Abstract
Sulfate-reducing bacteria (SRB) can affect metal mobility either directly by reductive transformation of metal ions, e.g., uranium, into their insoluble forms or indirectly by formation of metal sulfides. This study evaluated in situ and biostimulated activity of SRB in groundwater-influenced soils from a creek bank contaminated with heavy metals and radionuclides within the former uranium mining district of Ronneburg, Germany. In situ activity of SRB, measured by the 35SO42− radiotracer method, was restricted to reduced soil horizons with rates of ≤142 ± 20 nmol cm−3 day−1. Concentrations of heavy metals were enriched in the solid phase of the reduced horizons, whereas pore water concentrations were low. X-ray absorption near-edge structure (XANES) measurements demonstrated that ∼80% of uranium was present as reduced uranium but appeared to occur as a sorbed complex. Soil-based dsrAB clone libraries were dominated by sequences affiliated with members of the Desulfobacterales but also the Desulfovibrionales, Syntrophobacteraceae, and Clostridiales. [13C]acetate- and [13C]lactate-biostimulated soil microcosms were dominated by sulfate and Fe(III) reduction. These processes were associated with enrichment of SRB and Geobacteraceae; enriched SRB were closely related to organisms detected in soils by using the dsrAB marker. Concentrations of soluble nickel, cobalt, and occasionally zinc declined ≤100% during anoxic soil incubations. In contrast to results in other studies, soluble uranium increased in carbon-amended treatments, reaching ≤1,407 nM in solution. Our results suggest that (i) ongoing sulfate reduction in contaminated soil resulted in in situ metal attenuation and (ii) the fate of uranium mobility is not predictable and may lead to downstream contamination of adjacent ecosystems.
Dissimilatory sulfate-reducing bacteria (SRB) play an important role in the sulfur cycle and the mineralization of organic matter in anoxic marine and freshwater environments (53). In addition, sulfate reduction can occur in oxygenated habitats where anoxic niches (8) and the expression of superoxide reductase activity (34) provide protection for SRB against oxygen toxicity. The rate-limiting step of sulfate reduction is catalyzed by the dissimilatory (bi-)sulfite reductase (encoded by the dsrAB gene). Phylogenetic investigations have shown that this key enzyme for sulfate and sulfite respiration was present in early ancestors of modern Bacteria and Archaea (66).
Dissimilatory sulfate reduction has been shown to be a terminal-electron-accepting process (TEAP) in acid mine drainage (AMD)-impacted and radionuclide- and metal-contaminated environments. Sulfate-reducing activities as well as SRB abundances show a wide range in these habitats (24, 29, 69). SRB are able to reductively transform metal ions, e.g., uranium and chromium, into insoluble and chemically inert forms via direct enzymatic reduction (41, 42). Sulfide, the end product of microbial sulfate reduction, may further contribute to metal attenuation through reduction of metal oxycations and oxyanions, such as those of uranium and chromium (4, 19), or through precipitation of metal cations as sulfides (15, 20). In addition, SRB have the potential to enhance metal retention via extracellular binding, cellular uptake and accumulation of metals, oxidation/reduction processes, and surface-mediated mineral precipitation (20, 52). Metal stress for SRB in uranium-contaminated sediments (48, 63) and biofilms from Pb-Zn deposits (39) can be reduced by the formation of uraninite and metal sulfides.
Previous work in uranium-contaminated environments has emphasized the role of biostimulated SRB in mediating uranium and/or technetium reduction (3, 46, 63), although other metal contaminants are present (55, 63). The long-term stability of immobilized, reduced contaminants is a concern due to the potential for remobilization after carbon addition is stopped. Therefore, it is important to understand alternative remediation processes, such as those involved in natural attenuation. In the former uranium mining district of Ronneburg (Thuringia, Germany), leaching of low-grade black shale by acid mine drainage and sulfuric acid and pyrite oxidation resulted in serious large-scale contamination with heavy metals and radionuclides (28). Metal- and sulfate-enriched seepage waters and surface runoff infiltrated adjacent soils and surface waters, leading to elevated concentrations of sulfate, nickel, copper, cadmium, zinc, arsenic, and uranium in creek bank soils (9). At the Ronneburg site, the presence of high levels of mixed contaminants provides a unique environment to look at complex processes involved in natural attenuation of contaminants. It is hypothesized that resident SRB contribute to natural uranium and heavy metal attenuation at the Ronneburg site. Thus, the objective of this study was to resolve the potential importance of SRB in contaminated creek bank soils both in situ and in biostimulated soil microcosms using stable isotope probing (SIP).
MATERIALS AND METHODS
Site description.
The study site is located on the bank of Gessen Creek, one of the main drainage systems for the former leaching heaps within the former uranium mining site near Ronneburg (Thuringia, Germany; location E 4510121, N 5635807, Gauss-Krueger Potsdam coordinate system). In the luvic gleysol, two iron-rich groundwater-influenced oxidized horizons (BElc and Btlc) and two groundwater-influenced reduced horizons (Br1 and Br2) could be distinguished by color and texture below the humus top horizon (Ah) and a yellowish horizon (BEw/Ah) (6). The reduced horizons were gray (upper Br1, 82 to 103 cm in depth) and black (lower Br2, 102 to 110 cm in depth), respectively. The solid phase of Br2 had a high total sulfur and carbon content (up to 2.0% and 3.4%, respectively) and showed a low redox potential (Eh) of −30 mV (37). In contrast, Br1 had a sulfur content of only 0.4% and a redox potential of 60 mV.
Sampling procedure.
Soil was aseptically sampled and stored in plastic bags in the dark at 4°C for transport and until further processing on the following day. For determination of the acid volatile sulfur (AVS) fraction, the soil was frozen at −20°C. Soil samples for total extraction were stored at 4°C in the dark prior to analysis. Pore water samples for determination of pH and redox potential and measurements of nitrate, sulfate, and soluble heavy metal concentrations were taken with Rhizon suction samplers (Eijkelkamp, Giessbeek, Netherlands) monthly from June to November 2007 from the soil profile at ∼10-cm-depth intervals. The redox potential of pore water was measured directly after sampling, and soil waters were stored at 4°C overnight prior to subsequent analyses.
Determination of total soil metal concentrations.
Soils from the Br1 and Br2 horizons were air dried and then finely ground to <63-μm sieve size. The ground Br1 and Br2 soils were digested with concentrated hydrofluoric acid, nitric acid, and perchloric acid at 150 to 170°C in a pressure digestion system (DAS; PicoTrace, Bovenden, Germany). Single elements were measured in the resulting solutions using inductively coupled plasma-optical emission spectroscopy (ICP-OES; Spectroflame P FAV05; Spectro Analytical Instruments, Kleve, Germany) for Fe and inductively coupled plasma-mass spectrometry (ICP-MS; X-SeriesII; ThermoFisher Scientific, Bremen, Germany) for As, Cd, Co, Cu, Ni, U, and Zn.
X-ray absorption spectroscopy for analysis of solid uranium.
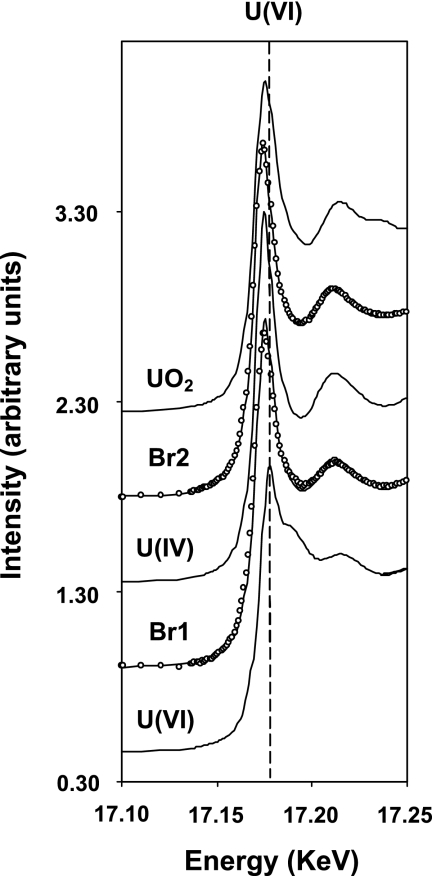
X-ray absorption near-edge structure (XANES) spectra were collected for soils of Br1 and Br2 at the Rossendorf Beamline (Grenoble, France). Samples were prepared under an anoxic atmosphere. Uranium LIII-edge spectra were collected in fluorescence mode at 15 K using a closed-cycle He cryostat. In order to determine the relative proportions of U(IV) and U(VI) in these samples, linear combination fits of the XANES region of the spectra using reference spectra of U(IV) and U(VI) were performed. A reference spectrum of U(IV) aqueous complex was chosen for the U(IV) component, whereas a spectrum of U(VI) sorbed on clay mineral was selected for U(VI). Relative proportions of U(VI) and U(IV) obtained from the fits were within ±1% of the reported values. Additional details are described in a previous study (6).
Determination of AVS.
Acid volatile sulfur (AVS) was determined by suspending 10 g of soil in 50% ethanol under a nitrogen flow. AVS was liberated as hydrogen sulfide by cold acid distillation for 1 h after addition of 8 ml of 30% (wt/vol) HCl. The released H2S was collected in 2% (wt/vol) zinc acetate and measured spectrophotometrically according to the method described by Cline (12).
SRR.
Sulfate reduction rates (SRR) were determined using the 35SO42− radiotracer technique (17). Rate determinations were conducted on all soil horizons in October 2007 and April 2008 in replicates of three and five, respectively. Soil (3 g) was transferred to sterile 7.5-ml serum bottles, which were sealed with butyl rubber stoppers; flushed with sterile argon; and then diluted with 3 ml of sterile, anoxic water. Soil suspensions were amended with H235SO4 (Hartmann Analytics, Braunschweig, Germany) to a final activity of 100 kBq cm soil−3 and incubated for 2 h at 15°C. Incubations were stopped by transferring the soil suspensions to plastic bottles containing 10 ml of 20% (wt/vol) zinc acetate. The samples were stored frozen at −20°C until further processing. The formation of sulfide was analyzed by combined chromium and acid distillation as described by Fossing and Jørgensen (17). Dry weight of soil was determined after the soil was dried at 105°C for 24 h to a constant weight.
Enumeration of sulfate-reducing bacteria.
A three-tube most probable number (MPN) technique (14) using 10-fold serial dilutions was used to enumerate cultivable sulfate-reducing bacteria in soils from the reduced horizons. For selective growth, a modified Postgate C medium (7) was used with a final sulfate concentration of 10 mM and pH 6.2. The medium was amended with sodium acetate or lactate (final concentrations, 5 mM) as an electron donor. From both horizons, an additional MPN series was prepared to which heavy metals (0.3 μM CdCl2, 38.6 μM ZnCl2, 1.5 μM CuCl2, 16.4 μM NiCl2, 0.6 μM CoCl2, and 23.2 μM AlCl3) were added, reflecting the maximum pore water concentrations in the soil profile. The culture tubes were incubated at 16°C in the dark for 5 months. Positive growth was scored by a decrease in sulfate concentrations, and MPN values with 95% confidence limits were calculated according to the method of de Man (13). When the ratio between MPN values was above 8.87, the abundance of organisms was considered significant (2).
Soil microcosms.
Anoxic soil incubation experiments were performed (i) to study microbial activity under anoxic conditions (soil from horizon Br1 or Br2) and (ii) to investigate active sulfate-reducing bacteria by using stable isotope probing (SIP; soil from Br2 only). Soil samples were collected in May and June 2007 from the Br2 horizon and in February and October 2008 from the Br1 horizon. With the exception of the second experiment using Br1 soil, microcosms were constructed by loading 20 g (fresh weight) of soil into 150-ml incubation bottles under a sterile argon atmosphere, and then the bottles were sealed with rubber stoppers and secured with aluminum caps. A mineral solution (50 μM NaNO3, 70 μM NH4Cl, and 10 mM MgSO4, reflecting in situ soil water concentrations) was added in a ratio of 1:5 (weight of soil/volume) in a mineral solution. Seventy-five grams (fresh weight) of soil was prepared in 500-ml incubation bottles as described above for the second Br1 horizon experiment (experiment 2) with the sulfate concentrations again adjusted to match in situ conditions. The soil was diluted with mineral solution in a ratio of 1:4 (weight of soil/volume) containing 6 mM MgSO4 only. Triplicate bottles for all experiments were amended to a final concentration of 5 mM glucose, 10 mM lactate, 10 mM acetate, or 10 mM ethanol as electron donor. Glucose and ethanol were not supplied in the SIP or the second Br1 experiment. 13C-labeled acetate or lactate (10 mM final concentration; >99 atom% 13C; Cambridge Isotopes) was used for the SIP microcosms. Reduced carbon concentrations, 3 mM acetate or lactate, were used for the second set of Br1 microcosms. Three replicates without an added carbon source served as a control. The pH of the microcosms was adjusted to 6.3 and 6.1 for Br1 and Br2, respectively, and the microcosms were then shaken for 1 h prior to incubation in the dark at 16°C. Microcosms were regularly sampled over a period of 31 to 37 days using anoxic, aseptic techniques to measure SO42−, NO3−, Fe(II), and Mn (except for the Br1 microbial activity experiment). Soluble metal concentrations, pH, and redox potential were determined at the beginning and end of the anoxic incubation.
Analytical techniques.
Pore water redox potential, pH, Fe(II), total Fe, nitrate, ammonia, sulfate, metals, and arsenic were measured as described elsewhere (6). In the microcosms, pH and Eh were measured in the soil suspension immediately after sampling. Soil suspensions were centrifuged at 2,300 × g for 3 min, and the supernatant was filtered through an 0.2-μm nylon membrane for further analysis. Nitrate and sulfate were analyzed by ion chromatography. Soluble manganese, assumed as Mn(II), was determined by the formaldoxime method (31). Soluble metal and arsenic concentrations in microcosms were determined as described for pore water samples. Concentrations of aliphatic fatty acids, sugars, and alcohols were analyzed using high-performance liquid chromatography (HPLC) according to the work of Reiche et al. (54). Headspace methane concentrations were determined by gas chromatography (Hewlett-Packard Company 5980 series II) as previously described (36). Gas pressures in the microcosms were measured with a TensioCheck TC 1066 needle manometer (Tensio-Technik, Geisenheim, Germany). Sulfate concentrations in MPN cultures were analyzed in the supernatant after centrifugation at 8,400 × g for 2 min using the barium chloride method (64).
Analysis of dsrAB functional genes. (i) DNA extraction and PCR amplification.
Soil samples (Br2) for DNA extraction were collected from the Gessen Creek bank in August 2007 and stored at −20°C. Genomic DNA was isolated and purified using the Power Soil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's instructions. Amplification of the dissimilatory sulfite reductase (DSR) gene (dsrAB) was performed using a forward and reverse DSR primer mix targeting a variety of sulfate-reducing bacteria (65; M. Pester, N. Bittner, P. Deevong, M. Wagner, and A. Loy, submitted for publication). The PCR mixture contained 1× MasterAmp PCR Premix D (Epicentre Biotechnologies, Madison, WI), 2.5 U Taq polymerase (Jena Bioscience, Jena, Germany), DSR primer mix with equimolar concentrations of 0.1 μM for each primer variant, and 3.5 ng soil DNA. Amplification was performed in three replicates under the following conditions: initial denaturation step for 3 min at 94°C, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 48°C for 40 s, and elongation at 72°C for 90 s and a final elongation step for 10 min at 72°C (65). The ∼1.9-kb dsrAB amplicon was purified with gel extraction using the agarose gel extraction kit (Jena Bioscience, Jena, Germany) according to the manufacturer's protocol. Purified dsrAB amplicons were cloned into the TOPO TA cloning vector pCR4 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Plasmids of clones containing the dsrAB gene were extracted with the plasmid miniprep kit (Jena Bioscience, Jena, Germany) according to the manufacturer's instructions. dsrAB fragments were sequenced by Macrogen (Seoul, South Korea).
(ii) dsrAB phylogenetic analysis.
Phylogenetic analyses were performed using the ARB program (44), where all nucleotide sequences (forward reads) were imported into a dsrAB/DsrAB-ARB database for SRB maintained at the Department of Microbial Ecology, University of Vienna (43), and aligned manually based on deduced amino acids. dsrAB clones with ≥90% nucleotide sequence identity to each other were grouped into an operational taxonomic unit (OTU). A consensus tree was based on neighbor-joining, maximum-parsimony (Phylip PROTPARS; 1,000 bootstraps), and maximum-likelihood methods using a DsrA filter without correction for partial dsrA sequences (245 amino acid positions) in the ARB program. Percent coverage was calculated as the ratio between observed and expected OTUs according to rarefaction analysis using Analytic Rarefaction 1.3 (22).
Stable isotope probing of 13C- and 12C-labeled small-subunit (SSU) rRNA genes.
Microcosm samples for stable isotope probing (SIP) were centrifuged for 3 min at 2,300 × g, and then the pellets were stored frozen at −20°C until nucleic acid extraction. Procedures used for DNA extraction, separation of 13C- and 12C-labeled DNA, SSU rRNA gene PCR amplification, and terminal restriction fragment length polymorphism (TRFLP) analysis were performed according to the method of Burkhardt et al. (6) with slight modifications. In brief, triplicate DNA extracts from solid-phase microcosm samples were combined, and then 13C- and 12C-labeled DNA were separated by density gradient centrifugation. After centrifugation the DNA was retrieved by the gradient fractionation method, cleaned, and precipitated. SSU rRNA genes were amplified using the general domain Bacteria SSU rRNA gene primers 27F and 1492R, where for TRFLP analysis, the 27F primer was 5′ end labeled with 6-carboxyfluorescein. For TRFLP, digested and purified DNA samples were run on the ABI 3730 genetic analyzer using GeneMapper software. The pairwise similarities between profiles were calculated from a matrix of the presence/absence of TRF using product-moment correlation distance matrix. Cluster analysis based on this similarity matrix was performed by UPGMA (unweighted-pair group method with arithmetic mean).
For SSU rRNA gene clone library construction and phylogenetic analyses, purified PCR products were ligated into the TOPO TA cloning vector pCR XL according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Ligations were shipped to the Genome Sequencing Center at Washington University (St. Louis, MO) for transformation and bidirectional sequencing with vector-specific primers (M13F/R). Sequences were assembled, and vector sequences flanking the SSU rRNA gene inserts were removed using Geneious Pro version 4.6.4 (Biomatters, Auckland, New Zealand). Clones were grouped into phylotypes based on a sequence similarity cutoff of 97%, and previously identified sequences with high sequence similarity to the clones obtained in this study were determined using the BLAST algorithm against the GenBank database available from the National Center for Biotechnology Information (NCBI). All clone sequences were aligned with the alignment tool by Greengenes, and the nearest neighbors were identified using the Classify tool against the Greengenes database.
Nucleotide sequence accession numbers.
Sequences from this study were published in the GenBank database under the accession numbers GU233963 to GU234006. Sequences generated in this study were also deposited in the GenBank database under the accession numbers GU235998 to GU236099.
RESULTS
Soil geochemistry.
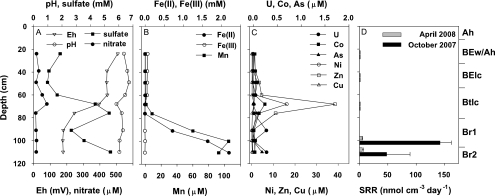
The average pH of the soil water measured monthly was neutral to slightly acidic, ranging from 5.8 to 7.0 over the whole profile (Fig. 1A). In general, the redox potential was low in both reduced horizons. Sulfate was enriched in the pore water (Fig. 1A) with average concentrations of 3.4 ± 3.2 mM and 3.1 ± 2.3 mM for Br1 and Br2, respectively. However, the range of sulfate concentrations for the reduced horizons was from 0.3 mM to 12.4 mM over the sampling period. Nitrate (Fig. 1A) was negligibly low in the reduced horizons (<20 μM), whereas it reached up to 1.2 mM in the upper horizons during the sampling period (data not shown). Both manganese and Fe(II) increased with depth, reaching highest concentrations in the horizon Br2 at 0.1 mM Mn and 2.2 mM Fe(II), while Fe(III) was negligible and declined within the oxidized horizons (Fig. 1B). Soluble heavy metal concentrations (U, Co, Ni, Zn, and Cu) peaked in the oxidized Btlc horizon but were low in the deeper reduced horizons (Fig. 1C). The maximum uranium concentration of 278 nM was observed in horizon Br2 in June 2007. In general, arsenic concentrations were higher in the reduced horizons, reaching up to 180 nM, compared to the upper, oxidized horizons (Fig. 1C).
FIG. 1.
Geochemistry (A to C) of the soil profile in June 2007 and depth profile of sulfate reduction rates (SRR) (D) at the bank of the contaminated Gessen Creek. SRR values are averages ± standard deviations (October 2007, n = 3; April 2008, n = 5). The corresponding soil horizons are given at the right side of the graphs.
Acid volatile sulfur (AVS) was higher in the solid phase of the reduced horizons (52 and 126 mmol kg [dry weight] soil−1 at Br1 and Br2, respectively), whereas much lower concentrations were observed in the upper soil horizons (AVS ≤ 0.2 mmol kg [dry weight] soil−1). Creek bank soil contained high contents of metals in the solid phase and exceeded background levels in the majority of cases (Table 1). Br2 soil accumulated total uranium, zinc, nickel, and copper to a higher extent than did Br1 soil (Table 1). According to XANES measurement, soil of both horizons was highly enriched in reduced uranium species, with Br2 (83.8%) containing a slightly larger amount of U(IV) than Br1 (79.5%). In addition, a comparison of samples Br1 and Br2 to a uraninite (UO2) reference spectrum showed a lack of features in the postedge region of the two samples, suggesting that U(IV) in these samples did not occur in a crystalline form similar to UO2 but was more likely to be present as a sorbed complex (Fig. 2).
TABLE 1.
Total metal content of Br1 and Br2 solid phase
| Horizon | Metal content (μg g [dry wt] soil −1)a |
||||||
|---|---|---|---|---|---|---|---|
| U | Zn | Ni | Cu | Co | As | Cd | |
| Br1 | 343 ± 4 | 425 ± 10 | 170 ± 1 | 289 ± 2 | 31 ± 1 | 37 ± 1 | 5 ± 0 |
| Br2 | 959 ± 3 | 565 ± 11 | 229 ± 1 | 325 ± 3 | 43 ± 0 | 55 ± 1 | 5 ± 0 |
| BBodSchVb | NDd | 60-200 | 15-70 | 20-60 | ND | 25 | 0.4-1.5 |
| IAEAc | 2 | ND | ND | ND | ND | ND | ND |
Values represent metal content in μg g−1 (dry weight) of Br1 or Br2 soil and background soil levels.
Bundes-Bodenschutz- und Altlastenverordnung (http://www.gesetze-im-internet.de/bundesrecht/bbodschv/gesamt.pdf; accessed 30 November 2009). BBodSchV values range from limits in sand to limits in clay material.
IAEA, International Atomic Energy Agency (http://www.iaea.org/NewsCenter/Features/DU/du_qaa.shtml; accessed 30 November 2009). Background soil level is given.
ND, standard values are not given.
FIG. 2.
XANES. Fitted XANES spectra of the Br1 and Br2 samples with reference spectra of U(IV) and U(VI), used for the fitting, as well as that of UO2. In the spectra of samples Br1 and Br2, the open circles represent data points and the solid line is the fit to the data obtained from linear combination fitting using U(IV) and U(VI) components. The vertical dashed line indicate the U(VI) peak position, which is clearly separated from U(IV).
SRR.
Sulfate-reducing activity was restricted to the reduced horizons (Fig. 1D) with sulfate reduction rates (SRR) of 142 ± 20 nmol cm−3 day−1 and 48 ± 41 nmol cm−3 day−1 for Br1 and Br2 in October 2007, respectively. Sulfate reduction rates determined in April 2008 showed a similar pattern, but total rates were only 5 nmol cm−3 day−1. In contrast, activity was below detection in the upper, oxidized horizons.
Enumeration of sulfate-reducing bacteria.
The abundances of cultivated SRB (Table 2) were similar in horizons Br1 and Br2 but differed slightly according to the amended electron donor in the enrichments. The highest abundances were observed in lactate-amended cultures of the Br2 horizon. SRB abundance in both horizons was slightly lower in the presence of heavy metals (Table 2). The difference was significant only in the lactate treatment for Br2.
TABLE 2.
Enumeration of sulfate-reducing bacteria in reduced soil horizons in the bank of Gessen Creek with and without metals
| Treatment and electron donor | MPN (cells g [fresh wt] soil−1)a |
|
|---|---|---|
| Br1 | Br2 | |
| Without metals | ||
| Acetate | 2.0 × 104 (0.4 × 104-9.3 × 104) A | 9.3 × 103 (1.9 × 103-4.2 × 104) A |
| Lactate | 2.0 × 104 (0.4 × 104-9.3 × 104) A | 7.0 × 105 (1.5 × 105-3.3 × 106) A |
| With metals | ||
| Acetate | 4.0 × 104 (0.8 × 104-1.9 × 105) A | 4.0 × 103 (0.8 × 103-1.9 × 104) A |
| Lactate | 2.0 × 104 (0.4 × 104-9.3 × 104) A | 4.0 × 104 (0.8 × 104-1.9 × 105) B |
Values represent abundances in cells g (fresh weight) of Br1 or Br2 soil−1, determined in triplicate MPN serial dilutions after 5 months of incubation at 16°C. Values in parentheses represent the ranges of MPN values within 95% certainty. Capital letters after values represent significant differences of MPN values between metal treatments for the different soil horizons and electron donors.
dsrAB soil community.
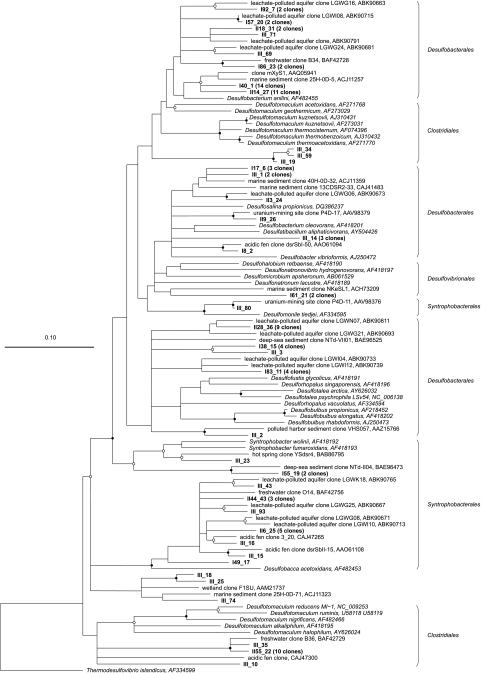
Partial dsrAB sequences were analyzed to identify soil-associated SRB. A clone library was constructed with 109 clones screened, resulting in a coverage of 76%. Sequences grouped into 39 OTUs, and all were closely related to uncultured SRB from pristine environments as well as a uranium mining site, a leachate-polluted aquifer, and sediment from a polluted harbor. dsrAB clones grouped within the families Desulfobacterales (64% of total clones), Desulfovibrionales (2% of total clones), and Syntrophobacterales (17% of total clones) within the Deltaproteobacteria and the Clostridiales (15% of total clones) within the Firmicutes (Fig. 3). The phylogenetic affiliation of a few clones (3% of total clones) remained uncertain (Fig. 3). With dsrAB primers, the non-SRB Carboxydothermus hydrogenoformans, which is known for sulfite reduction (23), was also detected.
FIG. 3.
Phylogenetic tree indicating the relationship of deduced DsrAB amino acid sequences retrieved from Gessen Creek bank soils (indicated by bold type) to those of cultivated sulfate-reducing bacteria and other environmental clone sequences. The tree represents a consensus of the phylogeny determined using neighbor-joining, maximum-parsimony, and maximum-likelihood methods on an alignment of 245 amino acids of the dsrA gene product. Parsimony bootstrap values (1,000 data resamplings) of ≥90% are indicated by closed circles, and values of ≥70% are indicated by open circles. The scale bar indicates the estimated number of amino acid changes per amino acid sequence position.
Reductive redox processes in anoxic soil microcosms.
Microbial activity was stimulated via carbon amendment in four experiments. In all of the microcosms, the pH increased over the incubation period and was slightly higher in biostimulated treatments (pH 7.3) than in the unamended controls (pH 6.6). The redox potential (Eh) declined in the biostimulated microcosms down to −305 mV, whereas changes in Eh were negligible for the controls (data not shown).
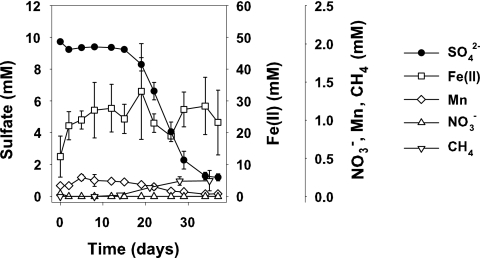
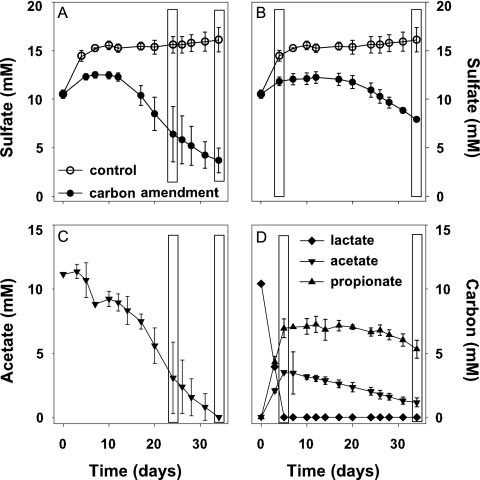
Nitrate was rapidly reduced in all treatments within the first 2 to 4 days with negligible nitrate reduction rates (Fig. 4) due to low in situ concentrations. Manganese was reduced in micromolar-scale range within the first 10 to 16 days in Br1 soil and showed a slight decrease after a plateau phase (Fig. 4). In contrast, manganese concentrations of Br2 microcosms did not change significantly (data not shown). Formation rates of Fe(II) were not consistent throughout the different experiments, but some Fe(III) was reduced within the first 5 to 16 days for both soil horizons in two of four nonbiostimulated and in 5 of 12 biostimulated treatments if the initial Fe(II) soil content was low (Fig. 4 and data not shown). Net sulfate reduction was the dominant TEAP, and trends were similar in all experiments (Fig. 4 and data not shown). Net sulfate reduction began only after a 12- or 17-day lag phase, after slight Fe(III) reduction had ceased (Fig. 4). Due to an accumulation of sulfate in the Br2 control microcosms, a slight decreasing trend in sulfate was observed in the carbon-amended treatments after 5 days (Fig. 5A and B), but depletion was below detection. Kinetics of sulfate reduction were similar for Br1 and Br2 soil, and the highest stimulation was observed with the addition of acetate (rates up to 30.4 μmol g [fresh weight] soil−1 day−1), followed by lactate and ethanol. Methane was formed simultaneously as sulfate concentrations decreased in soil microcosms after 16 to 20 days with rates of ≤0.3 μmol g (fresh weight) soil−1 day−1 (Fig. 4) in acetate- or lactate-amended treatments.
FIG. 4.
Electron-accepting processes represented by the Br1 microcosms amended with acetate. Values are means of triplicates ± standard deviations.
FIG. 5.
Electron acceptor and donor utilization in the SIP microcosms, amended with [13C]acetate (A and C) and [13C]lactate (B and D). Open boxes indicate sampling points for SIP. Values are means of triplicates ± standard deviations.
Characterization of active microbial communities in anoxic soil microcosms.
Addition of supplemental acetate and lactate stimulated SRB best, and consumption rates of these substrates were similar in all microcosm experiments (Fig. 5C and D for the SIP experiment). [13C]acetate was consumed slowly during the incubation, and a near-linear decrease was observed from days 18 to 34 (Fig. 5C). Most of the acetate was consumed concomitantly with sulfate reduction, and all acetate was consumed by day 34 (Fig. 5A and C). In the [13C]lactate-amended microcosms, lactate was consumed by day 6 of the incubation, yielding acetate and propionate as end products (Fig. 5D). Accumulated acetate was consumed slowly and appeared to be in parallel with sulfate reduction (Fig. 5B and D).
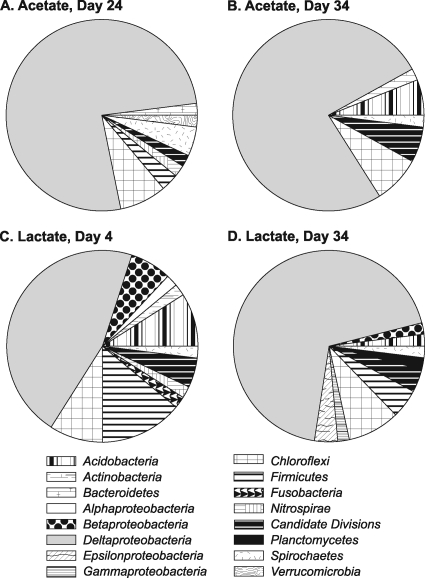
DNA was detected in gradient fractions with buoyant densities ranging from 1.52 to 1.62 g ml−1 (data not shown). The highest-density DNA fractions represented the microbial community with the highest 13C incorporation. Comparison of TRFLP patterns generated for each of the DNA fractions revealed a distinct clustering of the heavy fractions from days 4 and 34 of the [13C]lactate treatment (data not shown). The majority of heavy fractions clustered together for the [13C]acetate treatment; however, we observed no distinct clustering as a function of incubation time. Cloning and sequencing were performed for the heavy fractions of each treatment for each time point. A total of 4 clone libraries were constructed, and 62 and 50 clones were screened for the [13C]acetate day 24 and 34 libraries, respectively, and 56 clones were screened for the [13C]lactate libraries. Rarefaction analysis revealed that the diversity was not exhausted (data not shown). Phylotypes related to members of the Deltaproteobacteria, Chloroflexi, Spirochaetes, and candidate divisions OP8, OP10, and OP11 were detected in all treatments and at all time points. In addition, members of the Actinobacteria, Bacteroidetes, Firmicutes, Acidobacteria, Fusobacteria, Nitrospirae, Planctomycetes, and Verrucomicrobia were detected. Members of the Deltaproteobacteria dominated all clone libraries and represented ∼77% of all clones in the acetate treatment (Fig. 6). In the lactate treatment, the abundance of Deltaproteobacteria-related phylotypes increased from 44 to 70% of the total clones from day 4 to day 34. The majority of Deltaproteobacteria clones detected in all treatments were related to Fe(III)-reducing bacteria within the genus Geobacter (>80% of Deltaproteobacteria clones; data not shown). Additional Deltaproteobacteria clones were related to members of the sulfate-reducing genera Desulfobacca and Desulfocapsa. The increase in Deltaproteobacteria at day 34 in the lactate treatment was due to the appearance of phylotypes related to SRB within the Desulfobacteriaceae, Desulfovibrionaceae, and Desulfobulbaceae families that are known to utilize acetate (10). Phylotypes related to the Chloroflexi phylum were equally abundant at the two time points, representing 8 and 9% of acetate and lactate clone libraries, respectively (Fig. 6). Members of the Firmicutes were detected only at day 24 in the acetate treatment but at both time points in the lactate treatment (Fig. 6). Firmicutes were in higher abundance at day 4 in the lactate treatment (14% of total clones) than at day 34 (5% of total clones). The majority of Firmicutes phylotypes were related to Pelosinus fermentans, which is known to ferment lactate and reduce Fe(III), and to Desulfotalea psychrophila, which reduces sulfate and utilizes lactate as a carbon substrate.
FIG. 6.
Frequencies of active bacterial phylogenetic lineages detected in SSU rRNA gene clone libraries from [13C]acetate (A and B)- and [13C]lactate (C and D)-amended microcosms. Calculations were made based on the total number of clones associated with phylotypes of sequenced representatives.
Effect of anoxic incubation on dissolved metal concentrations.
Surprisingly, uranium was released to solution at the end of incubation in all carbon-amended treatments (Table 3 and data not shown). Concentrations of U reached up to 1,407 nM in the Br2 soil suspension amended with acetate (235-fold increase) and 165 nM in the Br1 soil suspension amended with lactate (6.6-fold increase). Nickel and cobalt were immobilized in all treatments, with up to 100% of the soluble metals removed from solution (Table 3 and data not shown). The addition of carbon led to a 3.0-fold-higher reduction in nickel concentrations at the end of the incubation. Similarly, cobalt concentrations were reduced to a 2.1-fold-higher extent than in the unamended controls. The dynamics of soluble zinc were not consistent among the treatments and experiments. A decline in soluble zinc, up to 95% of the starting concentration, was observed in soil microcosms for the Br1 and Br2 horizons (Table 3 and data not shown), whereas the soluble zinc concentrations in the second Br1 experiment declined in a similar manner for both the control and carbon-amended treatments (Table 3). The low soluble copper concentrations were constant or even declined in Br1 soil microcosms (Table 3 and data not shown). While little to no change in soluble arsenic concentrations was observed in the second Br1 experiment, arsenic concentrations were up to 5.7-fold higher at the end of incubation in the other microcosm experiments (Table 3).
TABLE 3.
Soluble metal concentration at the beginning (T0) and end (Tend) of incubation of Br1 and Br2 soil suspensions
| Expt | Soluble metal concn (nM)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U |
Ni |
Co |
Zn |
Cu |
As |
|||||||
| Ctr | Ace | Ctr | Ace | Ctr | Ace | Ctr | Ace | Ctr | Ace | Ctr | Ace | |
| Br1 | ||||||||||||
| T0 | 9 ± 3 | 14 ± 8 | 538 ± 393 | 508 ± 215 | 188 ± 128 | 194 ± 47 | 266 ± 79 | 196 ± 31 | 22 ± 6 | 17 ± 6 | 520 ± 46 | 365 ± 46 |
| Tend | 35 ± 13 | 148 ± 48 | 91 ± 19 | 40 ± 8 | 32 ± 8 | 8 ± 0 | 118 ± 23 | 78 ± 9 | 5 ± 4 | 26 ± 3 | 498 ± 20 | 190 ± 57 |
| Br2 | ||||||||||||
| T0 | 1 ± 0 | 1 ± 0 | 422 ± 46 | 422 ± 46 | 92 ± 8 | 92 ± 8 | 277 ± 309 | 277 ± 309 | 5 ± 2 | 5 ± 2 | 417 ± 30 | 417 ± 30 |
| Tend | 9 ± 2 | 256 ± 60 | 71 ± 10 | 42 ± 4 | 18 ± 1 | 6 ± 1 | 53 ± 14 | 36 ± 36 | 10 ± 2 | 16 ± 2 | 1,444 ± 38 | 1,629 ± 76 |
| Br2 (SIP)b | ||||||||||||
| T0 | 6 ± 1 | 6 ± 1 | 1,210 ± 402 | 1,210 ± 402 | 133 ± 67 | 133 ± 67 | 34 ± 33 | 34 ± 33 | 31 ± 0 | 31 ± 0 | 343 ± 37 | 343 ± 37 |
| Tend | 8 ± 1 | 1,407 ± 332 | 839 ± 156 | 69 ± 6 | 74 ± 8 | 9 ± 1 | 234 ± 102 | 15 ± 0 | 31 ± 0 | 85 ± 15 | 248 ± 18 | 560 ± 81 |
Values represent soluble average metal concentrations (nM) of Br1 or Br2 soil suspension at the beginning (T0) and end (Tend) of the microcosm experiments (n = 3) treated as a control (Ctr) or amended with acetate (Ace).
Second experiment with soil of Br2 horizon.
DISCUSSION
Ongoing anaerobic microbial activities in Gessen Creek bank soil.
Pore water profiles suggested that reduction of nitrate occurred mainly in the upper oxidized horizons, whereas soluble manganese and Fe(II) increased primarily in the reduced soil horizons Br1 and Br2. These horizons contained the highest concentrations of sulfate, which were highly variable with time. The accumulation of acid volatile sulfur and high in situ sulfate reduction rates in Br1 and Br2 soils indicated ongoing dissimilatory sulfate reduction in the creek bank soil. The in situ sulfate-reducing activity was slightly higher than what was previously found in contaminated soils and sediments of the Norilsk mining area (29) and was in the range of rates reported from unpolluted freshwater (25, 70) or marine (32, 57) ecosystems. A long, heavy rainfall prior to our measurements diluted sulfate concentrations, oxygenated the soil, and was likely responsible for the lower reduction rates observed in April 2008.
The detection of a diverse sulfate-reducing community via analysis of the functional marker gene dsrAB in the Br2 soil horizon supported our findings of ongoing sulfate reduction. The majority of clones were related to the Desulfobacterales, and the closest relatives of our clones were freshwater-associated SRB that were found in uranium mining tailings (11), metal-contaminated aquifers (21), or intertidal river soil (47). SRB abundance was within the same range as that in samples from other uranium-contaminated subsurface sediments (48), deciduous forest soils (59), or lake sediments (35, 38). Abundances of resident SRB were only slightly inhibited in the presence of metals at maximum in situ pore water concentrations. This indicates that the resident, cultivatable SRB community is adapted to the level of metal stress in the Gessen Creek bank soils.
Microbial activities during anoxic soil incubation.
Sulfate reduction was an important TEAP in biostimulated, anoxic microcosms after 12 to 17 days of incubation, with amendment by acetate and lactate resulting in the highest sulfate reduction rates. In situ SRR were in the nanomole range, suggesting that sulfate reducers were active at a low level also during the first days of incubation but masked due to the high sulfate concentrations in the treatments. The increase in sulfate concentrations that was observed in the Br2 control microcosms at the beginning of incubation may have been caused by the desorption of sulfate bound or adsorbed to Fe(III)-(hydr)oxides (56). Because nitrate and manganese were present at low concentrations, nitrate reduction and manganese reduction were likely not substantial energy-generating processes. Microbial Fe(III) reduction was a significant TEAP next to sulfate reduction, although not steadily since Fe(II) formation rates were low and highly variable among treatments and replicates. However, Fe(II) may also have been formed from abiotic reduction coupled to sulfide oxidation, since a high reduced sulfur content was present in the Br2 soil. Although we did not detect significantly higher Fe(II)-forming activities compared to the unamended control (data not shown), Geobacter was active in the lactate- and acetate-amended treatments. In Btlc soil, stimulated Fe(III) reduction yielded the presence of the genera Geobacter, Geothrix, and Pelosinus (6). This observation was similar to previous field and laboratory experiments at other uranium-contaminated sites, where Fe(III)- and metal-reducing bacteria were of great importance for bioremediation (26, 27).
Active biostimulated sulfate-reducing bacteria were shown to be related to SRB known for complete or incomplete acetate utilization (10) and the capability of oxidizing propionate, which was observed in [13C]lactate treatments (68). Desulfobacca acetoxidans- and Desulfocapsa thiozymogenes-related clones have been previously isolated from uranium and Pb/Zn mining sites (11, 62), indicating the presence of two metal- and radionuclide-tolerant species. The high abundance of Geobacter spp. found in the active communities may also have been involved in reduction of humic substances and elemental sulfur (40, 61), since soil showed high carbon and sulfur contents.
Metal retention potential.
An accumulation of metals in the solid phase was not associated with high soluble concentrations of metals, indicating an additional capacity for metal attenuation for the inherent soil-associated microorganisms and/or different soil properties. A decrease in soluble nickel and cobalt was observed independently of biostimulation, as expected based on previous work on sulfate reduction in batch experiments (7, 33). It is hypothesized that the decrease in nickel and cobalt was related to the formation of metal sulfides in the soil incubation experiments. Therefore, ongoing, in situ sulfate-reducing activity would be sufficient to precipitate nickel and cobalt as sulfides (16, 33). However, passive mechanisms, such as metal binding to sites on bacterial cell surfaces and to metal-complexing groups of extrapolymeric substances (20), cannot be ruled out. Metal dynamics for zinc, copper, and arsenic were not uniform throughout the anoxic incubations of the soil and were therefore in contrast to previous work that showed metal sulfide formation with these cations and anions under sulfate-reducing conditions (39, 50, 60). As(VI)-reducing bacteria, such as Desulfosporosinus auripigmenti, likely contributed to the observed increase in soluble arsenic (51). Therefore, sulfate-reducing bacteria have the potential to attenuate metal cations in the investigated field site, which was indicated by the metal geochemistry.
In contrast to what other studies have shown (e.g., references 1 and 71), we observed that soluble uranium concentrations increased under anoxic, sulfate-reducing conditions. This was unexpected because anaerobic metabolism of bacteria has been shown to promote reduction in soluble U by indirect and direct mechanisms (5, 42, 45). However, as was seen in this study, an increase in uranium concentration was previously observed during sulfate reduction in uranium-contaminated aquifers (3) or in pure cultures of Desulfovibrio desulfuricans G20 (58). It is also possible that bacteria from the soil produced siderophores, thereby promoting the dissolution of UO2 (18), or that microbially mediated formation of carbonates could have resulted in highly stable carbonate-U(VI) complexes (67). However, carbonate-U(VI) complexes are unlikely to have been formed, due to a lack of carbonates in the soil, and also XANES data did not indicate the presence of carbonate-U(VI) complexes in the soil. Surface-bound uranyl ions [U(IV)], ∼20% of the uranium in the soil solid phase, may have been detached from Fe(III)-mineral phases, which have an affinity for uranium (49) and which were bacterially reduced upon amendment of the microcosms. For example, the Fe(III)-mineral phase illite was likely to be present in Br1 and Br2 and could provide sorption surfaces for hexavalent actinides (30).
Conclusions.
Active SRB were identified in reduced soils within the bank of Gessen Creek in the former uranium mining site of Ronneburg. SRB were shown to be adapted to the presence of metals and radionuclides and to influence the retention of contaminants. In particular, nickel and cobalt, but occasionally zinc and copper, were retained during sulfate reduction, indicating that soil-associated SRB contributed to in situ metal attenuation, which could explain the high solid metal contents and the low concentrations of metals in the pore water. Uranium was released during anaerobic microbial activities acting as potential sources of contaminants for downstream ecosystems. The increase in soluble uranium concentrations is in contrast to what has been seen during biostimulation of iron- and sulfate-reducing bacteria at other uranium-contaminated sites (3, 27). Our results show that site-specific geochemistry and variable in situ microbial communities can affect the success of biostimulation as a strategy for the enhancement of metal retention. In reduced soils of the Gessen Creek bank, ongoing sulfate reduction is providing a means for natural attenuation of nickel and cobalt contaminants but does not lessen the risk of downstream uranium contamination.
Acknowledgments
This project is part of the graduate research school Alteration and Element Mobility at the Microbe-Mineral Interface, financially supported by the German Research Foundation DFG (1257).
We thank T. Wiegers, W. Fischer, and S. Löffler for technical assistance and T. Pfannschmidt (Institute of Plant Physiology, FSU Jena) for providing access to a radioisotope laboratory during the start of the SRR measurements.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Abdelouas, A., W. Lutze, W. Gong, E. H. Nuttall, B. A. Strietelmeier, and B. J. Travis. 2000. Biological reduction of uranium in groundwater and subsurface soil. Sci. Total Environ. 250:21-35. [DOI] [PubMed] [Google Scholar]
- 2.Alef, K. 1991. Methodenhandbuch der Bodenmikrobiologie. Aktivitäten, Biomasse, Differenzierung. Ecomed, Landsberg, Germany.
- 3.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyenal, H., R. K. Sani, B. M. Peyton, A. C. Dohnalkova, J. E. Amonette, and Z. Lewandowski. 2004. Uranium immobilization by sulfate-reducing biofilms. Environ. Sci. Technol. 38:2067-2074. [DOI] [PubMed] [Google Scholar]
- 5.Birch, L., and R. Bachofen. 1990. Complexing agents from microorganisms. Experientia 46:827-834. [Google Scholar]
- 6.Burkhardt, E., D. M. Akob, S. Bischoff, J. Sitte, J. E. Kostka, D. Banerjee, A. C. Scheinost, and K. Küsel. 2010. Impact of biostimulated redox processes on metal dynamics in an iron-rich creek soil of a former uranium mining area. Environ. Sci. Technol. 44:177-183. [DOI] [PubMed] [Google Scholar]
- 7.Cabrera, G., R. Perez, J. Gomez, A. Abalos, and D. Cantero. 2006. Toxic effects of dissolved heavy metals on Desulfovibrio vulgaris and Desulfovibrio sp. strains. J. Hazard. Mater. 135:40-46. [DOI] [PubMed] [Google Scholar]
- 8.Canfield, D. E., and D. J. Des Marais. 1991. Aerobic sulfate reduction in microbial mats. Science 251:1471-1473. [DOI] [PubMed] [Google Scholar]
- 9.Carlsson, E., and G. Büchel. 2005. Screening of residual contamination at a former uranium heap leaching site, Thuringia, Germany. Chem. Erde Geochem. 65:75-95. [Google Scholar]
- 10.Castro, H. F., N. H. Williams, and A. Ogram. 2000. Phylogeny of sulfate-reducing bacteria. FEMS Microbiol. Ecol. 31:1-9. [DOI] [PubMed] [Google Scholar]
- 11.Chang, Y., A. D. Peacook, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. M. A. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 13.de Man, J. C. 1983. MPN tables, corrected. Eur. J. Appl. Microbiol. Biotechnol. 17:301-305. [Google Scholar]
- 14.de Man, J. C. 1975. The probability of most probable numbers. Eur. J. Appl. Microbiol. 1:67-78. [Google Scholar]
- 15.Ehrlich, H. L. 1999. Microbes as geologic agents: their role in mineral formation. Geomicrobiol. J. 16:135-153. [Google Scholar]
- 16.Fortin, D., G. Southam, and T. J. Beveridge. 1994. Nickel sulfide, iron-nickel sulfide and iron sulfide precipitation by a newly isolated Desulfotomaculum species and its relation to nickel resistance. FEMS Microbiol. Ecol. 14:121-132. [Google Scholar]
- 17.Fossing, H., and B. B. Jørgensen. 1989. Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8:205-2088. [Google Scholar]
- 18.Frazier, S. W., R. Kretzschmar, and S. M. Kraemer. 2005. Bacterial siderophores promote dissolution of UO2 under reducing conditions. Environ. Sci. Technol. 39:5709-5715. [DOI] [PubMed] [Google Scholar]
- 19.Fude, L., B. Harris, M. M. Urrutia, and T. J. Beveridge. 1994. Reduction of Cr(VI) by a consortium of sulfate-reducing bacteria (SRB III). Appl. Environ. Microbiol. 60:1525-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadd, G. M. 2004. Microbial influence on metal mobility and application for bioremediation. Geoderma 122:109-119. [Google Scholar]
- 21.Geets, J., B. Borremans, L. Diels, D. Springael, J. Vangronsveld, D. van der Lelie, and K. Vanbroekhoven. 2006. DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J. Microbiol. Methods 66:194-205. [DOI] [PubMed] [Google Scholar]
- 22.Heck, K. L., G. van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 23.Henstra, A. M., and A. J. M. Stams. 2004. Novel physiological features of Carboxydothermus hydrogenoformans and Thermoterrabacterium ferrireducens. Appl. Environ. Microbiol. 70:7236-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herlihy, A. T., and A. L. Mills. 1985. Sulfate reduction in freshwater sediments receiving acid mine drainage. Appl. Environ. Microbiol. 49:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmer, M., and P. Storkholm. 2001. Sulphate reduction and sulphur cycling in lake sediments: a review. Freshw. Biol. 46:431-451. [Google Scholar]
- 26.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, C., W. Wu, T. Gentry, J. Carley, G. Corbin, S. Carroll, D. B. Watson, P. Jardine, J. Zhou, C. S. Criddle, and M. Fields. 2009. Bacterial community succession during in situ uranium bioremediation: spatial similarities along controlled flow paths. ISME J. 3:47-64. [DOI] [PubMed] [Google Scholar]
- 28.Jakubick, A., R. Gatzweile, D. Mager, A. Mac, and G. Robertson. 1997. Abstr. 4th Int. Conf. Acid Rock Drainage, Vancouver, BC, Canada, p. 1285-1301.
- 29.Karnachuk, O., N. Pimenov, S. Yusupov, Y. Frank, A. Kaksonen, J. Puhakka, M. Ivanov, E. Lindström, and O. Tuovinen. 2005. Sulfate reduction potential in sediments in the Norilsk mining area, northern Siberia. Geomicrobiol. J. 22:11-25. [Google Scholar]
- 30.Kim, J. I., and B. Grambow. 1999. Geochemical assessment of actinide isolation in a German salt repository environment. Eng. Geol. 52:221-230. [Google Scholar]
- 31.Kostka, J., and K. H. Nealson. 1998. Isolation, cultivation and characterization of iron- and manganese-reducing bacteria, p. 58-78. In R. S. Burlage, R. Atlas, D. Stahl, G. Geesey, and G. Sayler (ed.), Techniques in microbial ecology. Oxford University Press, New York, NY.
- 32.Kristensen, E., J. Bodenbender, M. H. Jensen, H. Rennenberg, and K. M. Jensen. 2000. Sulfur cycling of intertidal Wadden Sea sediments (Konigshafen, Island of Sylt, Germany): sulfate reduction and sulfur gas emission. J. Sea Res. 43:93-104. [Google Scholar]
- 33.Krumholz, L. R., D. A. Elias, and J. M. Suflita. 2003. Immobilization of cobalt by sulfate-reducing bacteria in subsurface sediments. Geomicrobiol. J. 20:61-72. [Google Scholar]
- 34.Kurtz, D. M., and E. D. Coulter. 2002. The mechanism(s) of superoxide reduction by superoxide reductases in vitro and in vivo. J. Biol. Inorg. Chem. 7:653-658. [DOI] [PubMed] [Google Scholar]
- 35.Küsel, K., and T. Dorsch. 2000. Effect of supplemental electron donors on the microbial reduction of Fe(III), sulfate, and CO2 in coal mining-impacted freshwater lake sediments. Microb. Ecol. 40:238-249. [DOI] [PubMed] [Google Scholar]
- 36.Küsel, K., and H. L. Drake. 1995. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl. Environ. Microbiol. 61:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Küsel, K., E. Ewald, and J. Sitte. 2008. Effect of metal-reducing microorganisms on element fluxes in a former uranium-mining district, p. 128-137. In S. J. Liu and H. L. Drake (ed.), Microbes in the environment: perspectives and challenge. Science Press, Beijing, China.
- 38.Küsel, K. A., U. Roth, T. Trinkwalter, and S. Peiffer. 2001. Effect of pH on the anaerobic microbial cycling of sulfur in mining-impacted freshwater lake sediments. Environ. Exp. Bot. 46:213-223. [Google Scholar]
- 39.Labrenz, M., G. K. Druschel, T. Thomsen-Ebert, B. Gilbert, S. A. Welch, K. M. Kemner, G. A. Logan, R. E. Summons, G. De Stasio, P. L. Bond, B. Lai, S. D. Kelly, and J. F. Banfield. 2000. Formation of sphalerite (ZnS) deposits in natural biofilms of sulfate-reducing bacteria. Science 290:1744-1747. [DOI] [PubMed] [Google Scholar]
- 40.Lovley, D. R., J. Coates, E. L. Blunt-Harris, E. J. P. Phillips, and J. C. Woodward. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445-448. [Google Scholar]
- 41.Lovley, D. R., and E. J. P. Phillips. 1994. Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome. Appl. Environ. Microbiol. 60:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lovley, D. R., and E. J. P. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loy, A., S. Duller, C. Baranyi, M. Mußmann, J. Ott, I. Sharon, O. Béjà, D. Le Paslier, C. Dahl, and M. Wagner. 2009. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 11:289-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig, W., O. Strunk, R. Westram, L. Richter, F. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Stehlow, A. Stamatakis, M. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macaskie, L., J. D. Blackmore, and R. M. Empson. 1988. Phosphates overproduction and enhanced uranium accumulation by a stable mutant of a Citrobacter sp. isolated by a novel method. FEMS Microbiol. Lett. 55:157-161. [Google Scholar]
- 46.Michalsen, M. M., B. A. Goodman, S. D. Kelly, K. M. Kemner, J. P. McKinley, J. W. Stucki, and J. D. Istok. 2006. Uranium and technetium bio-immobilization in intermediate-scale physical models of an in situ bio-barrier. Environ. Sci. Technol. 40:7048-7053. [DOI] [PubMed] [Google Scholar]
- 47.Miletto, M., P. Bodelier, and H. Laanbroek. 2007. Improved PCR-DGGE for high resolution diversity screening of complex sulfate-reducing prokaryotic communities in soils and sediments. J. Microbiol. Methods 70:103-111. [DOI] [PubMed] [Google Scholar]
- 48.Moreels, D., G. Crosson, C. Garafola, D. Monteleone, S. Taghavi, J. Fitts, and D. Lelie. 2008. Microbial community dynamics in uranium contaminated subsurface sediments under biostimulated conditions with high nitrate and nickel pressure. Environ. Sci. Pollut. Res. 15:481-491. [DOI] [PubMed] [Google Scholar]
- 49.Moyes, L. N., R. H. Parkman, J. M. Charnock, D. J. Vaughan, F. R. Livens, C. R. Hughes, and A. Braithwaite. 2000. Uranium uptake from aqueous solution by interaction with goethite, lepidocrocite, muscovite, and mackinawite: an X-ray absorption spectroscopy study. Environ. Sci. Technol. 34:1062-1068. [DOI] [PubMed] [Google Scholar]
- 50.Newman, D. K., T. J. Beveridge, and F. M. M. Morel. 1997. Precipitation of arsenic trisulfide by Desulfotomaculum auripigmentum. Appl. Environ. Microbiol. 63:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman, D. K., E. K. Kennedy, J. D. Coates, D. Ahmann, D. J. Ellis, D. R. Lovley, and F. M. M. Morel. 1997. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch. Microbiol. 168:380-388. [DOI] [PubMed] [Google Scholar]
- 52.Picardal, F., and D. C. Cooper. 2005. Microbially mediated changes in the mobility of contaminant metals in soils and sediment, p. 43-88. In I. Ahmad, S. Hayat, and J. Pichtel (ed.), Heavy metal contamination of soil: problems and remedies. Science Publishers, Enfield, NH.
- 53.Rabus, R., T. A. Hansen, and F. Widdel. 2006. Dissimilatory sulfate- and sulfur-reducing prokaryotes. Prokaryotes 2:659-768. [Google Scholar]
- 54.Reiche, M., G. Torburg, and K. Küsel. 2008. Competition of Fe(III) reduction and methanogenesis in an acidic fen. FEMS Microbiol. Ecol. 65:88-101. [DOI] [PubMed] [Google Scholar]
- 55.Riley, R. G., and J. Zachara. 1992. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface research. U.S. Department of Energy, Washington, DC.
- 56.Rose, S., and A. M. Ghazi. 1997. Release of sorbed sulfate from iron oxyhydroxides precipitated from acid mine drainage associated with coal mining. Environ. Sci. Technol. 31:2136-2140. [Google Scholar]
- 57.Sahm, K., B. J. MacGregor, B. B. Jørgensen, and D. A. Stahl. 1999. Sulphate reduction and vertical distribution of sulphate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ. Microbiol. 1:65-74. [DOI] [PubMed] [Google Scholar]
- 58.Sani, R. K., B. M. Peyton, A. Dohnalkova, and J. E. Amonette. 2005. Reoxidation of reduced uranium with iron(III) (hydr)oxides under sulfate-reducing conditions. Environ. Sci. Technol. 39:2059-2066. [DOI] [PubMed] [Google Scholar]
- 59.Schnurr-Pütz, S., E. Bååth, G. Guggenberger, H. Drake, and K. Küsel. 2006. Compaction of forest soil by logging machinery favours occurrence of prokaryotes. FEMS Microbiol. Ecol. 58:503-516. [DOI] [PubMed] [Google Scholar]
- 60.Southam, G. 2000. Bacterial surface-mediated mineral formation, p. 257-276. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washington, DC.
- 61.Straub, K. L., and B. E. E. Buchholz-Cleven. 2001. Geobacter bremensis sp. nov. and Geobacter pelophilus sp. nov., two dissimilatory ferric-iron-reducing bacteria. Int. J. Syst. Evol. Microbiol. 51:1805-1808. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2005. Direct microbial reduction and subsequent preservation of uranium in natural near-surface sediment. Appl. Environ. Microbiol. 71:1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl. Environ. Microbiol. 69:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tabatabai, M. A. 1974. Determination of sulfate in water samples. Sulphur Inst. J. 10:11-13. [Google Scholar]
- 65.Wagner, M., A. Loy, M. Klein, N. Lee, N. B. Ramsing, D. A. Stahl, and M. Friedrich. 2005. Functional marker genes for identification of sulfate-reducing prokaryotes. Nucleic Acids Res. 29:469-489. [DOI] [PubMed] [Google Scholar]
- 66.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wall, J. D., and L. R. Krumholz. 2006. Uranium reduction. Annu. Rev. Microbiol. 60:149-166. [DOI] [PubMed] [Google Scholar]
- 68.Widdel, F., and N. Pfennig. 1982. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch. Microbiol. 131:360-365. [DOI] [PubMed] [Google Scholar]
- 69.Wielinga, B., J. K. Lucy, J. N. Moore, O. F. Seastone, and J. E. Gannon. 1999. Microbiological and geochemical characterization of fluvially deposited sulfidic mine tailings. Appl. Environ. Microbiol. 65:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wind, T., and R. Conrad. 1997. Localization of sulfate reduction in planted and unplanted rice field soil. Biogeochemistry 37:253-278. [Google Scholar]
- 71.Wu, W., J. Carley, J. Luo, M. A. Ginder-Vogel, E. Cardenas, M. B. Leigh, C. Hwang, S. D. Kelly, C. Ruan, L. Wu, J. Van Nostrand, T. Gentry, K. Lowe, T. Mehlhorn, S. Carroll, W. Luo, M. Fields, B. Gu, D. Watson, K. M. Kemner, T. Marsh, J. Tiedje, J. Zhou, S. Fendorf, P. K. Kitanidis, P. M. Jardine, and C. S. Criddle. 2007. In situ bioreduction of uranium (VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ. Sci. Technol. 41:5716-5723. [DOI] [PubMed] [Google Scholar]