Abstract
In this work, we report the expression and secretion of the leaderless two-peptide (EntL50A and EntL50B) bacteriocin enterocin L50 from Enterococcus faecium L50 by the methylotrophic yeast Pichia pastoris X-33. The bacteriocin structural genes entL50A and entL50B were fused to the Saccharomyces cerevisiae gene region encoding the mating pheromone α-factor 1 secretion signal (MFα1s) and cloned, separately and together (entL50AB), into the P. pastoris expression and secretion vector pPICZαA, which contains the methanol-inducible alcohol oxidase promoter (PAOX1) to express the fusion genes. After transfer into the yeast, the recombinant plasmids were integrated into the genome, resulting in three bacteriocinogenic yeast strains able to produce and secrete the individual bacteriocin peptides EntL50A and EntL50B separately and together. The secretion was efficiently directed by MFα1s through the Sec system, and the precursor peptides were found to be correctly processed to form mature and active bacteriocin peptides. The present work describes for the first time the heterologous expression and secretion of a two-peptide non-pediocin-like bacteriocin by a yeast.
Bacteriocins are a heterogeneous group of ribosomally synthesized antimicrobial peptides or proteins produced by Gram-negative and Gram-positive bacteria (39). They are highly potent, acting at nanomolar concentrations, and normally kill target cells by interfering with the integrity of their inner membrane. Many bacteriocins, especially those produced by lactic acid bacteria (LAB), target important pathogens and/or food spoilage bacteria, and they therefore have great potential application as food biopreservatives or pharmaceuticals and nutraceuticals in veterinary and human medicine (13, 28, 35, 36, 46). LAB bacteriocins have been classified into three main classes: class I, the lantibiotics containing thioether rings that are composed of either lanthionine or methyllanthionine; class II, the nonmodified, small, and heat-stable peptides; and class III, the large and heat-labile protein bacteriocins (16, 19, 39). The class II bacteriocins are quite diverse in terms of structure, amino acid sequence, and mode of action and have been further divided into five subclasses. (i) Subclass IIa comprises bacteriocins containing a conserved N-terminal motif (YGNGVxC); these are often called pediocin-like bacteriocins because pediocin PA-1 was the first bacteriocin to be characterized from this group. (ii) Subclass IIb is made up by bacteriocins whose full activity is dependent on the presence of two different peptides (thus often referred to as two-peptide bacteriocins). (iii) Subclass IIc includes the leaderless bacteriocins as opposed to other bacteriocins, which need an N-terminal extension (leader sequence or signal peptide) for secretion. (iv) Subclass IId is made up by the circular bacteriocins. (v) Subclass IIe includes one-peptide, non-pediocin-like, and noncircular bacteriocins.
The production of most class II bacteriocins relies on a well-conserved genetic organization of at least four genes, which are closely associated in one or two operon-like structures: (i) the structural gene encoding the prebacteriocin; (ii) a gene encoding the dedicated protein, which confers producer self-protection (immunity) against the toxicity of the bacteriocin (15); (iii) a gene encoding a dedicated ATP-binding cassette (ABC) transporter required for the processing and transport of the bacteriocin; and (iv) a gene encoding an accessory protein required for proper bacteriocin externalization (29, 38). Some bacteriocin systems involve a quorum-sensing mechanism to regulate gene expression mediated by three different gene products: a secreted pheromone peptide (the induction factor), a histidine protein kinase (HPK) that serves as a sensor for the pheromone, and a response regulator (RR) that triggers the expression of a set of genes after it is activated by the cognate HPK (14).
All lantibiotics and most class II bacteriocins are synthesized as biologically inactive precursors containing an N-terminal extension required for the secretion of the mature bacteriocin. This secretory signal extension, which can be of the so-called double-glycine-type leader sequence or the Sec-dependent signal peptide, is cleaved off concomitantly with the secretion of the active bacteriocin (11, 16, 29). Interestingly, a few LAB bacteriocins (those belonging to subclass IIc) are synthesized without an N-terminal extension; these LAB bacteriocins include enterocin L50 (L50A and L50B) (8), enterocin Q (10), enterocin EJ97 (43), and the bacteriocin LsbB (20). For most leaderless bacteriocins, it has been shown that secretion is mediated by ABC transporters (12, 20, 43); however, the nature of the signal that conveys their secretion is still elusive.
Enterococci produce a large variety of bacteriocins (commonly referred to as enterocins), which belong mostly to class II, except for the two-peptide lantibiotic cytolysin (21), the lytic protein enterolysin A (40), and the lantibiotic columbicin A (37). Enterococcus faecium L50 is a multiple-bacteriocin producer strain isolated from a Spanish dry-fermented sausage (7), which inhibits several spoilage and food-borne pathogenic bacteria as well as clinical human and animal pathogens (2, 8, 9, 10). E. faecium L50 produces three bacteriocins belonging to different subclasses of class II. (i) Enterocin P is a pediocin-like bacteriocin (subclass IIa) synthesized with a sec-dependent N-terminal extension (signal peptide). (ii) Enterocin L50 (L50A and L50B) is an unusual bacteriocin in that it has features characteristic of both subclass IIb and subclass IIc bacteriocins; its full antimicrobial activity is dependent on the complementary action of two different peptides, EntL50A and EntL50B (a typical subclass IIb feature), and these two peptides are secreted without N-terminal extensions (a typical subclass IIc feature). (iii) Enterocin Q is a subclass IIc leaderless bacteriocin (8, 10, 12). While enterocin P is secreted by the Sec translocase (30), it has been suggested that the enterocin L50 peptides are secreted by a dedicated ABC transporter, as demonstrated for enterocin Q and other leaderless bacteriocins (12, 20, 43). Although individual EntL50A and EntL50B possess some antimicrobial activity on their own, with EntL50A being the most active, a clearly synergistic effect is observed when both peptides are combined (2, 3, 8).
Interestingly, enterocin L50 (L50A and L50B), similarly to most enterocins characterized to date, displays a broad antimicrobial spectrum and possesses adequate technological properties (e.g., bactericidal mode of action, small size, thermoresistance, stability over a wide range of pHs and storage conditions, sensitivity to most proteases, etc.) for being used as food biopreservatives or pharmaceutical antimicrobials (11, 19, 39). However, the use of enterococci as food biopreservatives is highly controversial and merits particularly careful premarket safety evaluation since many strains, mainly within the species Enterococcus faecalis, have emerged as opportunistic human pathogens encoding potential virulence factors and carrying antibiotic resistance genes (17, 18, 32, 41). To overcome the concerns related to the safety of enterococci, enterocin production by alternative and safer hosts, including industrially interesting food-grade LAB and yeast strains, has emerged as a suitable strategy, which may also lead to a strict control of enterocin gene expression at the transcriptional and/or translational level and/or optimization of enterocin production and purification. Furthermore, eukaryotic genetic tools developed for gene expression in yeasts have been shown to be adaptable to bacteriocin genes (3, 25, 26). Despite the high versatility and efficiency of Saccharomyces cerevisiae and Pichia pastoris for the large-scale heterologous production of a variety of functional proteins (4, 6, 27, 33, 44, 45), the heterologous production of enterocins by these yeast hosts has been addressed in only a few works. With regard to this, the heterologous production of biologically active enterocin L50 (L50A and L50B) by S. cerevisiae and enterocin P and hiracin JM79 by P. pastoris has demonstrated the possibility of developing bacteriocin-producing yeast strains (3, 23, 42).
We have previously reported the construction of segregationally stable bacteriocinogenic clones of S. cerevisiae that were capable of producing the individual enterocin L50 peptides independently by using the yeast factor MFα1s to mediate the Sec-dependent secretion of the mature peptides (3); however, we somehow failed to generate a yeast clone that produced both peptides simultaneously. In the present paper, we report the successful generation of three bacteriocinogenic P. pastoris strains heterologously producing, separately and together, the leaderless peptides EntL50A and EntL50B directed by MFα1s through the yeast Sec system.
MATERIALS AND METHODS
Microorganisms, plasmids, media, and culture conditions.
The microorganisms and plasmids used in this work are listed in Table 1. The enterocin L50-producing strain E. faecium L50 (8) and the indicator strain Pediococcus damnosus CECT4797 (EntL50A and EntL50B sensitive [EntL50s]) (2) were grown aerobically in MRS broth (pH 6.2; Oxoid Ltd., Basingstoke, United Kingdom) at 30°C. Escherichia coli JM109 (Promega Corporation, Madison, WI) and E. coli TOP10 (Invitrogen Life Technologies, Carlsbad, CA) cells were propagated in Luria-Bertani (LB) broth (Sigma-Aldrich Inc., St. Louis, MO) at 37°C with shaking (200 to 250 rpm). Kanamycin (Kan) (50 μg/ml) (Sigma-Aldrich) and zeocin (Zeo) (25 μg/ml) (Invitrogen) were added to LB medium for the selection of E. coli transformants. P. pastoris X-33 (Invitrogen) strains were cultured in YPD medium (10 g/liter yeast extract [Oxoid], 20 g/liter peptone [Oxoid], 20 g/liter glucose [Panreac Química S.A., Barcelona, Spain]) (4) at 30°C with shaking (200 to 250 rpm). Solid media contained 1.5 or 2% (wt/vol) agar (Oxoid) for LAB and E. coli or P. pastoris, respectively, while soft MRS agar contained 0.8% (wt/vol) agar.
TABLE 1.
Microorganisms and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or referenceb |
|---|---|---|
| Bacterial strains | ||
| E. faecium L50 | EntL50 (EntL50A and EntL50B), EntP, and EntQ producer | DNBTA |
| Pc. damnosus 4797 | Indicator microorganism; EntL50s-EntPs-EntQr | CECT |
| E. coli high-efficiency JM109 | Host strain; F endA1 recA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) F′ traD36 proAB lacIZΔM15 | Promega |
| E. coli TOP10 | Host strain; F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Yeast (P. pastoris) strains | ||
| X-33 | Host strain; wild-type strain for selection on Zeo; Mut+ | Invitrogen |
| X-33C | P. pastoris X-33 derivative carrying pPICZαA; Zeor | This work |
| X-33A | P. pastoris X-33 derivative carrying pBAS01; EntL50A producer; Zeor | This work |
| X-33B | P. pastoris X-33 derivative carrying pBAS02; EntL50B producer; Zeor | This work |
| X-33AB | P. pastoris X-33 derivative carrying pBAS03; EntL50A and EntL50B producer; Zeor | This work |
| Plasmids | ||
| pPICZαA | P. pastoris 3.6-kb protein expression and secretion vector carrying a methanol-inducible promoter (PAOX1), 5′ AOX1 region, MFα1s; Zeor | Invitrogen |
| pTBS02 | PCR2.1-TOPO derivative carrying entL50A | 3 |
| pTBS03 | PCR2.1-TOPO derivative carrying entL50B | 3 |
| pBAS01 | pPICZαA derivative carrying entL50A fused in frame to MFα1s | This work |
| pBAS02 | pPICZαA derivative carrying entL50B fused in frame to MFα1s | This work |
| pBAS03 | pBAS01 derivative carrying entL50A fused in frame to MFα1s and entL50B fused in frame to MFα1s | This work |
MFα1s, yeast gene region encoding the mating pheromone α-factor 1 secretion signal (MFα1s).
Abbreviations: CECT, Colección Española de Cultivos Tipo (Valencia, Spain); DNBTA, Departamento de Nutrición, Bromatología y Tecnología de los Alimentos, Facultad de Veterinaria, Universidad Complutense de Madrid (Madrid, Spain).
Molecular techniques and enzymes.
Established protocols were employed for all DNA manipulations, including PCR amplifications, restriction endonuclease digestions, ligations, and transformations, as previously described (3, 24). Oligonucleotide primers (Table 2) were obtained from Sigma-Genosys Ltd. (Cambridge, United Kingdom). Nucleotide sequencing of both strands of purified PCR products was done at the DNA Sequencing Service of Sistemas Genómicos (Valencia, Spain). P. pastoris X-33 competent cells were obtained and transformed by use of the EasySelect Pichia expression kit (Invitrogen).
TABLE 2.
Primers and PCR products used in this study
| Primer or PCR product | Nucleotide sequence (5′→3′)a or PCR product descriptionb | Fragment(s) amplified |
|---|---|---|
| Primers | ||
| 5AOX1 | GACTGGTTCCAATTGACAAGC | α-L50A-α-L50B |
| 3AOX1 | GCAAATGGCATTCTGACATCC | α-L50A-α-L50B |
| Alfa5-XbaI | AATTATATCTAGAATTCGAAACGATGAGATTTCCTTCAATTTTTACTG | α-L50B |
| L50B9-SalI | ATAAGTTGTCGACAACATTAATGTCTTTTTAGCCATTTTTCAATTTGATC | α-L50B |
| AlfaF | TACTATTGCCAGCATTGCTGC | FA and FB |
| L50A3 | ATGGGAGCAATCGCAAAATTAGTAGCAAAG | entL50A |
| L50A4 | ATTTTAAATATGTTTTTTAATCCACTCAATG | FA and entL50A |
| L50B3 | ATGGGAGCAATCGCAAAACTAGTGAC | entL50B |
| L50B4 | AACATTAATGTCTTTTTAGCCATTTTTCAATTTG | FB and entL50B |
| PCR products | ||
| Fragment entL50A | 138-bp fragment containing structural gene of EntL50A (entL50A) | |
| Fragment entL50B | 136-bp fragment containing structural gene of EntL50B (entL50B) | |
| Fragment FA | 190-bp fragment containing MFα1s fused in frame to entL50A | |
| Fragment FB | 188-bp fragment containing MFα1s fused in frame to entL50B | |
| Fragment α-L50B | 427-bp fragment containing MFα1s fused in frame to entL50B | |
| Fragment α-L50A-α-L50B | 1,030-bp fragment containing MFα1s fused in frame to entL50A and MFα1s fused in frame to entL50B |
Cleavage sites for restriction enzymes are underlined. Boldface type indicates nucleotide tails added to the specific sequences to ensure the proper functioning of the restriction enzymes.
MFα1s refers to the yeast gene region encoding MFα1s.
Cloning of the bacteriocin structural genes entL50A and entL50B.
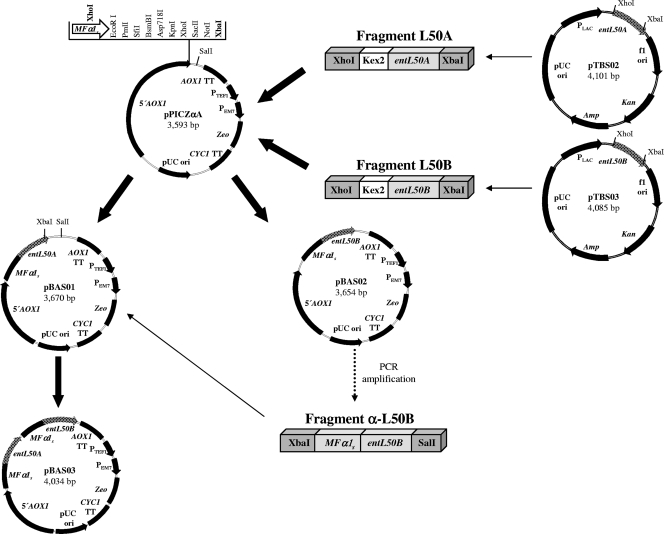
The strategy employed for the cloning of entL50A and entL50B, separately and together, in pPICZαA is summarized in Fig. 1. DNA fragments containing entL50A or entL50B fused to the nucleotide sequence (AAAAGA) encoding the Kex2 cleavage site were isolated from pTBS02 and pTBS03 (3) by using the restriction enzymes XhoI and XbaI and then cloned into expression vector pPICZαA, generating recombinant plasmids pBAS01 and pBAS02, respectively. In these plasmids, entL50A and entL50B are fused in frame to MFα1s, and gene expression is controlled by the inducible promoter PAOX1 (being induced by methanol and repressed by glycerol). MFα1s and the Kex2 sequence are required for the proper processing and secretion of the mature bacteriocin peptides. For the construction of pBAS03, which coexpresses MFα1s-Kex2-entL50A and MFα1s-Kex2-entL50B, the latter gene was obtained from pBAS02 by means of PCR and then cloned into pBAS01 between the restriction sites XbaI and SalI. Following amplification in E. coli JM109 cells, the recombinant plasmids as well as empty plasmid pPICZαA were linearized with SacI prior to electroporation into P. pastoris X-33 cells to obtain the clones P. pastoris X-33A (to produce EntL50A), P. pastoris X-33B (to produce EntL50B), P. pastoris X-33AB (to produce both EntL50A and EntL50B), and P. pastoris X-33C (used as a bacteriocin-negative control), which were selected on YPD plates supplemented with Zeo (100 μg/ml) and sorbitol (1 M) at 30°C for 96 h. Plasmids pBAS01, pBAS02, and pBAS03 integrated into the genome of P. pastoris X-33A, P. pastoris X-33B, and P. pastoris X-33AB transformants, respectively, were confirmed by PCR, restriction analysis, and DNA sequencing.
FIG. 1.
Construction of recombinant plasmids pBAS01, pBAS02, and pBAS03, derived from the P. pastoris expression and secretion vector pPICZαA, containing the yeast gene region encoding the mating pheromone α-factor 1 secretion signal (MFα1s), including the nucleotides encoding the Kex2 signal cleavage site, fused in frame to the entL50A and/or entL50B structural gene and under the control of the methanol-inducible alcohol oxidase promoter (PAOX1). Plasmid sizes are given in base pairs. Only relevant restriction enzymes sites are indicated. 5′ AOX1, promoter region; AOX1 TT, transcription termination; PTEF1, transcription elongation factor 1 that drives the expression of the Sh ble gene in Pichia; PEM7, constitutive promoter driving the expression of the Sh ble gene in E. coli; Zeo gene, zeocin resistance (Sh ble gene); CYC1 TT, transcription terminator; pUC ori, maintenance and high-copy replication in E. coli; entL50A, structural gene of EntL50A; entL50B, structural gene of EntL50B; PLAC, constitutive promoter driving the expression of the lacZ gene in E. coli; f1 ori, rescue of single-stranded DNA; Kan gene, kanamycin resistance; Amp gene, ampicillin resistance.
To determine if the differences in recombinant EntL50A and EntL50B amounts found in supernatants from P. pastoris X-33A and P. pastoris X-33B were due to differences in the copy numbers of the integrated genes, both transformants were spotted onto YPD plates supplemented with increasing Zeo concentrations (from 100 to 3,000 μg/ml) and grown at 30°C for 48 h. In this respect, it is known that P. pastoris is capable of integrating multiple copies of heterologous DNA, which leads to higher Zeo resistance and hyperexpression of recombinant proteins (1, 6, 33).
Detection and quantification of EntL50A and EntL50B heterologous production by antimicrobial and immunochemical assays.
The antimicrobial activities of cultures from several P. pastoris X-33A, P. pastoris X-33B, and P. pastoris X-33AB transformants were screened by a spot-on-agar test (SPAT) essentially as previously described (7). Briefly, transformants were first grown on YPD plates supplemented with Zeo (100 μg/ml) at 30°C for 48 h and then transferred onto BMMY (10 g/liter yeast extract, 20 g/liter peptone, 100 mM potassium phosphate [pH 6; Merck Farma y Química S.A., Barcelona, Spain], 1.34% [wt/vol] yeast nitrogen base without amino acids [Invitrogen], 4 × 10−5% biotin [Sigma-Aldrich], 0.5% [vol/vol] methanol [Merck]) buffered methanol complex medium plates and incubated further at 30°C for 5 days. During the incubation, methanol was added daily to attain a 0.5% (vol/vol) final concentration to maintain the induction. After this period, 40 ml of MRS soft agar containing about 1 × 105 CFU/ml of the indicator microorganism Pc. damnosus CECT4797 was poured over the plates, which were incubated at 30°C overnight for the development of inhibition zones.
In order to determine bacteriocin production kinetics, the clones P. pastoris X-33A, P. pastoris X-33B, and P. pastoris X-33AB were precultured in the buffered glycerol complex medium BMGY (10 g/liter yeast extract, 20 g/liter peptone, 100 mM potassium phosphate [pH 6], 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, 1% [vol/vol] glycerol [Sigma-Aldrich]) (23) at 30°C until the cell density reached an optical density at 600 nm (OD600) of approximately 2 to 6. Cells were harvested by centrifugation (5,000 × g at 4°C for 10 min), washed with BMGY (without glycerol), and resuspended to an OD600 of 1 in both the buffered methanol complex medium BMMY and the buffered methanol minimal medium BMM (100 mM potassium phosphate [pH 6], 1.34% [wt/vol] yeast nitrogen base without amino acids, 4 × 10−5% biotin, 0.5% [vol/vol] methanol) (23). Cultures were incubated at 30°C for 9 days with shaking. During this incubation, samples were collected periodically for determinations of overnight yeast growth (OD600), cell dry weight (CDW), and bacteriocin activity and concentration in duplicates. Bacteriocin activity was quantified by an agar well diffusion test (ADT) (7) and a microtiter plate assay (MPA) (10, 31) using Pc. damnosus CECT4797 as the indicator microorganism. Quantification of EntL50A and EntL50B heterologous production by a noncompetitive indirect enzyme-linked immunosorbent assay (NCI-ELISA) was performed as previously described (3). To measure the synergistic activity of heterologously produced EntL50A and EntL50B, supernatants from P. pastoris X-33A and P. pastoris X-33B were challenged against Pc. damnosus CECT4797, separately and combined, to achieve a 1:1 bacteriocin peptide ratio by SPAT and MPA.
Other methods.
The yeast-produced bacteriocin peptides EntL50A and EntL50B were purified and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis was performed essentially as previously described (3, 7). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blotting of the purified peptides using anti-LR1-keyhole limpet hemocyanin (KLH) (specific for EntL50A) and anti-LR2-KLH (specific for EntL50B) antibodies and detection of the bacteriocin activity after gel electrophoresis were performed essentially as previously described (3, 22).
RESULTS
Biologically active EntL50A and EntL50B produced separately or together by P. pastoris.
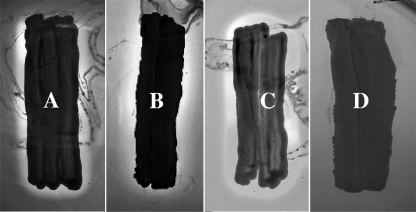
Three yeast clones were constructed, P. pastoris X-33A and P. pastoris X-33B to produce EntL50A and EntL50B, respectively, and P. pastoris X-33AB to produce both EntL50A and EntL50B, in addition to the bacteriocin-negative control clone P. pastoris X-33C. In the bacteriocinogenic clones, MFα1s and the Kex2 sequence were fused in frame to the bacteriocin genes to guide the proper processing and secretion of the mature bacteriocin peptides (Fig. 1). As shown in Fig. 2, the three bacteriocinogenic clones showed antimicrobial activity against the indicator microorganism Pc. damnosus CECT4797, while the control clone did not. To further confirm that the antimicrobial compounds had been secreted into the medium, the antimicrobial activities in the supernatants of liquid cultures were tested, and as shown in Table 3, only supernatants from cultures containing the bacteriocin genes could display antimicrobial activity, thus ruling out the possibility that the antimicrobial activity exerted by P. pastoris X-33A, P. pastoris X-33B, and P. pastoris X-33AB was due to metabolites other than bacteriocins.
FIG. 2.
Direct antimicrobial activity of P. pastoris X-33A (A), P. pastoris X-33B (B), P. pastoris X-33AB (C), and P. pastoris X-33C (D), grown in BMMY broth at 30°C after 5 days of incubation, as determined by a SPAT using Pc. damnosus CECT4797 as the indicator microorganism.
TABLE 3.
Production and antimicrobial activities of recombinant EntL50A and/or EntL50B from P. pastoris X-33A, P. pastoris X-33B, and P. pastoris X-33AB culturesa
| Medium and incubation time (h) |
P. pastoris X-33A |
P. pastoris X-33B |
P. pastoris X-33AB |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OD600b | EntL50A concnc (ng/mg CDW) | Antimicrobial activity assessed by: |
SAAf (BU/ng EntL50A) | OD600 | EntL50B concnc (ng/mg CDW) | Antimicrobial activity assessed by: |
SAAf (BU/ng EntL50B) | OD600 | Concnc (ng/mg [CDW]) |
Antimicrobial activity assessed by: |
|||||
| ADTd (mm) (inhibition zone) | MPAe (BU/mg CDW) | ADT (mm) (inhibition zone) | MPA (BU/mg CDW) | EntL50A | EntL50B | ADT (mm) (inhibition zone) | MPA (BU/mg CDW) | ||||||||
| BMM | |||||||||||||||
| 0 | 1.0 | ND | ND | ND | NE | 1.0 | ND | ND | ND | NE | 1.0 | ND | ND | ND | ND |
| 8 | 2.3 | 14.5 | ND | ND | NE | 2.4 | 70 | ND | ND | NE | 2.6 | 9.7 | 10.5 | ND | ND |
| 24 | 2.5 | 45.5 | 11.5 (s2) | 60 | 1.3 | 2.5 | 340 | ND | ND | NE | 2.6 | 24.0 | 32.1 | 11.0 (s2) | 40 |
| 48 | 3.4 | 56.0 | 14.7 (s2) | 115 | 2.1 | 3.2 | 350 | 8.0 (d) | ND | NE | 3.7 | 29.0 | 37.7 | 13.1 (s2) | 45 |
| 72 | 4.6 | 62.0 | 15.1 (s2) | 130 | 2.1 | 3.6 | 390 | 11.3 (s) | 40 | 0.10 | 4.5 | 30.0 | 39.3 | 13.6 (s2) | 60 |
| 96 | 5.3 | 70.5 | 15.8 (s2) | 150 | 2.1 | 4.6 | 430 | 12.2 (s2) | 45 | 0.10 | 5.3 | 32.8 | 28.4 | 14.1 (s2) | 65 |
| 120 | 5.7 | 84.5 | 16.0 (s2) | 160 | 1.9 | 4.8 | 475 | 12.8 (s2) | 50 | 0.10 | 5.6 | 29.2 | 18.9 | 13.4 (s2) | 55 |
| 144 | 6.3 | 89.0 | 16.4 (s2) | 175 | 2.0 | 5.5 | 405 | 11.1 (s2) | 30 | 0.07 | 6.2 | 20.1 | 7.1 | 13.2 (s2) | 40 |
| 168 | 6.9 | 125.5 | 16.7 (s2) | 190 | 1.5 | 6.3 | 335 | 10.3 (s) | 15 | 0.04 | 6.5 | 18.6 | 5.8 | 12.0 (s2) | 30 |
| 192 | 7.8 | 228.5 | 17.6 (s2) | 310 | 1.4 | 7.4 | 205 | 9.6 (s−) | 10 | 0.05 | 8.3 | 11.3 | 1.0 | 11.8 (s2) | 25 |
| 216 | 8.6 | 161.5 | 17.0 (s2) | 205 | 1.3 | 8.4 | 205 | 9.1 (s−) | 10 | 0.05 | 8.2 | 9.1 | 1.1 | 11.5 (s2) | 20 |
| BMMY | |||||||||||||||
| 0 | 1.0 | ND | ND | ND | NE | 1.0 | ND | ND | ND | NE | 1.0 | ND | ND | ND | ND |
| 8 | 3.5 | 5.0 | ND | ND | NE | 3.4 | 25 | ND | ND | NE | 3.7 | 2.4 | 4.5 | ND | ND |
| 24 | 4.7 | 7.5 | 10.2 (s) | ND | NE | 4.5 | 50 | ND | ND | NE | 4.9 | 2.5 | 5.1 | 10.7 (s2) | 10 |
| 48 | 5.2 | 12.5 | 11.7 (s2) | 10 | 0.8 | 5.2 | 70 | 8.5 (d) | ND | NE | 5.3 | 3.0 | 5.9 | 14.1 (s2) | 20 |
| 72 | 6.3 | 15.5 | 12.7 (s2) | 25 | 1.6 | 5.8 | 110 | 9.1 (d) | 10 | 0.09 | 6.1 | 2.9 | 5.4 | 11.4 (s2) | 10 |
| 96 | 6.8 | 16.5 | 13.3 (s2) | 30 | 1.8 | 6.2 | 170 | 9.5 (d) | 10 | 0.05 | 7.1 | 2.2 | 4.7 | 11.2 (s2) | 10 |
| 120 | 8.2 | 18.5 | 13.7 (s2) | 50 | 2.7 | 6.9 | 695 | 11.4 (s) | 45 | 0.06 | 7.7 | 2.1 | 4.1 | 11.0 (s2) | 10 |
| 144 | 9.3 | 21.0 | 13.9 (s2) | 55 | 2.6 | 7.2 | 1,240 | 14.0 (s2) | 95 | 0.08 | 7.9 | 2.0 | 4.0 | 10.8 (s2) | 10 |
| 168 | 9.5 | 25.0 | 14.2 (s2) | 70 | 2.8 | 7.8 | 950 | 13.0 (s) | 70 | 0.08 | 8.0 | 1.4 | 1.7 | 10.6 (s2) | 10 |
| 192 | 10.2 | 31.0 | 15.8 (s2) | 100 | 3.2 | 9.0 | 515 | 12.5 (s) | 45 | 0.09 | 8.6 | 0.4 | 0.2 | 10.5 (s2) | 10 |
| 216 | 10.9 | 12.0 | 13.3 (s2) | 15 | 1.2 | 9.8 | 415 | 12.1 (s) | 40 | 0.09 | 9.2 | 0.3 | 0.1 | 10.0 (s2) | 10 |
Cultures were grown in BMM and BMMY at 30°C.
Gene expression was induced at time zero.
Bacteriocin concentration calculated by using an NCI-ELISA using specific polyclonal antibodies. ND, no bacteriocin detected.
Antimicrobial activity against Pc. damnosus CECT4797 determined by an ADT. Inhibition zones are differentiated as follows: d, diffuse; s−, slightly sharp; s, sharp; s2, extremely sharp. ND, no inhibition zone detected using 50 ml of supernatant.
Antimicrobial activity against Pc. damnosus CECT4797 as determined by an MPA. ND, no inhibition detected using 100 ml of supernatant.
Specific antimicrobial activity, i.e., the antimicrobial activity (BU/mg CDW) calculated by an MPA, divided by the EntL50A or EntL50B concentration (ng/mg CDW). NE, not evaluable.
Recombinant yeasts grew much faster and to a higher cell density in the complex medium BMMY (OD600 of 9.2 to 10.9) than in the minimal medium BMM (OD600 of 8.2 to 8.6). Interestingly, the antimicrobial activities of the supernatants from cultures of P. pastoris X-33A and P. pastoris X-33AB were higher in BMM than in BMMY, whereas the opposite was observed for P. pastoris X-33B. Based on the MPA, the extracellular antimicrobial activity of P. pastoris X-33A was first detected at 24 h of incubation, and the maximum antimicrobial activity was found after incubation for 8 days (192 h) (310 and 100 bacteriocin units (BU)/mg CDW in BMM and BMMY, respectively) (Table 3). For P. pastoris X-33B, the extracellular antimicrobial activity was first detected at 72 h of incubation, and the maximum antimicrobial activity, which was lower than that of P. pastoris X-33A, was found after incubation for 120 and 144 h (50 and 95 BU/mg CDW in BMM and BMMY, respectively) (Table 3). The extracellular antimicrobial activity of P. pastoris X-33AB was first detected at 24 h of incubation, and the maximum antimicrobial activity was found after incubation for 96 and 48 h (65 and 20 BU/mg CDW] in BMM and BMMY, respectively) (Table 3). In both media, the antimicrobial activity of P. pastoris X-33AB was lower than that of P. pastoris X-33A. However, compared to that of P. pastoris X-33B, the antimicrobial activity of P. pastoris X-33AB was higher in BMM but lower in BMMY.
The presence of the recombinant bacteriocin peptides in the supernatants was also assessed by an NCI-ELISA using antibodies specific to the individual bacteriocin peptides. It was found that the amounts of the bacteriocin peptides measured by the immunoassay corresponded well with the antimicrobial activities in the supernatants (Table 3). Although the growth of P. pastoris X-33A in BMM and BMMY was slightly higher than that of P. pastoris X-33B, the maximum amounts of recombinant EntL50A found in supernatants from P. pastoris X-33A cultures were 2- and 40-fold lower, respectively, than those of EntL50B produced by P. pastoris X-33B grown in these media. However, the maximum specific activities of EntL50A in the supernatants from P. pastoris X-33A grown in BMM and BMMY were about 21- and 35-fold higher, respectively, than those of EntL50B in the supernatants from P. pastoris X-33B. Interestingly, we found significantly higher Zeo resistance for P. pastoris X-33B (up to 2,500 μg Zeo/ml) than for P. pastoris X-33A (up to 1,000 μg Zeo/ml).
The maximum antimicrobial activity of P. pastoris X-33AB was found in the supernatants containing both bacteriocin peptides at an approximately 1:1 ratio, and the specific activity (BU/ng total bacteriocin peptides) appeared to be higher than that of P. pastoris X-33A or P. pastoris X-33B (Table 3), suggesting that the recombinant bacteriocin peptides might act synergistically. To assess this synergistic effect further, supernatants from P. pastoris X-33A and P. pastoris X-33B grown in BMM and BMMY were challenged against Pc. damnosus CECT4797, independently or in combination, in a 1:1 bacteriocin peptide ratio. As expected, the equimolar mixture of EntL50A and EntL50B in BMM and BMMY displayed a greater antimicrobial activity (1,707 and 143 BU/mg CDW, respectively) than the additive effect of the peptides acting independently (316 and 0 BU/mg CDW in BMM and 35 and 16 BU/mg CDW in BMMY, respectively), thus being the synergism degree of approximately 5.4- and 2.8-fold in BMM and BMMY, respectively.
Purification and characterization of EntL50A and EntL50B produced by P. pastoris.
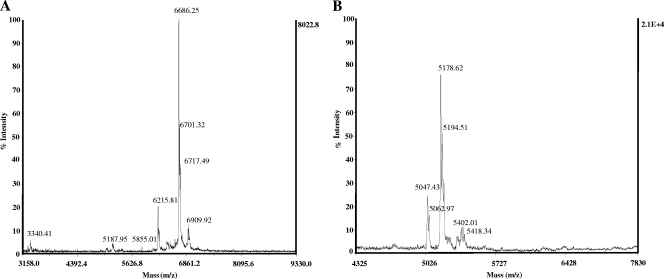
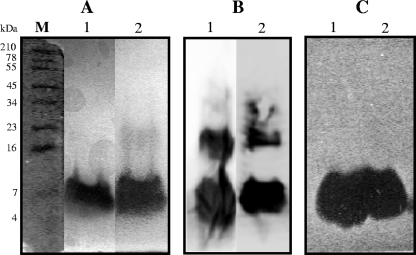
The bacteriocin peptides were purified from the different yeast recombinants in order to compare their biochemical natures to those produced by the wild bacterial producer. At the final step of purification (reversed-phase chromatography), the specific antimicrobial activities of purified EntL50A and EntL50B from P. pastoris X-33A and P. pastoris X-33B were about 322,000 BU/A254 and 20,000 BU/A254, respectively, which corresponded to 154- and 161-fold increases from their respective conditioned medium (after growth in BMMY broth at 30°C for approximately 192 h for P. pastoris X-33A and 48 h for P. pastoris X-33B). From 400-ml cultures, the final yields of purified EntL50A and EntL50B were 24 and 6.5 μg, which correspond to 21 and 7.2% recoveries, respectively, as determined by the immunoassay approach or 43 and 5% recoveries, respectively, by the antimicrobial microtiter plate assay approach (Table 4). The purity and molecular mass of recombinant EntL50A and EntL50B were also analyzed by MALDI-TOF MS. The results obtained for recombinant EntL50A revealed a minor peptide peak with a molecular mass (5,188 Da) highly similar to that of natural EntL50A (5,190 Da) (Fig. 3A) as well as multiple peptide peaks with molecular masses ranging from 6.2 to 6.9 kDa, while the results for recombinant EntL50B identified a major peptide peak with a molecular mass identical to that of natural EntL50B (5,178 Da) as well as a second major peptide peak with a molecular mass of 5,194 Da (Fig. 3B). As shown by the silver-stained Tricine-SDS-PAGE gels in Fig. 4A, purified recombinant EntL50A and EntL50B each gave rise to a major band of the expected size, in addition to a faint band of a larger peptide (16 to 23 kDa) from the EntL50B sample; this band was detected by Western blotting using antibodies specific for EntL50B, suggesting that this recombinant peptide could form aggregates. Similarly, it was demonstrated by immunodetection using an antibody specific for EntL50A that purified recombinant EntL50A also appeared to form aggregates of sizes similar to those of the EntL50B aggregates (Fig. 4B). However, only the monomer forms of these peptides could exert antimicrobial activity when assessed by a gel overlay assay (Fig. 4C).
TABLE 4.
Purification of recombinant EntL50A and EntL50B produced by P. pastoris X-33A and P. pastoris X-33B, respectivelya
| Strain and purification stage | Volume (ml) | Total A254b | Total activity (103 BU)c | Sp act (BU/A254)d | Increase in sp act (fold)e | Total activity (%) | Enterocin yield (ng)f | Enterocin yield (%) | Enterocin sp act (BU/ng)g |
|---|---|---|---|---|---|---|---|---|---|
| P. pastoris X-33A | |||||||||
| Culture supernatant | 400 | 320.8 | 670 | 2,100 | 1 | 100 | 114,500 | 100 | 5.9 |
| Ammonium sulfate precipitation | 40 | 29.2 | 865 | 29,600 | 14 | 129 | 74,100 | 65 | 11.7 |
| Gel filtration chromatography | 80 | 10.9 | 550 | 50,500 | 24 | 82 | 68,000 | 60 | 8.1 |
| Cation-exchange chromatography | 50 | 2.3 | 490 | 213,000 | 102 | 73 | 57,300 | 50 | 8.6 |
| Hydrophobic-interaction chromatography | 10 | 0.9 | 450 | 500,000 | 239 | 67 | 46,200 | 41 | 9.7 |
| Reversed-phase chromatography | 3.5 | 0.9 | 290 | 322,200 | 154 | 43 | 24,000 | 21 | 12.1 |
| P. pastoris X-33B | |||||||||
| Culture supernatant | 400 | 321.2 | 40 | 124 | 1 | 100 | 89,600 | 100 | 0.4 |
| Ammonium sulfate precipitation | 40 | 29.3 | 100 | 3,400 | 27 | 250 | 60,900 | 68 | 1.6 |
| Gel filtration chromatography | 80 | 15.2 | 24 | 1,600 | 12 | 60 | 57,300 | 64 | 0.4 |
| Cation-exchange chromatography | 50 | 2.1 | 3 | 1,400 | 12 | 7.5 | 49,300 | 55 | 0.06 |
| Hydrophobic-interaction chromatography | 10 | 1.0 | 6 | 6,000 | 48 | 15 | 29,600 | 33 | 0.2 |
| Reversed-phase chromatography | 1.6 | 0.1 | 2 | 20,000 | 161 | 5 | 6,500 | 7.2 | 0.3 |
Cultures were grown in BMMY broth at 30°C.
Absorbance at 254 nm multiplied by the volume in milliliters.
Antimicrobial activity against Pc. damnosus CECT4797 in bacteriocin units per milliliter (BU/ml), as determined by an MPA, multiplied by the total volume.
Specific activity expressed as the total activity (BU) divided by the total A254.
The specific activity of a fraction (BU/A254) divided by the specific activity of the culture supernatant (BU/A254).
EntL50A and EntL50B concentration as determined by an NCI-ELISA using specific polyclonal antibodies for EntL50A or EntL50B.
Specific activity expressed as the total activity (BU) divided by the enterocin yield (ng).
FIG. 3.
Mass spectrometry analysis of recombinant EntL50A (A) and EntL50B (B) purified from P. pastoris X-33A and P. pastoris X-33B-33 cultures, respectively, grown in BMMY broth at 30°C.
FIG. 4.
(A) Tricine-SDS-PAGE of purified recombinant EntL50A and EntL50B after silver staining. (B) Western blotting using rabbit polyclonal antibodies with specificity for EntL50A (anti-LR1-KLH) and EntL50B (anti-LR2-KLH). (C) Antimicrobial activity after gel overlay with the indicator strain Pc. damnosus CECT4797. Lane 1, purified EntL50A; lane 2, purified EntL50B. SeeBlue prestained standard molecular mass marker (Invitrogen) band sizes (M) are indicated on the left.
DISCUSSION
Herein, we describe for the first time the heterologous expression and secretion of a two-peptide non-pediocin-like bacteriocin, enterocin L50 (L50A and L50B), by a yeast. For this purpose, the expression and secretion vector pPICZαA from the methylotrophic yeast P. pastoris was selected, since it contains three important elements for successful heterologous expression in the yeast host: (i) the inducible promoter PAOX1 for the controlled expression of the bacteriocin structural genes, (ii) MFα1s including the nucleotide sequence (AAAAGA) encoding the Kex2 signal cleavage site required for the processing of fusion proteins during MFα1s-directed secretion through the Sec system, and (iii) the AOX1 gene, which drives the integration of this vector into the P. pastoris genome and thus maximizes the stability of foreign protein production and/or allows the generation of multicopy strains (6, 23). The generated recombinant P. pastoris strains showed bacteriocinogenic activity both in solid and in liquid complex (BMMY) and minimal (BMM) media. Notwithstanding the finding that yeast growth in the complex medium BMMY was better, the extracellular antimicrobial activity of P. pastoris X-33A (producing only EntL50A) and P. pastoris X-33AB (producing both EntL50A and EntL50B) was significantly lower than that obtained when these strains were grown in the minimal medium BMM; thus, the amounts of the different bacteriocin peptides produced by these two strains in BMMY varied from 10 to 15% of those produced in BMM. These results are in striking contrast to observations for P. pastoris X-33B (producing only EntL50B), whose maximum extracellular antimicrobial activity was found in BMMY, corresponding to a bacteriocin peptide concentration 2.6-fold higher than that in BMM. The nature that caused differential peptide concentrations with the different medium compositions and different levels of bacteriocin peptide production by the bacteriocinogenic recombinant yeasts might be complex but could possibly be ascribed to one or more of the following factors: (i) (higher) aggregation of bacteriocin peptides to form oligomers and/or complexes with medium constituents that results in reduced antigen epitope recognition and, thus, also reduced antimicrobial activity (4, 12, 23, 33, 44, 45, 47); (ii) higher C-terminal proteolytic degradation due a high concentration of vacuolar proteases resulting from higher cell density and lysis (4, 12, 23); and (iii) higher recombinant gene expression due to multiple integration events (1, 6, 33). With regard to the latter, the higher Zeo resistance of P. pastoris X-33B than that of P. pastoris X-33A favors the possibility that the higher bacteriocin peptide concentration found in supernatants from P. pastoris X-33B may be ascribed to a higher-level multiple-integration event of entL50B, in effect resulting in a higher recombinant bacteriocin gene expression level.
To date, MFα1s-directed bacteriocin secretion by P. pastoris has been described for only three pediocin-like bacteriocins (subclass IIa), namely, enterocin P from E. faecium P13 (23), pediocin PA-1 from Pediococcus acidilactici PAC1.0 (4), and hiracin JM79 from Enterococcus hirae DCH5 (42). The heterologous production of pediocin PA-1 by P. pastoris has been shown to result in the secretion of recombinant bacteriocin tightly associated with “collagen-like” material that lacks antimicrobial activity. On the other hand, P. pastoris has been shown to be able to heterologously produce high levels of biologically active enterocin P and hiracin JM79 in BMM and BMMY (23, 42). Irrespective of the growth medium, bacteriocinogenic P. pastoris strains producing enterocin P, hiracin JM79, EntL50A, EntL50B, and EntL50A and EntL50B showed similar growth rates; however, the maximum concentrations of enterocin P and hiracin JM79 were obtained at the beginning of the exponential growth phase (OD600 of 1.7 to 3.3), while EntL50A and EntL50B were maximally found at the beginning of the stationary growth phase (OD600 of 7.8 to 10.2) and the middle-late exponential growth phase (OD600 of 4.8 to 7.2), respectively. Strikingly, the concentrations and biological activities of recombinant enterocin P and hiracin JM79 decreased rapidly, and no antimicrobial activity (in BMM) or only a very low level of antimicrobial activity (1.5 or 45% of the maximum value in BMMY) was found after the incubation of cultures for 10 and 12 h (23, 42); however, recombinant EntL50A and EntL50B showed a higher stability, especially in BMM.
In contrast to our previous work (3), in which the paired production of EntL50A and EntL50B by a single recombinant S. cerevisiae strain was not achievable, we succeeded in the development of a P. pastoris recombinant strain with the ability to produce both biologically active peptides simultaneously. However, levels of EntL50A and EntL50B production by P. pastoris X-33AB were 7- to 10- and 12- to 210-fold lower than those by P. pastoris X-33A and P. pastoris X-33B, respectively. Interestingly, the maximum amount of recombinant EntL50B produced by P. pastoris X-33B (1,240 ng/mg CDW in BMMY at 30°C) represented a 5.9-fold increase over the maximum EntL50B production by wild-type strain E. faecium L50 (210 ng/mg CDW in MRS broth at 25°C) (12). Moreover, although recombinant EntL50B production by P. pastoris X-33B grown in BMM (475 ng/mg CDW) was 2.6-fold lower than that in BMMY (1,240 ng/mg CDW), this bacteriocin peptide amount still represents a 2.3-fold increase over the maximum production by the wild-type strain. However, the maximum amount of recombinant EntL50A produced by P. pastoris X-33A (228.5 ng/mg CDW in BMM at 30°C) is closely similar to the maximum level of EntL50A production by E. faecium L50 (217 ng/mg CDW in MRS broth at 25°C) (12). Moreover, some important differences in bacteriocin production and purification were found when comparing P. pastoris and S. cerevisiae as heterologous hosts for the production of EntL50A and EntL50B. Interestingly, the maximum concentrations of EntL50A and EntL50B in the supernatants from P. pastoris were 27- and 52-fold higher, respectively, than those in the supernatants from S. cerevisiae. On the other hand, the yields of EntL50A and EntL50B purified from P. pastoris supernatants were approximately 2- and 6-fold lower, respectively, than those of the recombinant peptides purified from S. cerevisiae supernatants; however, the specific antimicrobial activities of EntL50A and EntL50B purified from P. pastoris cultures were 60-fold and 6-fold higher, respectively (3). With regard to this finding, the specific antimicrobial activities of EntL50A and EntL50B purified from P. pastoris were 205 and 75%, respectively, of those found in the culture supernatants, while the specific antimicrobial activities of EntL50A and EntL50B purified from S. cerevisiae drastically decreased during the purification process, to 2.6 and 5.5%, respectively (3).
The MALDI-TOF MS analysis of purified recombinant EntL50B showed a major peptide with the same molecular mass as that of natural EntL50B (5,178 Da) (8), demonstrating that the EntL50B precursor was correctly processed by the Kex2 enzyme. The presence of a second major peptide with a molecular mass of 5,194 Da (16 Da more than EntL50B) in the purified fraction probably resulted from the spontaneous oxidation of one of the methionine residues of EntL50B (Met1 or Met24), giving rise to a methionine sulfoxide (MetSO), which is 16 Da more than the nonoxidized form. Such an oxidation of methionine residues during bacteriocin purification is in fact relatively common, as it was described previously for several other bacteriocins (5, 8, 10, 34). For EntL50A, the results from MALDI-TOF MS analyses revealed a minor peptide with a molecular mass (5,188 Da) highly similar to that of natural EntL50A (5,190 Da) (8), demonstrating that the EntL50A precursor was correctly processed by the Kex2 enzyme. However, there were several other major peptides, in the range of 6.2 to 6.9 kDa, also present in the purified fraction. At first glance, this result could indicate that most EntL50A precursors were not correctly processed by the Kex2 enzyme as a consequence of a reduced recognition of the cleavage site (Glu-Lys-Arg) due to a conformational interference exerted by its N terminus; however, the high level of similarity between EntL50A and EntL50B (8, 10) does not favor this possibility. More likely, recombinant EntL50A may be associated with a hitherto unknown compound, as previously suggested for the biologically inactive pediocin PA-1 heterologously produced by P. pastoris (4) and the biologically active EntL50A purified from recombinant S. cerevisiae strains (3).
The results presented in this work demonstrate that the cloning of entL50A and entL50B fused to MFα1s is enough for the efficient production of biologically active EntL50A and EntL50B, separately or together, by P. pastoris through the Sec system. This yeast host is more suitable than S. cerevisiae for the heterologous production of these antimicrobial bacteriocin peptides, especially for gene coexpression. We show herein that it is possible to use the nonpathogenic yeast P. pastoris to produce bacteriocins originally derived from potentially pathogenic microorganisms (e.g., Enterococcus species); this is especially relevant for bacteriocins from multibacteriocin producers, where purification of the bacteriocin of interest is often problematic due to their relatively similar physicochemical properties (being small, cationic, and amphiphilic), and for bacteriocins with a barely understood mechanism of secretion, such as enterocin L50 (L50A and L50B) and other leaderless bacteriocins. This approach can thus be an important foundation for the future production of leaderless bacteriocins at industrial scales for use in the pharmaceutical and food industries.
Acknowledgments
This work was partially supported by grants PR248/02-11688 from the Fundación Danone/Complutense (Madrid, Spain); PR41/06-15051 from the Grupo Santander Central Hispano/Universidad Complutense de Madrid (Madrid, Spain); AGL2003-01508, AGL2006-01042, and AGL2009-08348 from the Ministerio de Educación, Cultura y Deporte (MECD), Spain; and S-0505/AGR/0265 from the Comunidad de Madrid (CAM), Spain. A.B. was a recipient of an FPI fellowship from CAM, Spain. B.G.-S. holds a contract from the company Innaves S.A. (Vigo, Spain). J.S. was a recipient of an FPU fellowship from MECD, Spain.
We thank Juan José Jiménez for helpful discussions. We also recognize the help of Jorge Gutiérrez and Raquel Criado. We are grateful to the anonymous reviewers for their constructive comments and very valuable suggestions for improvement of the manuscript.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Apte-Deshpnade, A., G. Mandal, S. Soorapaneni, B. Prasad, J. Kumar, and S. Padmanabhan. 2009. High-level expression of non-glycosylated and active staphylokinase from Pichia pastoris. Biotechnol. Lett. 31:811-817. [DOI] [PubMed] [Google Scholar]
- 2.Basanta, A., J. Sánchez, B. Gómez-Sala, C. Herranz, P. E. Hernández, and L. M. Cintas. 2008. Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 175:293-307. [DOI] [PubMed] [Google Scholar]
- 3.Basanta, A., C. Herranz, J. Gutiérrez, R. Criado, P. E. Hernández, and L. M. Cintas. 2009. Development of bacteriocinogenic strains of Saccharomyces cerevisiae heterologously expressing and secreting the leaderless enterocin L50 peptides L50A and L50B from Enterococcus faecium L50. Appl. Environ. Microbiol. 75:2382-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaulieu, L., D. Groleau, C. B. Miguez, J. F. Jetté, H. Aomari, and M. Subirade. 2005. Production of pediocin PA-1 in the methylotrophic yeast Pichia pastoris reveals unexpected inhibition of its biological activity due to the presence of collagen-like material. Protein Expr. Purif. 43:111-125. [DOI] [PubMed] [Google Scholar]
- 5.Casaus, M. P., T. Nilsen, L. M. Cintas, I. F. Nes, and P. E. Hernández. 1997. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 143:2287-2294. [DOI] [PubMed] [Google Scholar]
- 6.Cereghino, J. L., and J. M. Cregg. 2000. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 24:45-66. [DOI] [PubMed] [Google Scholar]
- 7.Cintas, L. M., J. M. Rodríguez, M. F. Fernández, K. Sletten, I. F. Nes, P. E. Hernández, and H. Holo. 1995. Isolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory spectrum. Appl. Environ. Microbiol. 61:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cintas, L. M., P. Casaus, H. Holo, P. E. Hernández, I. F. Nes, and L. S. Håvarstein. 1998. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J. Bacteriol. 180:1988-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cintas, L. M., P. Casaus, M. F. Fernández, and P. E. Hernández. 1998. Comparative antimicrobial activity of enterocin L50, pediocin PA-1, nisin A and lactocin S against spoilage and foodborne pathogenic bacteria. Food Microbiol. 15:289-298. [Google Scholar]
- 10.Cintas, L. M., P. Casaus, C. Herranz, L. S. Håvarstein, H. Holo, P. E. Hernández, and I. F. Nes. 2000. Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182:6806-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cintas, L. M., P. Casaus, C. Herranz, I. F. Nes, and P. E. Hernández. 2001. Bacteriocins of lactic acid bacteria. Food Sci. Technol. Int. 7:281-305. [Google Scholar]
- 12.Criado, R., J. Gutiérrez, M. Martín, C. Herranz, P. E. Hernández, and L. M. Cintas. 2006. Immunochemical characterization of temperature-regulated production of enterocin L50 (EntL50A and EntL50B), enterocin P, and enterocin Q by Enterococcus faecium L50. Appl. Environ. Microbiol. 72:7634-7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deegan, L. H., P. D. Cotter, C. Hill, and P. Ross. 2006. Bacteriocins: biological tools for bio-preservation and shelf-life extension. Int. Dairy J. 16:1058-1071. [Google Scholar]
- 14.Diep, D. B., L. S. Håvarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diep, D. B., M. Skaugen, Z. Salehian, H. Holo, and I. F. Nes. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drider, D., G. Fimland, Y. Héchard, L. M. McMullen, and H. Prévost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton, T. J., and M. J. Gasson. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franz, C. M. A. P., M. E. Stiles, K. H. Schleifer, and W. H. Holzapfel. 2003. Enterococci in foods—a conundrum for food safety. Int. J. Food Microbiol. 88:105-122. [DOI] [PubMed] [Google Scholar]
- 19.Franz, C. M. A. P., M. J. van Belkum, W. H. Holzapfel, H. Abriouel, and A. Gálvez. 2007. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 31:293-310. [DOI] [PubMed] [Google Scholar]
- 20.Gajic, O., G. Buist, M. Kojic, L. Topisirovic, O. P. Kuipers, and J. Kok. 2003. Novel mechanism of bacteriocin secretion and immunity carried out by lactococcal multidrug resistance proteins. J. Biol. Chem. 278:34291-34298. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore, M. S., R. A. Segarra, M. C. Booth, C. P. Bogie, L. R. Hall, and D. B. Clewell. 1994. Genetic structure of Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176:7335-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutiérrez, J., R. Criado, R. Citti, M. Martín, C. Herranz, M. F. Fernández, L. M. Cintas, and P. E. Hernández. 2004. Performance and applications of polyclonal antipeptide antibodies specific for the enterococcal bacteriocin enterocin P. J. Agric. Food Chem. 52:2247-2255. [DOI] [PubMed] [Google Scholar]
- 23.Gutiérrez, J., R. Criado, M. Martín, C. Herranz, L. M. Cintas, and P. E. Hernández. 2005. Production of enterocin P, an antilisterial pediocin-like bacteriocin from Enterococcus faecium P13, in Pichia pastoris. Antimicrob. Agents Chemother. 49:3004-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutiérrez, J., D. Bourque, R. Criado, Y. J. Choi, L. M. Cintas, P. E. Hernández, and C. B. Míguez. 2005. Heterologous extracellular production of enterocin P from Enterococcus faecium P13 in the methylotrophic bacterium Methylobacterium extorquens. FEMS Microbiol. Lett. 248:125-131. [DOI] [PubMed] [Google Scholar]
- 25.Gutiérrez, J., R. Criado, R. Citti, M. Martín, C. Herranz, I. F. Nes, L. M. Cintas, and P. E. Hernández. 2005. Cloning, production and functional expression of enterocin P, a sec-dependent bacteriocin produced by Enterococcus faecium P13, in Escherichia coli. Int. J. Food Microbiol. 103:239-250. [DOI] [PubMed] [Google Scholar]
- 26.Gutiérrez, J., R. Larsen, L. M. Cintas, J. Kok, and P. E. Hernández. 2006. High-level heterologous production and functional expression of the sec-dependent enterocin P from Enterococcus faecium P13 in Lactococcus lactis. Appl. Microbiol. Biotechnol. 72:41-51. [DOI] [PubMed] [Google Scholar]
- 27.Han, X., L. B. Ye, B. Z. Li, G. Bo, W. J. Cai, Z. Hong, Y. L. She, Y. Li, L. B. Kong, and Z. H. Wu. 2006. Expression, purification and characterization of the hepatitis B virus entire envelope large protein in Pichia pastoris. Protein Expr. Purif. 49:168-175. [DOI] [PubMed] [Google Scholar]
- 28.Hancock, R. E. W., and H.-G. Sahl. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551-1557. [DOI] [PubMed] [Google Scholar]
- 29.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 30.Herranz, C., and A. J. M. Driessen. 2005. Sec-mediated secretion of bacteriocin enterocin P by Lactococcus lactis. Appl. Environ. Microbiol. 71:1959-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holo, H., Ø. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hummel, A. S., W. H. Holzapfel, and C. M. A. P. Franz. 2007. Characterisation and transfer of antibiotic resistance genes from enterococci isolated from food. Syst. Appl. Microbiol. 30:1-7. [DOI] [PubMed] [Google Scholar]
- 33.Ilgen, C., J. L. Cereghino, and J. M. Cregg. 2005. Pichia pastoris, p. 143-162. In G. Gellissen (ed.), Production of recombinant proteins: novel microbial and eucaryotic expression systems. Wiley-Vch Verlag GmbH & Co. KGaA, Weinheim, Germany.
- 34.Johnsen, L., G. Fimland, V. Eijsink, and J. Nissen-Meyer. 2000. Engineering increased stability in the antimicrobial peptide pediocin PA-1. Appl. Environ. Microbiol. 66:4798-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkup, B. C., Jr. 2006. Bacteriocins as oral and gastrointestinal antibiotics: theoretical considerations, applied research, and practical applications. Curr. Med. Chem. 13:3335-3350. [DOI] [PubMed] [Google Scholar]
- 36.Lawton, E. M., R. P. Ross, C. Hill, and P. D. Cotter. 2007. Two-peptide lantibiotics: a medical perspective. Mini Rev. Med. Chem. 7:1236-1247. [DOI] [PubMed] [Google Scholar]
- 37.Martín, M. 2006. Ph.D. thesis. Universidad Complutense de Madrid, Madrid, Spain.
- 38.Nes, I. F., D. B. Diep, L. S. Håvarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 39.Nes, I. F., D. B. Diep, and H. Holo. 2007. Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsen, T., I. F. Nes, and H. Holo. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69:2975-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogier, J.-C., and P. Serror. 2008. Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 126:291-301. [DOI] [PubMed] [Google Scholar]
- 42.Sánchez, J., J. Borrero, B. Gómez-Sala, A. Basanta, C. Herranz, L. M. Cintas, and P. E. Hernández. 2008. Cloning and production of hiracin JM79, a Sec-dependent bacteriocin produced by Enterococcus hirae DCH5, in heterologous lactic acid bacteria and Pichia pastoris. Appl. Environ. Microbiol. 74:2471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sánchez-Hidalgo, M., M. Maqueda, A. Gálvez, H. Abriouel, E. Valdivia, and M. Martínez-Bueno. 2003. The genes coding for enterocin EJ97 production by Enterococcus faecalis EJ97 are located on a conjugative plasmid. Appl. Environ. Microbiol. 69:1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoeman, H., M. A. Vivier, M. du Toit, L. M. T. Dicks, and I. S. Petrorius. 1999. The development of bactericidal yeast strains by expressing the Pediococcus acidilactici pediocin gene (pedA) in Saccharomyces cerevisiae. Yeast 15:647-656. [DOI] [PubMed] [Google Scholar]
- 45.Schuster, M., A. Einhauer, E. Wasserbauer, F. Süßenbacher, C. Ortner, M. Paumann, G. Werner, and A. Jungbauer. 2000. Protein expression in yeast; comparison of two expression strategies regarding protein maturation. J. Biotechnol. 84:237-248. [DOI] [PubMed] [Google Scholar]
- 46.Wu, J., S. Hu, and L. Cao. 2007. Therapeutic effect of nisin Z on subclinical mastitis in lactating cows. Antimicrob. Agents Chemother. 51:3131-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, R., M. C. Johnson, and B. Ray. 1992. Novel method to extract large amounts of bacteriocins from lactic acid bacteria. Appl. Environ. Microbiol. 58:3355-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]