllBac was approximately 450 copies/5 μl, or about 5.6 × 104 copies/100 ml, and the LOD for E. coli was 1 MPN/100 ml. All HF marker measurements were at or near the LOD under each treatment by day 5. AllBac was detectable up to day 11 in all treatments. E. coli was detectable up to day 5 in all treatments except light and in reduced predation up to day 11. In all but the reduced predation treatment, E. coli density was below the U.S. EPA single-sample water quality criterion (235 MPN/100 ml) (42) by day 3.
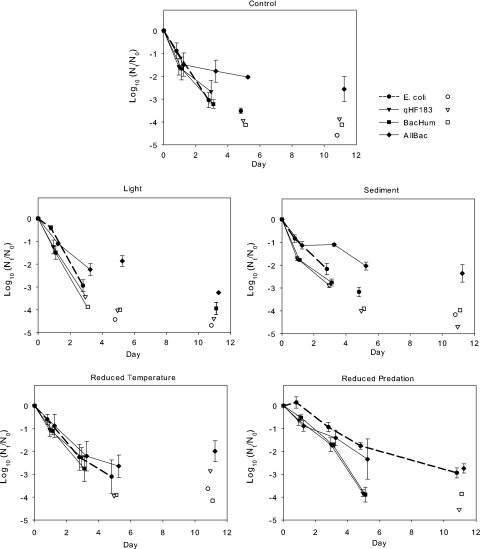
Decay curves.
Fig. 1 shows the mean decay curves for the four targets in each treatment. Statistical differences evaluated by mean separation of the rates of target decay within and across each treatment and each sample time are shown in Tables 2 and 3, to be used in conjunction with Fig. 1. There were no differences in decay among any targets in control and reduced temperature treatments on the first day (Table 2). Among the other three treatments, decay of E. coli at day 1 was always the same or less than decay of the other targets. Decay of the two human markers was not different under any treatments on day 1. The decay response of AllBac compared with the other targets varied on day 1.
FIG. 1.
Mean decay curves for E. coli and markers in microcosms exposed to different conditions, measured as the remaining concentration [log10(Nt/N0)] over time (t = days). Data points represent the mean values obtained among triplicate microcosms. Error bars indicate standard deviations. Filled symbols represent data points for which the target could be measured in all three microcosms. Open symbols represent data points for which at least one observation was below the limit of detection. Points used for the calculation of decay constants are connected by lines.
By the third day of the experiment, the decay of AllBac was always the same or less than that of the other targets. E. coli decay remained the same as or less than the decay of the HF markers. The reduced temperature treatment was the only one for which there remained no difference among the decay of targets. The decay of the two HF targets was not significantly different for any of the treatments. The HF markers dropped to below the LOD in the light treatment, and the decay was greater than those of E. coli and AllBac in the sediment treatment.
By the fifth day of the experiment, one or both HF markers were below the LOD in all but the reduced predation treatment. E. coli and AllBac remained similar in decay under reduced temperature, and E. coli had greater decay than AllBac under control conditions and with sediment and showed less decay than AllBac under reduced predation.
When decay data among the treatments were evaluated (Table 3), the reduced predation treatment resulted in less decay of targets (E. coli, qHF183, and BacHum) than the control at day 1 and day 3. The only other treatment effect noted was less decay of BacHum at a reduced temperature than that under the control conditions on day 1. No differences in decay for AllBac among the treatments up to day 5 were observed.
Evaluation of time needed to produce 99% decay.
Changes in the slopes of the curves suggestive of biphasic decay occurred in all treatments (per Bae and Wuertz [3]) and were most pronounced for AllBac (Fig. 1). Nonlinear target reduction over time, compounded by differences in curve shapes for various treatments and targets, indicated that a simple first-order decay model up to day 11 was not appropriate in most cases. However, 99% reduction was achieved within the near-linear area of the decay curve in most cases, so the decay constants and resulting t99 values were generated from the linear portion of each curve. In the light treatment, the HF markers had decayed to below the LOD by day 3 in some microcosms. In these microcosms, a value representing one-half of the LOD was used to estimate t99.
No significant differences were observed among t99 values for any target in the control or the reduced temperature treatments (Table 4). Similarly, t99 values for the HF markers were not significantly different from each other under any treatment. As might be expected from evaluation of the decay curves (Fig. 1), both HF markers reached t99 earlier than E. coli under conditions of light, sediment, and reduced predation. Under control, light, and reduced temperature treatments, the t99 values for AllBac were not significantly different from those of E. coli.
It was of special interest to note that AllBac in sediment had a t99 that was longer than that under any other condition and longer than those of the other targets in the sediment treatment. Following collection of the last sample on day 11, sediments were resuspended, and an additional sample was collected and analyzed from each treatment. The AllBac marker recovered nearly all of its initial concentration (0.28-log reduction from the initial log concentration of 8.54) in the sediment treatment, while the HF markers remained at or below the LOD after mixing (approximately 4-log reduction from the initial log concentration of 7.29) (data not shown). Cultivable E. coli remained 2.7 logs below their initial log concentration of 4.28 in the resuspended sample.
In order to evaluate these results in terms of the consequence for water quality monitoring, the time needed to achieve 235 E. coli MPN/100 ml was also calculated (Table 4). Because the starting concentration of E. coli in the microcosms was 2.10 × 104 MPN/100 ml, nearly 100 times the criterion, the time needed to achieve 235 MPN/100 ml was very similar to the corresponding t99 values. Except for grossly contaminated waters (beyond 105 MPN/100 ml), the t99 values presented are relevant to most scenarios encountered in recreational water monitoring.
DISCUSSION
A primary goal of this research was to evaluate the relative rates of decay of genetic markers and cultivated E. coli. Among other uses, this information is necessary in order to substitute a genetic marker for cultivated E. coli, the current standard for public health risk in recreational waters, as considered by USEPA (43). Alternately, analysis for human-associated genetic markers might be used in conjunction with E. coli to evaluate the relative contribution from humans to the overall level of E. coli. A major finding of this study was that HF marker decay was consistent with, or significantly faster than, that of E. coli under all treatments. This indicates that the HF markers might be useful as conservative estimators of human-origin E. coli even as fecal contamination ages in the environment.
The complex interactions of various environmental parameters with bacterial decay are difficult to model (11). The scope of relevance for the decay evaluations reported in this paper is limited to the time needed to produce 99% decay because it was impossible to extrapolate the first-order decay model beyond this point. A water body would have to be contaminated beyond a level of 23,500 MPN/100 ml to retain significant levels of contamination (above 235 MPN/100 ml) past the t99 level of decay. For this reason, interpretations from this experiment, while limited in scope, are applicable under conditions found in most environmental settings.
Plots of decay as a function of time indicated that E. coli and AllBac tended to decay with a biphasic pattern rather than a first-order model of continuous decay. The shapes of the curves varied, indicating that the various treatments affected subpopulations of targets differently. Possible explanations for this phenomenon include population heterogeneity or differences in growth phase at the time of inoculation (1, 22). In this study, since wastewater was introduced into river water, either factor could contribute to the exhibited patterns; potential rapid initial decay of newly introduced cells and DNA may have been followed by a reduction in the decay rate due to the presence of a more adapted or protected background population. In all cases, however, the biphasic nature of the decay curve came into effect below the 99% (2-log) reduction level.
In one epidemiological study, Wade et al. found that a marker of general fecal contamination was not consistently correlated with health risk (45). The general fecal contamination marker used in the present study (AllBac) exhibited a pronounced biphasic decay curve, likely due to the greater heterogeneity (the number of different phylotypes) this marker represents compared with that of the other targets. Some of these groups were more persistent than the HF markers or E. coli. The biphasic curve suggests that a reservoir of persistent AllBac markers could be present in previously contaminated waters, such that AllBac could not be used as an alternate indicator of health risk (in place of E. coli) or in ratio with the HF markers to estimate the source contribution. Indeed, when sediments were resuspended at the end of this experiment, the final concentration of the AllBac marker returned to about 50% of its original concentration, while the other markers all remained at less than 1% of the original concentration. The failure of the other general Bacteroidales marker (GenBac) to predict health risk under some conditions in the prior study (45) may be explained, in part, by these findings.
While it has been suggested that sunlight is the most important controlling factor for the survival of E. coli in water (29, 36), the present study, as in that of Walters and Field (47), found no significant effect of exposure to sunlight on either the survival of E. coli or the persistence of the HF DNA markers. One plausible explanation for the finding in the current study is the turbidity in the microcosm source water (102 NTU)—turbidity plays a role in limiting the effects of light on decay rates (8). Another explanation is that the source of artificial sunlight was not as strong as natural sunlight, providing only about one-fourth of the daily irradiance measured throughout the summer at a Lake Erie beach (our unpublished data). The source of sunlight, temperature, and likely other environmental conditions were quite different in the Walters and Field study (47), which took place outdoors in Oregon, thus making direct comparisons difficult. The current study showed more similarity in the decay of E. coli and the HF markers in the sunlight treatment than the Oregon study.
Microcosms containing sediment had significantly higher mean levels of turbidity than the other microcosms over the course of the 11-day experiment (P = 0.016); however, at each time step, the decay of all targets in the sediment microcosms was not significantly different from the decay observed in the control microcosms (Table 3). Reduced temperature, as in other studies, resulted in longer persistence for all targets, and t99 values showed no significant differences among the targets at 15°C. However, the t99 values for qHF183 were considerably lower than those reported by Seurinck et al. (35) (6 days at 28°C versus 2.16 days at 25°C in this study; 10 days at 12°C versus 2.53 days at 15°C in this study).
Although reduced predation had the greatest effect on decay for all of the targets, the significance of this parameter may not be great in fresh recreational water where this is not a typical state of the environment. The t99 value for E. coli under reduced predation was significantly higher than those for the Bacteroidales targets, which suggests that in applications such as drinking water management, the molecular markers may not correlate well with E. coli.
Better understanding of decay characteristics of the regulatory FIB, E. coli, and MST molecular markers of fecal contamination was needed to evaluate the markers as potential substitutes for, or complements to, E. coli density measurements in water quality management (43). If the marker-to-E. coli ratio is consistent over time, it may be possible to (i) use AllBac as a rapidly measured indicator in place of E. coli or (ii) use the human-associated markers to describe the relative amount of E. coli contamination due to human sources. In general, the genetic markers had t99 values that were similar to or higher than those of E. coli under all tested conditions. However, AllBac showed evidence of a persistent subpopulation below the 99% removal level; HF markers had 4-log or greater removal by day 5 of the experiment, while less than 3-log removal of AllBac had been achieved by day 11. This reservoir of persistent AllBac makes it unlikely that the AllBac marker could be used as a rapidly measured surrogate for E. coli contamination in recreational waters. The qHF183 and BacHum markers, however, may be useful as conservative measures to indicate the prevalence of human-origin E. coli in contaminated waterways. Further evaluation of the cooccurrence of HF markers and E. coli in sources of human and nonhuman fecal contamination is required to derive the necessary models.
Acknowledgments
Research funding was provided under an agreement with the Ohio River Valley Water Sanitation Commission (ORSANCO), Cincinnati, OH, the USGS cooperative water program, and The Ohio State University Research Foundation.
We are grateful to Jason Heath and Sam Dinkins of ORSANCO and Jorge Santo Domingo of USEPA for their contributions to this project. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.ASTM. 2009. Standard terrestrial solar spectral irradiance distributions for U.S. latitude 37°N. Active standard ASTM G173 developed by subcommittee G03.09. In Book of standards, vol. 14.04. ASTM, West Conshohocken, PA.
- 2.Bae, S., and S. Wuertz. 2009. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with propidium monoazide. Appl. Environ. Microbiol. 75:2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, S., and S. Wuertz. 2009. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res. 43:4850-4859. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, B., and D. M. Gordon. 2004. Coliform dynamics and the implications for source tracking. Environ. Microbiol. 6:501-509. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A., A. C. Layton, L. McKay, D. Williams, R. Gentry, and G. S. Sayler. 2009. Factors influencing the persistence of fecal Bacteroides in stream water. J. Environ. Qual. 38:1224-1232. [DOI] [PubMed] [Google Scholar]
- 6.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beversdorf, L. J., S. M. Bornstein-Forst, and S. L. McLellan. 2007. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. J. Appl. Microbiol. 102:1372-1381. [DOI] [PubMed] [Google Scholar]
- 8.Cantwell, R. E., and R. Hofmann. 2008. Inactivation of indigenous coliform bacteria in unfiltered surface water by ultraviolet light. Water Res. 42:2729-2735. [DOI] [PubMed] [Google Scholar]
- 9.Coplin, D. L., D. R. Majerczak, Y. Zhang, W.-S. Kim, S. Jock, and K. Geider. 2002. Identification of Pantoea stewartii subsp. stewartii by PCR and strain differentiation by PFGE. Plant Dis. 86:304. [DOI] [PubMed] [Google Scholar]
- 10.Craig, D. L., H. J. Fallowfield, and N. J. Cromar. 2004. Use of microcosms to determine persistence of Escherichia coli in recreational coastal water and sediment and validation with in situ measurements. J. Appl. Microbiol. 96:922-930. [DOI] [PubMed] [Google Scholar]
- 11.Crane, S. R., and J. A. Moore. 1986. Modeling enteric bacterial die-off: a review. Water Air Soil Pollut. 27:411-439. [Google Scholar]
- 12.Englebert, E. T., C. McDermott, and G. T. Kleinheinz. 2008. Impact of the alga Cladophora on the survival of E. coli, Salmonella, and Shigella in laboratory microcosm. J. Great Lakes Res. 34:377-382. [Google Scholar]
- 13.Field, K. G. 2004. Faecal source identification, p. 349-366. In J. A. Cotruvo (ed.), Waterborne zoonoses: identification, causes, and control. IWA Publishing, Essex, United Kingdom.
- 14.Fremaux, B., J. Gritzfeld, T. Boa, and C. K. Yost. 2009. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res. 43:4838-4849. [DOI] [PubMed] [Google Scholar]
- 15.García-Armisen, T., and P. Servais. 2009. Partitioning and fate of particle-associated E. coli in river waters. Water Environ. Res. 81:21-28. [PubMed] [Google Scholar]
- 16.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D. M., and F. FitzGibbon. 1999. The distribution of enteric bacteria from Australian mammals: host and geographical effects. Microbiology 145:2663-2671. [DOI] [PubMed] [Google Scholar]
- 18.Gourmelon, M., M. P. Caprais, R. Ségura, C. Le Mennec, S. Lozach, J. Y. Piriou, and A. Rincé. 2007. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl. Environ. Microbiol. 73:4857-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haile, R. W., J. S. Witte, M. Gold, R. Cressey, C. McGee, R. C. Millikan, A. Glasser, N. Harawa, C. Ervin, P. Harmon, J. Harper, J. Dermand, J. Alamillo, K. Barrett, M. Nides, and G. Y. Wang. 1999. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology 10:355-363. [PubMed] [Google Scholar]
- 20.Harwood, V. J., M. Brownell, S. Wang, J. Lepo, R. D. Ellender, A. Ajidahun, K. N. Hellein, E. Kennedy, X. Ye, and C. Flood. 2009. Validation and field testing of library-independent microbial source tracking methods in the Gulf of Mexico. Water Res. 43:4812-4819. [DOI] [PubMed] [Google Scholar]
- 21.Harwood, V. J., A. D. Levine, T. M. Scott, V. Chivukula, J. Lukasik, S. R. Farrah, and J. B. Rose. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71:3163-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellweger, F. L., V. Bucci, M. R. Litman, A. Z. Gu, and A. Onnis-Hayden. 2009. Biphasic decay kinetics of fecal bacteria in surface water not a density effect. J. Environ. Eng. 135:372-376. [Google Scholar]
- 23.Ishii, S., D. L. Hansen, R. E. Hicks, and M. J. Sadowsky. 2007. Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environ. Sci. Technol. 41:2203-2209. [DOI] [PubMed] [Google Scholar]
- 24.Kildare, B. J., C. M. Leutenegger, B. S. McSwain, D. G. Bambic, V. B. Rajal, and S. Wuertz. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701-3715. [DOI] [PubMed] [Google Scholar]
- 25.Kreader, C. A. 1998. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl. Environ. Microbiol. 64:4103-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Layton, A., L. McKay, D. Williams, V. Garrett, R. Gentry, and G. Sayler. 2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72:4214-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, C. M., T. Y. Lin, C. C. Lin, G. A. Kohbodi, A. Bhatt, R. Lee, and J. A. Jay. 2006. Persistence of fecal indicator bacteria in Santa Monica Bay beach sediments. Water Res. 40:2593-2602. [DOI] [PubMed] [Google Scholar]
- 28.Maïga, Y., K. Denyigba, J. Wethe, and A. S. Ouattara. 2009. Sunlight inactivation of Escherichia coli in waste stabilization microcosms in a sahelian region (Ouagadougou, Burkina Faso). J. Photochem. Photobiol. B 94:113-119. [DOI] [PubMed] [Google Scholar]
- 29.Muela, A., J. M. Garcia-Bringas, I. Arana, and I. Barcina. 2000. The effect of simulated solar radiation on Escherichia coli: the relative roles of UV-B, UV-A, and photosynthetically active radiation. Microb. Ecol. 39:65-71. [DOI] [PubMed] [Google Scholar]
- 30.Okabe, S., and Y. Shimazu. 2007. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl. Microbiol. Biotechnol. 76:935-944. [DOI] [PubMed] [Google Scholar]
- 31.Pote, J., L. Haller, R. Kottelat, V. Sastre, P. Arpagaus, and W. Wildi. 2009. Persistence and growth of faecal culturable bacterial indicators in water column and sediments of Vidy Bay, Lake Geneva, Switzerland. J. Environ. Sci. 21:62-69. [DOI] [PubMed] [Google Scholar]
- 32.Prüss, A. 1998. Review of epidemiological studies on health effects from exposure to recreational water. Int. J. Epidemiol. 27:1-9. [DOI] [PubMed] [Google Scholar]
- 33.Reischer, G. H., D. C. Kasper, R. Steinborn, A. H. Farnleitner, and R. L. Mach. 2007. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett. Appl. Microbiol. 44:351-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaub, S. A. 2004. A regulatory perspective on zoonotic pathogens in water, p. 439-470. In J. A. Cotruvo (ed.), Waterborne zoonoses: identification, causes, and control. IWA Publishing, Essex, United Kingdom.
- 35.Seurinck, S., T. Defoirdt, W. Verstraete, and S. D. Siciliano. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249-259. [DOI] [PubMed] [Google Scholar]
- 36.Sinton, L. W., R. K. Finlay, and P. A. Lynch. 1999. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl. Environ. Microbiol. 65:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoeckel, D. M., and V. J. Harwood. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73:2405-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoeckel, D. M., E. A. Stelzer, and L. K. Dick. 2009. Evaluation of two spike-and-recovery controls for assessment of extraction efficiency in microbial source tracking studies. Water Res. 43:4820-4827. [DOI] [PubMed] [Google Scholar]
- 40.Tambong, J. T., K. N. Mwange, M. Bergeron, T. Ding, F. Mandy, L. M. Reid, and X. Zhu. 2008. Rapid detection and identification of the bacterium Pantoea stewartii in maize by TaqMan real-time PCR assay targeting the cpsD gene. J. Appl. Microbiol. 104:1525-1537. [DOI] [PubMed] [Google Scholar]
- 41.USEPA. 1986. Ambient water quality criteria for bacteria, EPA/440584002. United States Environmental Protection Agency, Washington, DC.
- 42.USEPA. 2003. Bacterial water quality standards for recreational waters (freshwater and marine waters) status report, EPA/823/R-03/008. United States Environmental Protection Agency, Washington, DC.
- 43.USEPA. 2007. Critical path science plan for the development of new or revised recreational water quality criteria, EPA-823-R-08-002. United States Environmental Protection Agency, Washington, DC.
- 44.USEPA. 2007. Report of the experts scientific workshop on critical research needs for the development of new or revised recreational water quality criteria, EPA-823-R-07-007. United States Environmental Protection Agency, Washington, DC.
- 45.Wade, T. J., R. L. Calderon, E. Sams, M. Beach, K. P. Brenner, A. H. Williams, and A. P. Dufour. 2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ. Health Perspect. 114:24-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wade, T. J., N. Pai, J. N. S. Eisenberg, and J. M. Colford, Jr. 2003. Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 111:1102-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walters, S. P., and K. G. Field. 2009. Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ. Microbiol. 11:1410-1421. [DOI] [PubMed] [Google Scholar]
- 48.Walters, S. P., K. M. Yamahara, and A. B. Boehm. 2009. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: implications for their use in assessing risk in recreational waters. Water Res. 43:4929-4939. [DOI] [PubMed] [Google Scholar]
- 49.Whitman, R. L., D. A. Shively, H. Pawlik, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]