Abstract
A unique, coleopteran-active protein, termed eCry3.1Ab, was generated following variable-region exchange of a Bacillus thuringiensis lepidopteran-active protein, Cry1Ab, with a Cry3A region. Our results support the hypothesis that this variable-region exchange is responsible for imparting strong bioactivity against the larvae of western corn rootworm (WCR) (Diabrotica virgifera virgifera LeConte), a pest species which is not susceptible to either parent protein sequence. This study demonstrates the potential of successfully engineering a portion(s) of a lepidopteran-active B. thuringiensis sequence so that it has activity against coleopterans. Further elucidation of the eCry3.1Ab activity indicated the importance of variable regions 4 to 6 that were derived from Cry1Ab instead of Cry1Ac. There was some flexibility in making domain III of engineered hybrid insecticidal proteins even more Cry1Ab-like and retaining activity, while there was less flexibility in making domain III more Cry3A-like and retaining activity. In vitro binding studies with brush border membrane vesicles demonstrated that there was specific binding of chymotrypsin-processed modified Cry3A (mCry3A), which was not diminished by addition of a 100-fold molar excess of chymotrypsin-processed eCry3.1Ab or unprocessed eCry3.1Ab. In addition, in the converse experiment, specific binding of chymotrypsin-processed eCry3.1Ab was not diminished by the presence of a 75-fold molar excess of chymotrypsin-processed mCry3A. These data support the hypothesis that eCry3.1Ab can interact with different binding sites than the activated form of mCry3A in the WCR brush border and may provide a different mode of action from the standpoint of resistance management.
The three-dimensional structures of Bacillus thuringiensis δ-endotoxins indicate that the Cry1, Cry2, Cry3, and Cry4 types of proteins have similar three-domain organizations (2, 10, 16, 18). The three structural domains are N-terminal domain I comprised of 7 α-helices, domain II containing three β-sheets in a so-called Greek key conformation, and C-terminal domain III, which is a β-sandwich in a so-called jellyroll conformation. The active portions of B. thuringiensis Cry proteins are also characterized by having five conserved blocks in their amino acid sequences, which are designated CB1 to CB5 from the N terminus to the C terminus (11). CB1 comprises approximately 29 amino acids, CB2 comprises approximately 67 amino acids, CB3 comprises approximately 48 amino acids, CB4 comprises approximately 10 amino acids, and CB5 comprises approximately 12 amino acids. The sequences before and after these five conserved blocks are highly variable and thus are designated the “variable regions,” which are designated V1 toV6. Domain I of a B. thuringiensis δ-endotoxin typically comprises V1, CB1, V2, and the N-terminal 52 amino acids of CB2. Domain II typically comprises approximately the C-terminal 15 amino acids of CB2, V3, and approximately the N-terminal 10 amino acids of CB3. Domain III typically comprises approximately the C-terminal 38 amino acids of CB3, V4, CB4, V5, and CB5. While the role of each of the three domains has not been completely identified, much work has been done by a number of investigators, and our current understanding can be summarized as follows: domain I is critical for membrane insertion and subsequent generation of high-conductance, ion-selective pores; domain II is essential for specific binding events with target membrane receptors, usually correlated with the degree of toxicity; and domain III is also involved in receptor binding and influencing insect pest specificity (for reviews, see references 3 and 23). There is also some evidence that one domain influences the properties of another domain (e.g., binding interaction accessibility [4, 18] or alteration of pore properties [6, 24, 28]).
Lepidopteran-active δ-endotoxins have been engineered by exchanges of domain regions in attempts to improve the specific activity or to broaden the spectrum of insecticidal activity. For example, addition of the C terminus of domain III of Cry1C improved the activity against Spodoptera exigua (beet armyworm) when the protein was engineered with domains I and II derived from the moderately active protein Cry1Ab, and it also improved activity when the protein was engineered with domains I and II derived from biologically inactive proteins, such as Cry1Ac, or Cry1E (7, 8). Similarly, Cry1Ca, Cry1Fb, and Cry1Ba were more active against Heliothis virescens (tobacco budworm) when they were engineered to contain domain III of Cry1Ac in place of the native sequence (13); however, the same effect was not found when Cry1Da or Cry1Ea was engineered to contain domain III of Cry1Ac. In addition, two different engineered Cry1Ba-Cry1Ia hybrid proteins in which there were exchanges of domains of the two parent proteins exhibited increased biological activity against Leptinotarsa decemlineata (Colorado potato beetle), although they were still less active than Cry3A (19). All of the hybrids described above were constructed to include domains from lepidopteran-active Cry1 proteins to obtain new proteins with activity against lepidopterans or to improve the existing activity against coleopteran pests. Shadenkov et al. (25) made a hybrid protein by fusing amino acids 48 to 565 of a Cry3A protein to amino acids 526 to 725 of a Cry1Aa protein, which resulted in a crossover between the Cry3A and Cry1Aa sequences in CB4 located in domain III. Cry3A is very active against the Colorado potato beetle (L. decemlineata) (22); however, the Cry3A-Cry1Aa hybrid protein was not active against Colorado potato beetle even though more than 75% of the hybrid protein was made up of Cry3A sequences. Also, Liu and Dean (17) demonstrated that a hybrid Cry1Aa toxin with novel mosquitocidal activity (and a concomitant loss of lepidopteran activity) could be obtained by using selected deletions, substitutions, and insertions (only 14 residues were involved) of corresponding domain II loop 1 and loop 2 structures from Cry4Ba. While the final construct exhibited activity against members of a different insect order, the process could be described as more of a targeted mutagenesis and not an exchange of domains, blocks, or variable-region sequences from the parent molecules.
Therefore, although biologically active hybrid proteins have been constructed by using domain exchanges in the Cry1 class of Cry proteins, no examples of successful domain exchanges across classes of Cry proteins have been described. In addition, engineering hybrid proteins by using a variable-region exchange strategy has not been documented. Here we report a unique, coleopteran-active protein, termed eCry3.1Ab (GenBank accession number GU327680), and related constructs that were generated following essentially a variable-region exchange involving a lepidopteran-active protein, Cry1Ab, and modified Cry3A (mCry3A) (27). Furthermore, our findings support the hypothesis that the exchanged sequence region was responsible for imparting strong bioactivity against a pest species, the western corn rootworm (WCR) (Diabrotica virgifera virgifera LeConte), which had very limited susceptibility to the parent Cry3A sequence.
MATERIALS AND METHODS
Cloning of engineered hybrid insecticidal protein genes.
The genes encoding all engineered hybrid insecticidal proteins were cloned into a modified pUC-derived vector (as described previously 27) for expression in Escherichia coli DH5α or TOP10 cells (Invitrogen).
(i) eCry3.1Ab gene.
For the initial engineered hybrid insecticidal protein, designated eCry3.1Ab (GenBank accession number GU327680), the portion of the mCry3A gene encoding the N terminus (27) through CB3 was PCR amplified from its plasmid using primers 5′3A-1-bam and C3-3A-6 (Table 1). It was noted that this PCR introduced a point mutation by deleting nucleotide 28 of the originally desired amplicon coding sequence. In addition, the resulting amplicon did not contain the 5′ BamHI site present in the primer but started with the ATG codon of the mCry3A gene. A second nucleic acid fragment encoding a C-terminal portion of a Cry1Ab protein comprising V4 through V6 was PCR amplified from a plasmid containing the maize-optimized Cry1Ab gene (15) using primers C3-1Ab-3 and 1Ab-6-Sac (Table 1). The first and second nucleic acids described above were connected by using them as templates in an overlap PCR (12) with primers 5′3A-1-bam and 1Ab-6-Sac (Table 1). The resulting amplicon was ligated into a pCR2.1-TOPO vector (Invitrogen) and transformed into E. coli; this was followed by ligation of a BamHI-SacI fragment to pET21a (EMD Biosciences, Inc.), which was cut with BamHI-SacI, and retransformation into E. coli. A preparation for the final eCry3.1Ab gene coding sequence was then constructed by placing a Kozak sequence (ACC) (14) and a start codon (ATG) immediately downstream of the N-terminal BamHI site of the pET21a vector sequence in a PCR using primers 8a-atg-delRI and C2-3A-4 (Table 1). The product was then ligated into a pCR2.1-TOPO vector (Invitrogen), which followed by BamHI-SacI subcloning into a modified pUC-derived vector (27).
TABLE 1.
Primers used for engineered hybrid insecticidal protein construct design
| Primer | Sequence (5′ to 3′) |
|---|---|
| 5′3A-1-bam | CCGGATCCATGACGGCCGACAACAACACCGAGGC |
| C3-3A-6 | CAGGGGCAGCTGGGTGATCT |
| C3-1Ab-3 | AGATCACCCAGATCCCCCTG |
| 1Ab-6-Sac | CCGAGCTCAGCTCCTACACCTGATCGATGTGGTAGTCGG |
| 8a-atg-delRI | CCGGATCCACCATGACTAGTAACGGCCGCCAGTGTGCTGGTATTCGCCCTTATGAC |
| C2-3A-4 | GTCCAGCACGGTCAGGGTCA |
| CMS96 | CCTGAACACCATCTGGCCCA |
| CMS97 | CTGGCTGCTGGGGATGATGTTGTTGAAGTCGACGCTCTT |
| CMS98 | GAGCTCTTAGGTCACCTCGGC |
| CMS99 | AAGAGCGTCGACTTCAACAACATCATCCCCAGCAGCCAG |
| CMS100 | GAAGTACCGCGCCCGCATCCGCTACGCCAGCACCACCAAC |
| CMS101 | GTTGGTGGTGCTGGCGTAGCGGATGCGGGCGCGGTACTTC |
| C2-3A-3 | TGACCCTGACCGTGCTGGAC |
| 1Ac-OL-1 | ACCCAGCTGC CCCTGGTGAAGGGAAACTTTCTTTTTA |
| 1Ac-OL-2 | TAAAAAGAAAGTTTCCCTTCACCAGGGGCAGCTGGGT |
| 1Ac-3′sac | GAGCTCCTATGTTGCAGTAACTGGAATAAA |
| FWa | GAATTTATTCCAGTTACGGTGACCTAGGAGCTCGAATTC |
| FWb | GAATTCGAGCTCCTAGGTCACCGTAACTGGAATAAATTC |
Thus, the eCry3.1Ab engineered hybrid insecticidal protein comprises (from the N terminus to the C terminus) (i) a peptidyl cap portion consisting of the amino acid sequence MTSNGRQCAGIRPYDGRQQHRG, (ii) amino acids 10 to 468 of mCry3A, which comprise V1, CB1, V2, CB2, V3, and the N-terminal 24 amino acids of CB3, and (iii) amino acids 477 to 648 of a Cry1Ab protein, which comprise the C-terminal 24 amino acids of CB3, V4, CB4, V5, CB5, V6, and 38 amino acids of the Cry1Ab protoxin tail region (Fig. 1 and 2).
FIG. 1.
Sequence alignments of (A) the N-terminal region of eCry3.1Ab and variant engineered hybrid insecticidal proteins and (B) the C-terminal region of variant engineered hybrid insecticidal proteins. Solid underlining indicates “cap sequence” residues, gray shading indicates the engineered cathepsin G site, boxes indicate the conserved sequence blocks indicated beneath the boxes, the dotted vertical line indicates the junction between domain II (D II) and domain III (D III), and dashed underlining indicates the C-terminal Cry1Ab 38 amino acid “tail sequence.” Conserved blocks were defined as described by Höfte and Whiteley (11). Note that subscript designations of engineered hybrid insecticidal proteins are used to distinguish different proteins in the present work and do not correspond to any other sequence numbering convention.
FIG. 2.
Relationship of eCry3.1Ab to other engineered hybrid insecticidal protein variants. Identical sequence regions are indicated by the same pattern. The presence of bioactivity against first-instar WCR larvae is indicated on the right.
(ii) eCry3.1Ab1 gene.
For construction of the eCry3.1Ab1 gene, the portion of the mCry3A gene (27) encoding the N terminus through CB3 was PCR amplified from its plasmid using primers 5′3A-1-bam and C3-3A-6 (Table 1). The resulting amplicon had an intact 5′ BamHI site that was present in the primer. A second nucleic acid fragment encoding a C-terminal portion of a Cry1Ab protein comprising V4 through V6 was PCR amplified from a plasmid that included the maize-optimized Cry1Ab gene (15) using primers C3-1Ab-3 and 1Ab-6-Sac (Table 1). The first and second nucleic acids described above were connected by using them as templates in an overlap PCR (12) with primers 5′3A-1-bam and 1Ab-6-Sac (Table 1) and were subcloned using steps similar to those described above for the eCry3.1Ab gene.
Thus, the eCry3.1Ab1 engineered hybrid insecticidal protein comprises (from the N terminus to the C terminus) (i) amino acids 1 to 468 of mCry3A, which comprise V1, CB1, V2, CB2, V3, and the N-terminal 24 amino acids of CB3, and (ii) amino acids 477 to 648 of a Cry1Ab protein, which comprise the C-terminal 24 amino acids of CB3, V4, CB4, V5, CB5, V6, and 38 amino acids of the Cry1Ab protoxin tail region (Fig. 1 and 2).
(iii) eCry3.1Ab1 gene-CATG.
The eCry3.1Ab1 gene-CATG construct was made by subcloning a BamHI/SalI fragment encoding an N-terminal sequence from a Cry3A gene in the modified pUC-derived vector (27) into a BamHI/SalI vector fragment from a plasmid containing the eCry3.1Ab gene sequence. The eCry3.1Ab1-CATG engineered hybrid insecticidal protein is identical to eCry3.1Ab1 except for amino acid residues 108 to 111, where it has the Cry3A sequence (Fig. 1 and 2).
(iv) eCry3.1Ab gene-CATG.
The eCry3.1Ab gene-CATG construct was made by subcloning a PpuMI/SacI fragment from the eCry3.1Ab1 gene-CATG plasmid into a PpuMI/SacI vector fragment from a plasmid containing the eCry3.1Ab gene sequence. The eCry3.1Ab-CATG engineered hybrid insecticidal protein is identical to eCry3.1Ab except for amino acid residues 121 to 124, where it has the Cry3A sequence (Fig. 1 and 2).
(v) eCry3.1Ab2 gene.
A portion of the eCry3.1Ab gene encoding the N terminus through the end of the domain II sequence was PCR amplified from its plasmid using primers CMS96 and CMS97 (Table 1). A second nucleic acid fragment encoding a domain III portion of a Cry1Ab protein was PCR amplified from a plasmid containing the maize-optimized Cry1Ab gene (15) using primers CMS98 and CMS99 (Table 1). The resulting two amplicons were then used as templates in an overlap PCR with primers CMS96 and CMS98. The resulting amplicon was cloned into pCR4 Blunt (Invitrogen). A StuI/SacI fragment of the cloned amplicon and a StuI/SacI fragment of the mCry3A gene from the modified pUC-derived vector (27) were then ligated. The eCry3.1Ab2 hybrid protein consists of (from the N terminus to the C terminus) amino acids 1 to 454 of mCry3A, which comprise domains I and II, and amino acids 463 to 610 of a Cry1Ab protein, which comprise all of domain III containing the C-terminal 38 amino acids of CB3, V4, CB4, V5, CB5, and V6 (Fig. 2).
(vi) eCry3.1Ab3 gene.
The eCry3.1Ab3 gene construct was made by subcloning a PmlI/SacI fragment of the eCry3.1Ab2 gene into the PmlI/SacI vector fragment from the eCry3.1Ab gene in the modified pUC-derived vector (27). The eCry3.1Ab2 hybrid protein consists of (from the N terminus to the C terminus) amino acids 1 to 454 of mCry3A, which comprise domains I and II, and amino acids 463 to 648 of a Cry1Ab protein, which comprise all of domain III containing the C-terminal 38 amino acids of CB3, V4, CB4, V5, CB5, V6, and 38 amino acids of the Cry1Ab protoxin tail region (Fig. 1 and 2).
(vii) eCry3.1Ab4 gene.
The eCry3.1Ab4 gene construct was made by PCR amplification of a nucleic acid fragment encoding the N-terminal portion of mCry3A into CB4 using primers CMS96 and CMS101 and a second nucleic acid fragment encoding part of domain III of Cry1Ab using primers CMS98 and CMS100 (Table 1). The two resulting amplicons were used as templates in an overlap PCR with primers CMS96 and CMS98. The resulting amplicon was then cloned into pCR4 Blunt (Invitrogen), and a SalI/SacI fragment then ligated with a SalI/SacI vector fragment from the eCry3.1Ab2 gene. Thus, the eCry3.1Ab4 hybrid protein consists of (from the N terminus to the C terminus) amino acids 1 to 519 of mCry3A into CB4 in domain III and amino acids 528 to 610 of a Cry1Ab protein, which comprise the remainder of domain III, CB4, V5, CB5, and V6 (Fig. 2).
(viii) eCry3.1Ab5 gene.
The eCry3.1Ab5 gene construct was made by subcloning a PmlI/SacI fragment of the eCry3.1Ab4 gene into the PmlI/SacI vector fragment of the eCry3.1Ab gene in the modified pUC-derived vector (27). Thus, the eCry3.1Ab4 hybrid protein consists of (from the N terminus to the C terminus) amino acids 1 to 519 of mCry3A into CB4 in domain III and amino acids 528 to 648 of a Cry1Ab protein, which comprise the remainder of domain III, CB4, V5, CB5, V6, and 38 amino acids of the Cry1Ab protoxin tail region (Fig. 1 and 2).
(ix) eCry3.1Ac1 gene.
The eCry3.1Ac1 gene construct was made by PCR amplification of a nucleic acid fragment encoding the N-terminal portion of eCry3.1Ab using primers C2-3A-3 and 1Ac-OL-2 and another nucleic acid fragment encoding part of domain III of a Cry1Ac sequence (sequence 79 [H. Hart, J. S. Chen, C. Stacy, and F. Walters, U.S. patent application WO 08121633A1]) using primers 1Ac-OL-1 and 1Ac-3′sac (Table 1). The two PCR products were used as templates in an overlap PCR with primers C2-3A-3 and 1Ac-3′sac (Table 1). The overlap PCR product was cloned into pCR2.1-TOPO (Invitrogen). A BamHI/MluI fragment from a plasmid containing the eCry3.1Ab gene, the MluI/SacI fragment from the overlap PCR product in pCR2.1, and a BamHI/SacI fragment of pET21a were then ligated. Finally, a BamHI/SacI fragment was subcloned into the BamHI/SacI vector fragment from the eCry3.1Ab gene in the modified pUC-derived vector (27) to create the eCry3.1Ac1 gene. The eCry3.1Ac1 protein comprises (from the N terminus to the C terminus) a peptidyl fragment comprising the amino acid sequence MTSNGRQCAGIRPYDGRQQHRG, amino acids 10 to 468 of mCry3A, and amino acids 477 to 610 of a Cry1Ac protein which comprises the remainder of domain III, CB4, V5, CB5, and V6 (Fig. 2).
(x) eCry3.1Ac gene.
The eCry3.1Ac gene construct was made by adding a BstEII site to the eCry3.1Ac1 gene sequence described above by QuikChange site-directed mutagenesis using primers FWa and FWb (Table 1), and a BamHI/BstEII fragment was subcloned into the BamHI/BstEII vector fragment from the eCry3.1Ab gene in the modified pUC-derived vector (27). The eCry3.1Ac protein comprises (from the N terminus to the C terminus) a peptidyl fragment comprising the amino acid sequence MTSNGRQCAGIRPYDGRQQHRG, amino acids 10 to 468 of mCry3A, and amino acids 477 to 610 of a Cry1Ac protein, which comprise the remainder of domain III, CB4, V5, CB5, V6, and 38 amino acids of the Cry1Ab protoxin tail region. This construct contains a mutation corresponding to A608V of the Cry1Ac component (Fig. 1 and 2).
Expression of engineered hybrid insecticidal proteins.
E. coli cultures were grown for 48 h at 37°C in 50 ml Luria-Bertani broth plus 100 μg/ml ampicillin and centrifuged at 4,000 rpm for 10 min, and the pellets were resuspended in 500 μl of 50 mM NaCl-50 mM Tris (pH 8.5) buffer. The resuspended cultures were then probe sonicated using three cycles with 5-s bursts over ice to lyse the cells. The cell lysates were then centrifuged at 25,000 × g for 10 min, and the resulting supernatants were used for subsequent bioassays. The expression of each engineered hybrid insecticidal protein was confirmed by Western blot analysis. In brief, blots were blocked and then incubated with rabbit anti-Cry3A immunoaffinity-purified polyclonal antibodies, followed by a secondary antibody (donkey anti-rabbit IgG linked to alkaline phosphatase [Jackson ImmunoResearch Laboratories, Inc.]). Blots were visualized by development with an alkaline phosphatase substrate solution (5-bromo-4-chloro-3-indolylphosphate [BCIP]-nitroblue tetrazolium [NBT]) (Moss, Inc.). Cell lysates contained approximately 10 to 20 μg/ml of engineered hybrid insecticidal protein, based on Western blot visualization. To obtain protein for in vivo processing and brush border membrane vesicle (BBMV) binding studies, the eCry3.1Ab gene was cloned into a modified pET24 vector (Novagen) to obtain an N-terminal His6-tagged form of the eCry3.1Ab protein, which was purified by immobilized metal affinity chromatography.
Insect bioassay of engineered hybrid insecticidal proteins.
The bioactivities of cell lysate extracts were determined using a diet incorporation assay and neonate WCR larvae as described previously (27). In brief, 200 μl of each cell lysate was mixed with an equal volume of 2× molten diet, which was then cooled and transferred into the bottom of a 47-mm sterilized petri dish (Millipore, Billerica, MA). Replicates of individual types of samples were obtained from separate culture lysates and generally bioassayed on separate days with different hatches of larvae. Between 12 and 19 neonate WCR larvae were added to each dish and kept at room temperature in the dark. The corrected mortality (expressed as a percentage) after 144 h was calculated by comparison to the results for cell extracts lacking an engineered hybrid insecticidal protein using Abbott's correction (1), and data were transformed prior to analysis of variance (ANOVA) and the Student-Newman-Keuls mean separation procedure. Corrected mortality values were transformed using the arcsine square root of the proportion procedure. An adjustment of 1/(4n), where n is the number of insects, was used for each sample with between 0 and 14% corrected mortality, while an adjustment of 100 − (1/4n) was used for samples with between 86 and 100% mortality. Also, as the sample sizes for some comparisons were not equal, each least significant range (LSR) was determined by using the following recommended procedure described by Sokal and Rohlf (26): LSR = Qα[k,v]·0.05 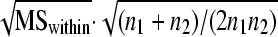 . Qα[k,v] is the critical value statistic for the Studentized range at probability level α, with k being the number of groups and v being the degrees of freedom. MSwithin is the error mean square from the ANOVA, and n1 and n2 are the sample sizes of the compared means.
. Qα[k,v] is the critical value statistic for the Studentized range at probability level α, with k being the number of groups and v being the degrees of freedom. MSwithin is the error mean square from the ANOVA, and n1 and n2 are the sample sizes of the compared means.
In vivo processing and binding of eCry3.1Ab protein to first-instar WCR.
To determine the processing of eCry3.1Ab in neonate WCR larvae, insects were allowed to feed for 24 h on the artificial diet containing a biotinylated N-terminally His6-tagged version of eCry3.1Ab and then recovered and treated as described previously (27) for Western blot analysis of the supernatant and pellet fractions.
BBMV binding assays.
Chymotrypsin processing, biotin labeling, and BBMV competition binding assays with chymotrypsin-processed mCry3A and eCry3.1Ab were carried out as described previously for mCry3A (27). BBMV preparations were obtained using first-instar larval whole-body homogenates and the differential magnesium precipitation method (29).
RESULTS
Insecticidal activity of eCry3.1Ab.
eCry3.1Ab was consistently highly active against neonate WCR larvae (Table 2). Importantly, the removal of the engineered cathepsin G recognition sequence also led to a consistently active protein, eCry3.1Ab-CATG (Fig. 1 and 2). Similarly, removal of the cathepsin G recognition sequence did not significantly impact the strong biological activity of the related construct eCry3.1Ab1 (Table 2 and Fig. 1 and 2). Recently, it was shown that the engineered cathepsin G recognition site (as present in mCry3A) was essential to obtain >90% mortality with WCR larvae (27). Therefore, together, these data support the conclusion that novel activity against coleopterans was generated in eCry3.1Ab by exchange of the Cry1Ab sequence (starting in the second half of CB3 in domain III and continuing through the end of the molecule, including the C-terminal 38-amino-acid addition) with the corresponding region of the original Cry3A C-terminal sequence (Fig. 1 and 2). Further elucidation of the sequence responsible for imparting the eCry3.1Ab activity indicated the importance of the fact that the exchanged region was derived from Cry1Ab rather than Cry1Ac, since neither eCry3.1Ac nor eCry3.1Ac1 exhibited activity in the bioassay (Table 2). The hypothesis that the Cry1Ab sequence was the sequence imparting the potential for WCR bioactivity was also supported by the results of a comparison of the eCry3.1Ab2 and eCry3.1Ab3 proteins with the eCry3.1Ab4 and eCry3.1Ab5 proteins. The latter two proteins have an extension of the Cry3A sequence through V4 and into CB4, whereas in eCry3.1Ab2 and eCry3.1Ab3 the Cry1Ab sequence for V4 is retained and the N-terminal Cry3A domain III sequence is exchanged with the corresponding sequence of Cry1Ab (Fig. 1 and 2). Only eCry3.1Ab3 showed activity in the WCR bioassay (Table 2), indicating that there is some flexibility in domain III for making it even more Cry1Ab-like and retaining activity, while there is less flexibility for making domain III more Cry3A-like and retaining activity. The apparent requirement of the Cry1Ab 38-amino-acid tail sequence for WCR activity in eCry3.1Ab3 also highlights the role that this sequence may play in some constructs but not in others (e.g., eCry3.1Ab5 and eCry3.1Ac).
TABLE 2.
Insecticidal activities of eCry3.1Ab and related proteins with first-instar WCR larvae
| Proteina | No. of replicationsb | % Mortality (mean ± SE)c |
|---|---|---|
| eCry3.1Ab | 12 | 93 ± 3 C |
| eCry3.1Ab-CATG | 5 | 69 ± 8 B |
| eCry3.1Ab1 | 9 | 87 ± 4 BC |
| eCry3.1Ab1-CATG | 4 | 94 ± 6 C |
| eCry3.1Ab2 | 6 | 13 ± 8 A |
| eCry3.1Ab3 | 5 | 77 ± 6 BC |
| eCry3.1Ab4 | 5 | 3 ± 3 A |
| eCry3.1Ab5 | 3 | 19 ± 19 A |
| eCry3.1Ac | 3 | 5 ± 3 A |
| eCry3.1Ac1 | 3 | 17 ± 4 A |
Each protein was added to the diet as a supernatant of the cell lysate in 50 mM NaCl-50 mM Tris (pH 8.5) buffer, as described in Materials and Methods.
Each replication consisted of 12 to 19 larvae per treatment.
Mean ± SE of the corrected mortality. Values were transformed using the arcsine square root of the proportion for analysis; actual means are shown. Means followed by the same letter are not significantly different (P > 0.05, Student-Newman-Keuls test).
eCry3.1Ab processing and binding in first-instar WCR larvae.
Following ingestion of biotinylated eCry3.1Ab, first-instar WCR larvae converted full-length eCry3.1Ab protein to an ∼50-kDa form (Fig. 3), which was observed in both supernatant and washed pellet fractions. It has been found previously that mCry3A is processed in a similar feeding assay, as well as in vitro, by the action of α-chymotrypsin to a stable ∼55-kDa form which can be considered an activated form as it exhibits specific binding to brush border membranes of first-instar WCR larvae (27).
FIG. 3.
In vivo processing and binding of eCry3.1Ab in first-instar WCR larvae: Western blot of recovered whole-body homogenate supernatant or pellet fractions derived after larval feeding on a diet with (+) or without (−) biotinylated eCry3.1Ab. b-eCry3.1Ab, biotinylated eCry3.1Ab marker protein; b-c-mCry3A, biotinylated chymotrypsin-treated mCry3A marker protein. The filled and open arrowheads indicate the positions of processed and unprocessed forms of eCry3.1Ab, respectively.
Chymotrypsin-processed mCry3A exhibited specific binding to WCR BBMV material, which was not diminished by addition of a 100-fold molar excess of chymotrypsin-processed eCry3.1Ab or unprocessed eCry3.1Ab (Fig. 4A). The increased signal recovered following heterologous competition may have been due to promotion of binding or aggregation of toxin at the membrane surface at a high competitor concentration; toxin precipitation is a less likely explanation based on the solubility characteristics of the processed toxins and the net dilution effect of the incubation conditions. In addition, in the converse experiment, specific binding of chymotrypsin-processed eCry3.1Ab was not diminished by the presence of a 75-fold molar excess of chymotrypsin-processed mCry3A (Fig. 4B).
FIG. 4.
Homologous and heterologous BBMV competition binding assay results obtained with (A) biotinylated chymotrypsin-treated mCry3A (c-mCry3A) and (B) biotinylated chymotrypsin-treated eCry3.1Ab (c-eCry3.1Ab) labeled proteins. The Western blots show the effects of using buffer alone (Ø) and a 100-fold molar excess of unlabeled proteins during BBMV incubation (A) or the effects of using buffer alone (Ø) and a 75-fold molar excess of unlabeled proteins (B).
DISCUSSION
We established that there was consistent eCry3.1Ab activity against neonate WCR larvae and that the engineered domain I cathepsin G recognition site was not sufficient to account for this activity of this Cry3A-derived sequence, although it clearly was responsible in mCry3A (27). Our data therefore support the conclusion that novel activity against coleopterans was generated in eCry3.1Ab by exchange of the Cry1Ab sequence (starting in the second half of CB3 in domain III and continuing through the end of the molecule, including the C-terminal 38-amino-acid addition) with the corresponding region of the original Cry3A C-terminal sequence. The eCry3.1Ab and Cry3A protein sequences are 67.4% divergent in this exchanged region. The variable-region sequence (V4, V5, and V6) and the C-terminal 38-amino-acid 1Ab tail sequence account for over 58% of this divergence, and only about 9% of the divergence is accounted for by nonidentical residues in the 1Ab and Cry3A CB regions used. Further elucidation of the sequence responsible for imparting the eCry3.1Ab activity pointed to the importance of the fact that the exchanged region was derived from Cry1Ab rather than Cry1Ac. In addition, other construct comparisons supported the conclusion that the activity was imparted by the Cry1Ab sequence, as more Cry1Ab residues could be used in the exchanged region with retention of activity, but loss of activity followed partial restoration of Cry3A residues in the exchanged region.
eCry3.1Ab protein is processed by neonate WCR larvae to a soluble ∼50-kDa form which can bind to WCR membranes. It has been found previously that mCry3A is processed in a similar feeding assay, as well as in vitro by the action of α-chymotrypsin, to a stable ∼55-kDa form, which can be considered an activated form as it exhibits specific binding to brush border membranes of first-instar WCR larvae (27). We found that in vitro chymotrypsin-processed eCry3.1Ab also exhibited specific binding to WCR brush border membranes. In heterologous BBMV competition experiments, processed forms of either toxin did not compete with binding of the other toxin. These data support the interpretation that eCry3.1Ab recognizes different binding sites than the activated form of mCry3A in the WCR brush border. For insect resistance management, it has been recommended that two or more insecticidal proteins that have different modes of action be used to minimize the potential for development of resistance in the pest population (5, 21). In the case of two B. thuringiensis delta-endotoxins, the interaction with unique binding sites is believed to be an important indication of the durability of a given insecticidal protein combination, as alteration of the binding step in the mode of action has often been implicated in the ability of a target pest to generate resistance to a single protein (5). In bioassays which use combinations of proteins which do not differ widely in their individual interactions with the target membrane, cross-resistance has been documented (9).
Our work demonstrates that using exchange of variable regions across classes of B. thuringiensis Cry proteins may result in novel bioactivity. This is completely in keeping with the present understanding that highly conserved block regions may be essential for structural integrity of the Cry proteins and their discrete domain functions (e.g., allowing membrane insertion and pore formation, providing oligomerization contact points and/or whole domain movements, and conserving structure necessary for presenting sequence motifs for membrane interaction or binding protein interactions) (23). At the same time, the data support the interpretation that alteration or manipulation of steps in the mode of action could occur through changes in the variable-region sequence. eCry3.1Ab most likely built on the potential that this strategy provides for novel binding interactions, as the interaction with the neonate WCR brush border differed from that of another WCR-active molecule, mCry3A. Clearly, not all variable-region exchanges result in a stable protein structure or retention or expansion of the spectrum of bioactivity (e.g., eCry3.1Ac is inactive, whereas eCry3.1Ab is active), but the data suggest new combinations which might be explored. As discovery of B. thuringiensis Cry proteins which target pests such as Diabrotica spp. has been challenging (20, 27), this approach provides a welcome new direction.
Acknowledgments
This work was supported by Syngenta Biotechnology, Inc.
We gratefully acknowledge the assistance of Jared Conville and Shank Palekar for help in maintaining the WCR larvae and bioassay supplies. We also thank the protein production team at the Syngenta Jealott's Hill Research Station for the eCry3.1Ab purified protein test substance used in binding studies.
Footnotes
Published ahead of print on 19 March 2010.
REFERENCES
- 1.Abbott, W. S. 1925. A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 18:265-267. [Google Scholar]
- 2.Boonserm, P., P. Davis, D. J. Ellar, and J. Li. 2005. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J. Mol. Biol. 348:363-382. [DOI] [PubMed] [Google Scholar]
- 3.Bravo, A., S. S. Gill, and M. Soberon. 2007. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49:423-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bravo, A., J. Sánchez, T. Kouskoura, and N. Crickmore. 2002. N-terminal activation is an essential early step in the mechanism of action of the B. thuringiensis Cry1Ac insecticidal toxin. J. Biol. Chem. 277:23985-23987. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, A., and M. Soberon. 2008. How to cope with insect resistance to Bt toxins? Trends Biotechnol. 26:573-579. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X. J., M. K. Lee, and D. H. Dean. 1993. Site-directed mutations in a highly conserved region of Bacillus thuringiensis δ-endotoxin affect inhibition of short circuit current across Bombyx mori midguts. Proc. Natl. Acad. Sci. U. S. A. 90:9041-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Maagd, R. A., M. S. G. Kwa, H. van der Klei, T. Yamamoto, B. Schipper, J. M. Vlak, W. J. Stiekma, and D. Bosch. 1996. Domain III substitution in Bacillus thuringiensis delta-endotoxin Cry1A(b) results in superior toxicity for Spodoptera exigua and altered membrane protein recognition. Appl. Environ. Microbiol. 62:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Maagd, R. A., M. Weemen-Hendriks, W. Stiekema, and D. Bosch. 2000. Bacillus thuringiensis delta-endotoxin Cry1C domain III can function as a specificity determinant for Spodoptera exigua in different, but not all, Cry1-Cry1C hybrids. Appl. Environ. Microbiol. 66:1559-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffitts, J. S., and R. V. Aroian. 2005. Many roads to resistance: how invertebrates adapt to Bt toxins. BioEssays 27:614-624. [DOI] [PubMed] [Google Scholar]
- 10.Grochulski, P., L. Masson, S. Borisova, M. Pusztai-Carey, J. L. Schwartz, R. Brousseau, and M. Cygler. 1995. Bacillus thuringiensis Cry1A(a) insecticidal toxin: crystal structure and channel formation. J. Mol. Biol. 254:447-464. [DOI] [PubMed] [Google Scholar]
- 11.Höfte, H., and H. R. Whiteley. 1989. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53:242-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 13.Karlova, R., M. Weemen-Hendriks, S. Naimov, J. Ceron, S. Dukiandjiev, and R. A. de Maagd. 2005. Bacillus thuringiensis δ-endotoxin Cry1Ac domain III enhances activity against Heliothis virescens in some, but not all Cry1-Cry1Ac hybrids. J. Invertebr. Pathol. 88:169-172. [DOI] [PubMed] [Google Scholar]
- 14.Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283-292. [DOI] [PubMed] [Google Scholar]
- 15.Koziel, M. G., N. M. Desai, K. S. Lewis, V. C. Kramer, G. W. Warren, S. V. Evola, L. D. Crossland, M. S. Wright, E. J. Merlin, K. L. Launis, S. J. Rothstein, C. G. Bowman, J. L. Dawson, E. M. Dunder, G. M. Pace, and J. L. Suttie. April 1997. Synthetic DNA sequence having enhanced insecticidal activity in maize. U.S. patent 5,625,136.
- 16.Li, J., J. Caroll, and D. J. Ellar. 1991. Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature 353:815-821. [DOI] [PubMed] [Google Scholar]
- 17.Liu, X. S., and D. H. Dean. 2006. Redesigning Bacillus thuringiensis Cry1Aa toxin into a mosquito toxin. Protein Eng. Des. Sel. 19:107-111. [DOI] [PubMed] [Google Scholar]
- 18.Morse, R. J., T. Yamamoto, and R. M. Stroud. 2001. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure 9:409-417. [DOI] [PubMed] [Google Scholar]
- 19.Naimov, S., M. Weemen-Hendriks, S. Dukiandjiev, and R. A. de Maagd. 2001. Bacillus thuringiensis delta-endotoxin Cry1 hybrid proteins with increased activity against the Colorado potato beetle. Appl. Environ. Microbiol. 67:5328-5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostlie, K. 2001. Crafting crop resistance to corn rootworms. Nat. Biotechnol. 19:624-625. [DOI] [PubMed] [Google Scholar]
- 21.Roush, R. T. 1998. Two-toxin strategies for management of insecticidal transgenic crops: can pyramiding succeed where pesticides mixtures have not? Philos. Trans. R. Soc. Lond. B Biol. Sci. 353:1777-1786. [Google Scholar]
- 22.Rupar, M. J., W. P. Donovan, R. Gene Groat, A. C. Slaney, J. W. Mattison, T. J. Johnson, J.-F. Charles, V. C. duManoir, and H. de Barjac. 1991. Two novel strains of Bacillus thuringiensis toxic to coleopterans. Appl. Environ. Microbiol. 57:3337-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Ziegler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz, J. L., L. Potvin, X. J. Chen, R. Brousseau, R. Laprade, and D. H. Dean. 1997. Single-site mutations in the conserved alternating-arginine region affect ionic channels formed by CryIAa, a Bacillus thuringiensis toxin. Appl. Environ. Microbiol. 63:3978-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shadenkov, A. A., R. M. Kadyrov, S. V. Uzbekova, E. V. Kuźmin, A. L. Osterman, G. G. Chestukhina, and M. F. Shemyakin. 1993. Construction of a hybrid gene from CryIIIA and CryIA(a) δ-endotoxin genes of Bacillus thuringiensis and expression of its derivatives in Escherichia coli cells. Mol. Biol. 27:586-591. [PubMed] [Google Scholar]
- 26.Sokal, R. R., and F. J. Rohlf. 1969. Single classification analysis of variance, p. 239-246. In J. Wilson and S. Cotter (ed.), Biometry, 1st ed. W. H. Freeman and Co., New York, NY.
- 27.Walters, F. S., C. M. Stacy, M. K. Lee, N. Palekar, and J. S. Chen. 2008. An engineered chymotrypsin/cathepsin G site in domain I renders Bacillus thuringiensis Cry3A active against western corn rootworm larvae. Appl. Environ. Microbiol. 74:367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfersberger, M. G., X. J. Chen, and D. H. Dean. 1996. Site-directed mutations in the third domain of Bacillus thuringiensis δ-endotoxin CryIAa affect its ability to increase the permeability of Bombyx mori midgut brush border membrane vesicles. Appl. Environ. Microbiol. 62:279-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfersberger, M., P. Luethy, A. Maurer, P. Parenti, F. V. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation of brush border membrane vesicles (BBMV) from larval lepidopteran midgut. Comp. Biochem. Physiol. Part A Physiol. 86:301-308. [Google Scholar]