Abstract
This study describes refined electroporation parameters for efficient transformation of Bacteroides fragilis by plasmids prepared from laboratory strains of Escherichia coli. Development of the method used included determination of the optimal growth conditions for competent cell preparation, selectable antimicrobial resistance markers, electric field strength, and postpulse incubation time. Of the four E. coli-Bacteroides shuttle plasmids tested (pVAL-1, pVAL-2, pNLY1, and pLYL05), pLYL05 containing the cefoxitin resistance marker was found to be the most suitable for B. fragilis transformation, and it generated 2- to 900-fold more transformants (about 104 transformants per μg pLYL05 DNA) than the other plasmids. For the 72-h cultivation period tested, B. fragilis cells harvested at 48 h yielded the highest numbers of transformants. The transformation efficiency of pLYL05 increased linearly with the electric field strength over a range from 5.0 to 12.5 kV/cm. At least 3 h of postpulse incubation was required to maximize the transformation efficiency. For deletion of B. fragilis genes by homologous recombination, competent cells grown to early exponential phase and 12 h of postpulse incubation were required for efficient integration of the pLYL05-based suicide vector into the target site. The expected integration was obtained in B. fragilis strain NCTC9343 only when a homologously prepared (i.e., in vivo methylated) suicide vector was used. Spontaneous resolution of the diploid successfully deleted the expected genetic region. Our simple and efficient plasmid transfer method enabled disruption of a B. fragilis gene using in vivo-methylated targeted vectors. Our optimized electroporation parameters provide a useful tool for genetic manipulation of Bacteroides species.
Numerous microbes inhabit the human intestine, and the number of microbial cells can be nearly 1011 cells per g of feces. The colon is the most densely populated environment in the human body, and the genus Bacteroides, which contains Gram-negative, obligate anaerobes, is one of the most abundant genera in the intestinal microbiota. In the clinical setting, Bacteroides strains are considered opportunistic pathogens that can occasionally cause infections, such as peritoneal abscesses, appendicitis, and septicemia (3, 4, 23). In the genus Bacteroides, B. fragilis is considered the most virulent species, and its capsular polysaccharides are especially linked to its pathogenesis (8, 13, 22). However, recent reports have highlighted the symbiotic properties of B. fragilis since the capsular polysaccharide displayed by this species can modify the human immune system (21). Another example of a host-Bacteroides interaction is a B. thetaiotaomicron interaction; this species can repress host inflammatory responses by promoting the binding of peroxisome proliferator-activated receptor gamma (PPAR-γ) to the NFκ-b subunit RelA (11). B. thetaiotaomicron is also known to induce production of antimicrobial peptides from Paneth cells in host intestinal epithelia (10).
The recent accumulation of genomic information about several Bacteroides species (2) has enabled us to search for the genes responsible for the symbiotic properties of intestinal Bacteroides described above. However, screening for genes involved in host-Bacteroides symbioses requires development of a suitable, simple, and efficient genetic manipulation system. Although transconjugation by filter mating can be performed with Bacteroides species (19, 24 ), this technique is labor-intensive and time-consuming. Electroporation is a simple and reproducible procedure that is widely used for introduction of foreign nucleic acids into various bacteria. While Smith et al. previously described an electroporation protocol for Bacteroides species that could transform Bacteroides with plasmids from homologous hosts, plasmids derived from Escherichia coli laboratory strains could be transformed into Bacteroides only with difficulty (20). Recently, Patrick et al. described an efficient electrotransformation method for B. fragilis using purified T7 antirestriction protein Ocr (15). While these workers also demonstrated that there was genetic disruption in B. fragilis strain NCTC9343, a transconjugation system was employed to integrate the suicide plasmid into the chromosome.
In the present study, we refined the electrotransformation method for Bacteroides species whose genomes have been sequenced, especially B. fragilis, that allows highly efficient transformation even for plasmids purified from E. coli laboratory strains. In addition, we performed deletion mutagenesis of a B. fragilis gene using an in vivo-methylated suicide vector introduced into the host strain by electroporation rather than by a conjugal DNA transfer system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. E. coli strains were grown aerobically in Luria-Bertani medium (LB) at 37°C. Bacteroides strains were grown anaerobically in Gifu anaerobic medium (GAM) (Nissui Pharmaceutical Co., Tokyo, Japan) at 37°C using the AnaeroPack system (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan). If necessary, antibiotics were added to the media at the following concentrations: ampicillin (Amp), 50 μg/ml; cefoxitin (Cfx), 50 μg/ml; chloramphenicol (Cm), 15 μg/ml; erythromycin (Em), 10 μg/ml; and tetracycline (Tc), 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| HB101 | supE44 Δ(mcrC-mrr) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1 | 14 |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| Bacteroides strains | ||
| B. fragilis NCTC9343 | Type strain, appendix abscess | NCTC |
| B. fragilis YCH46 | Clinical isolate, blood | 12 |
| B. thetaiotaomicron VPI-5482 | Human feces | VPI Anaerobe Laboratory |
| B. vulgatus ATCC8482 | Human feces | ATCC |
| Plasmids | ||
| pBlueScript KS II(+) | Cloning vector, Apr | Stratagene |
| pVAL-1 | Apr Tcr in E. coli, Emr in Bacteroides; Mob+ Rep+ | 24 |
| pVAL-2 | Apr in E. coli, Cfxr in Bacteroides; Mob+ Rep+ | This study |
| pLYL05 | Apr in E. coli, Cfxr in Bacteroides; Mob+ Rep+ | 18 |
| pNLY1 | Apr in E. coli, Cmr in Bacteroides; Mob+ Rep+ | 18 |
| pLYL0519 | Flanking region (2 kb each) of Tsr19 gene deletion site was cloned into pLYL05; Cfxr Mob+ Rep+ | This study |
| pLYL0520 | Targeting vector for deletion of Tsr19 gene in strain NCTC9343 | This study |
Abbreviations: Ap, ampicillin; Tc, tetracycline; Em, erythromycin; Cfx, cefoxitin; Mob, ability to be mobilized by a conjugative element; Rep, ability to replicate in Bacteroides.
Plasmid construction.
Plasmids used in this study are listed in Table 1. Three E. coli-Bacteroides shuttle vectors were kindly provided by Nadja B. Shoemaker. These plasmids were purified from E. coli host strains using a Hispeed plasmid midi kit (Qiagen) and were used for electrotransformation of Bacteroides strains. To change the selective marker of pVAL-1 from Emr to Cfxr, the Cfxr gene on pLYL05 was amplified with primers cfxA-F (5′-AAAATCAGTTCTTTAGCGA-3′) and cfxA-R (5′-ACACAGGCGGAACTTTGATA-3′). The resultant PCR product and the EcoRI/AflII fragment from pVAL-1 were purified, blunt ended, and phosphorylated using the blunting-kination enzyme mixture (TAKARA Co. Ltd., Otsu, Japan). These fragments were ligated together to construct pVAL-2.
For B. fragilis gene disruption, we constructed a suicide plasmid based on pLYL05. We selected the Tsr19 gene of B. fragilis strain NCTC9343 as the target for genetic deletion. This gene encodes a master tyrosine recombinase that mediates promoter inversions at two distant loci that are associated with the large encapsulation phenotype (6, 15, 17). Upstream and downstream DNA fragments (2 kb each) flanking the region deleted were amplified separately using primers H1 (5′-AATCCGGCTCCTGAGTAATCTC-3′) and H2 (5′-GTATACCCAGTGTGTTCAAGAACACGG-3′) and primers F1 (5′-AACACACTGGGTATACAACAAACGCCTC-3′) and F2 (5′-ATAATTGGCCATGCGAACCG-3′), respectively. These fragments were fused together by performing a second amplification with primers H1 and F2 through an overlapping region inserted into the sequences of primers H2 and F1 (underlined). The products obtained were phosphorylated at both ends and ligated into the blunt-ended PstI site of pLYL05. The ligated DNA was introduced into E. coli strain DH5α, generating pLYL0519. The pLYL0519 plasmid was digested by XbaI to remove a 2.7-kb fragment containing the Bacteroides replication origin. After separation of the XbaI-digested fragments on a 0.5% agarose gel, an 8.85-kb fragment was recovered using a QIAquick gel extraction kit (Qiagen) and self-ligated with T4 DNA ligase (Promega) to construct pLYL0520EC.
Ligated DNA was then purified using a QIAquick PCR purification kit (Qiagen), eluted into sterilized deionized water, and electroporated into B. fragilis strain NCTC9343 cells. E. coli strain DH5α was also transformed with the ligated DNA and plasmid pLYL0520EC purified using a Hispeed plasmid midi kit (Qiagen).
For in vivo methylation of the suicide plasmid, pLYL0519 was first introduced into B. fragilis strain NCTC9343 competent cells (prepared from a 48-h culture) by electroporation. Following purification of pLYL0519 from B. fragilis NCTC9343 cells, the Bacteroides replication origin was removed by XbaI digestion as described above, generating homologously modified suicide plasmid pLYL0520BF.
Optimization of the electroporation parameters for B. fragilis. (i) Plasmid selection.
Competent cells for electrotransformation were prepared as follows. A single colony of B. fragilis strain NCTC9343 or YCH46 grown on GAM agar plates was inoculated into 4 ml GAM broth and grown anaerobically overnight. Part of the overnight culture (0.1 ml) was inoculated into 10 ml freshly prepared GAM broth and incubated for 3.5 to 4.5 h until the optical density at 660 nm (OD660) reached 0.4 to 0.6. One milliliter of this culture was inoculated into 100-ml freshly prepared GAM broth and incubated. Cells were then harvested by centrifugation at 6,000 × g for 10 min at 4°C when the OD660 reached 0.4 to 0.6. Cell pellets were washed twice with 100 ml ice-cold 10% (wt/vol) glycerol. After centrifugation, cells were resuspended in 1.0 ml 10% glycerol.
Standard electroporation was performed as follows. To avoid arcing during the electric pulse, we performed electroporation in a room kept at 18°C and prepared input DNA solutions whose volumes were less than 10% of the volume of the competent cell solution. Aliquots (1 μg) of pVAL-1, pVAL-2, pNLY1, or pLYL05 DNA were mixed with 100 μl competent cells. The mixtures were then transferred into electroporation cuvettes purchased from Bio-Rad Laboratories (interelectrode distance, 0.2 cm), which were then electrically pulsed using the Gene Pulser II system (Bio-Rad Laboratories) and the following parameters: 12.5 kV/cm, 200 Ω, and 25 μF. We sometimes encountered arcing problems when electroporation cuvettes from other sources were used. Immediately after the electric pulse, 0.9 ml prewarmed GAM broth was added to each sample and incubated anaerobically for 12 h at 37°C. Culture aliquots were spread onto GAM agar plates containing the selective antibiotic corresponding to the plasmid used, and the plates were incubated anaerobically at 37°C for 48 h to recover the transformants. The plasmid that yielded the highest number of transformants was used to optimize the other parameters described below.
(ii) Growth phase.
Competent cells used for electroporation were prepared from cultures at various phases, from the early exponential phase (OD660, 0.2) to the stationary phase (OD660, 1.8). The transformation efficiencies for the culture conditions were compared to determine the optimal growth conditions for preparation of competent cells. Competent cells were divided into 100-μl aliquots, frozen in a dry ice-ethanol bath, and stored at −70°C until they were used.
(iii) Electric field strength.
To determine the optimal electric field strength for maximal recovery of transformants, we tested a range of electric field strengths from 5.0 to 12.5 kV/cm.
(iv) Postpulse incubation time.
We tested various postpulse incubation times (from immediately after the electric pulse to 12 h after the electric pulse) and evaluated the effect on transformation efficiency. Since prolonged postpulse incubation can generate sister clones that result in overestimation of the transformation efficiency, we also determined total numbers of viable cells using nonselective GAM agar plates after postpulse incubation and calculated a transformation index (Ti) using the following equation to allow comparisons of transformation efficiency: Ti = −1/log10(number of transformants/μg plasmid/viable cell).
B. fragilis gene deletion.
We attempted to delete the Tsr19 gene (6, 15, 17) of B. fragilis strain NCTC9343 by introducing the suicide plasmid pLYL0520EC (prepared in E. coli) or pLYL0520BF (prepared in B. fragilis). The suicide plasmids were electroporated into B. fragilis strain NCTC9343, and the resultant diploids with the suicide plasmid integrated were selected on GAM agar plates containing Cfx. The Cfx-resistant colonies included transformants that were generated by residual replicative pLYL0519 despite the XbaI digestion step to remove the Bacteroides replication origin. After exclusion of pLYL0519-borne transformants by PCR screening with primers M13M4 and M13RV, transformants in which the suicide plasmid pLYL0520 had integrated into the target site were selected by PCR. The diploids obtained were grown in GAM broth, spread onto nonselective GAM agar plates, and replica plated on GAM agar plates containing Cfx to screen for mutants that resolved the diploid state through a second homologous recombination. Cfx-sensitive colonies were selected, and genetic depletion of the Tsr19 gene was confirmed by PCR using the following primers flanking the deletion sites: D1 (5′-GCGATTGCTTTCTCAGTGGT-3′) and D2 (5′-GAGGTTGACCTTTTCGTTGC-3′). Disruption of the Tsr19 gene was also confirmed by the abrogation of promoter inversions in two distant regions. Promoter inversion was assessed by PCR using orientation-specific primer pairs as described by Roche-Hakansson et al. (17). Synthetic oligonucleotides were purchased from Sigma-Aldrich Japan Co., Ltd. (Tokyo, Japan). DNA sequencing was performed with an ABI PRISM 3100 genetic analyzer (Applied Biosystems) using an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (version 1.1; Applied Biosystems).
RESULTS
Plasmid selection.
The plasmid that produced the highest number of transformants following electroporation was selected from the four E. coli-Bacteroides shuttle plasmids listed in Table 1. B. fragilis strains NCTC9343 and YCH46, whose genomes have been sequenced, were employed as test strains to select the best plasmid for B. fragilis transformation. Of the plasmids tested, pLYL05 generated higher numbers of transformants of both strains (1.98 × 104 ± 0.60 × 104 transformants/μg pLYL05 for strain NCTC9343 and 2.01 × 104 ± 2.59 × 104 transformants/μg pLYL05 for strain YCH46) than the other three plasmids (Table 2). Plasmid pLYL05 generated around 700-fold more transformants than pNLY1 in strain YCH46 despite the fact that the sequence of pLYL05 differed from the sequence of pNLY1 only in the drug resistance determinant. This result suggested that the Cfxr marker was suitable for B. fragilis transformation. Similarly, plasmid pVAL-2 yielded higher numbers of transformants in both B. fragilis strains tested than pVAL-1, even though the plasmids differed from each other only in the drug resistance gene. Based on these results, we used pLYL05 for optimization of the other electroporation parameters.
TABLE 2.
Effect of plasmid on electrotransformation efficiency of B. fragilis calculated from the results of three independent experimentsa
| Plasmidb | Selection marker | Size (kb) | Log10 no. of transformants/μg plasmid DNAc |
|
|---|---|---|---|---|
| NCTC9343 | YCH46 | |||
| pLYL05 | Cfxr | 7.55 | 4.28 ± 0.13 | 4.01 ± 0.59 |
| pNLY1 | Cmr | 7.76 | 3.34 ± 0.61 | 1.22 ± 0.53 |
| pVAL-1 | Emr | 11 | 0.05 ± 0.09 | 1.66 ± 0.37 |
| pVAL-2 | Cfxr | 10.1 | 1.27 ± 1.10 | 2.83 ± 0.15 |
B. fragilis cells at mid-exponential growth phase were electroporated with 1.0 μg DNA of each plasmid indicated. After 12 h of postpulse incubation, cells were spread onto GAM agar plates containing the selective antibiotic corresponding to the plasmid used.
All plasmids were purified from E. coli laboratory strain HB101.
The data are means ± standard deviations.
In the following experiments, electrotransformations were performed using B. fragilis strain NCTC9343 as the test strain.
Optimal growth conditions.
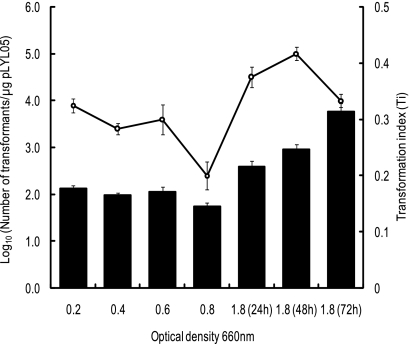
We examined the competence of B. fragilis strain NCTC9343 cells harvested at various growth stages from early exponential phase to stationary phase (OD660, 0.2, 0.4, 0.6, 0.8, and 1.8) and compared the results. Unexpectedly, stationary-phase cells showed a higher level of competence than exponentially growing cells (Fig. 1). Cells harvested after 48 h of cultivation (OD660, 1.8) yielded the largest absolute number of transformants, 9.34 × 104 ± 2.99 × 104 transformants/μg pLYL05 DNA, which was approximately 10- to 30-fold higher than the number of transformants for cells grown to early or mid-log phase (7.75 × 103 ± 2.62 × 103 and 2.65 × 103 ± 0.76 × 103 transformants/μg pLYL05 DNA, respectively). Furthermore, cells cultured for 72 h showed the highest transformation efficiency (Ti, 0.314 ± 0.014) even though the viability of the cells declined. We concluded that cells from a 48-h culture were the optimal cells for electrotransformation with pLYL05. In addition, competent cells stored at −70°C and prepared from a 48-h culture also yielded 2.62 × 106 ± 0.70 × 106 transformants/μg pLYL05 DNA, which indicated that competent cells prepared in this study exhibited a high level of competence even after storage at −70°C.
FIG. 1.
Electrocompetence of B. fragilis strain NCTC9343 cells harvested at various growth stages from early exponential phase to stationary phase. The open circles indicate the log10 number of transformants per μg pLYL05 DNA obtained with competent cells at different growth phases. Transformation indices (Ti) are indicated by bars. Cultivation periods are indicated in parentheses.
Optimal postpulse incubation time.
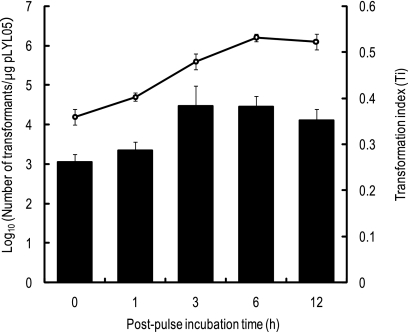
Postpulse incubation is an important step in the recovery of competent cells from electric shock and also allows expression of the selective marker gene on the plasmid. We determined the optimal postpulse incubation time for B. fragilis electroporation. Prolonged postpulse incubation can allow transformants to divide and generate large numbers of daughter cells from the original transformed cells, resulting in overestimation of the transformation efficiency. Therefore, we determined the total numbers of viable cells using nonselective GAM agar plates after postpulse incubation and calculated Ti values from the resulting values (see Materials and Methods) for samples obtained after various postpulse incubation times (0, 3, 6, and 12 h). As shown in Fig. 2, the peak Ti values were obtained with 3 h of postpulse incubation (Ti, 0.383 ± 0.044). Thereafter, the Ti values remained relatively stable until 12 h. For absolute counts of transformants, 6 h of postpulse incubation generated the highest numbers of transformants. These results indicated that 3 to 12 h of postpulse incubation was necessary to obtain efficient electrotransformation of B. fragilis.
FIG. 2.
Effect of postpulse incubation time on the recovery of pLYL05 electrotransformants of B. fragilis strain NCTC9343. The open circles indicate the log10 number of transformants per μg pLYL05 DNA at the postpulse incubation times indicated. Transformation indices (Ti) are indicated by bars.
Optimal electric field strength.
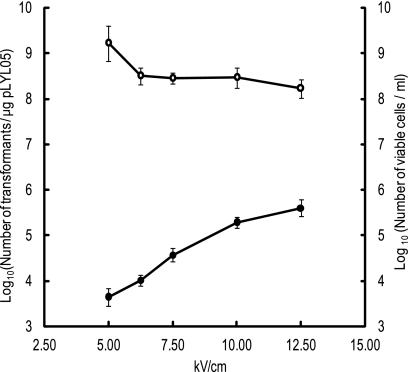
The optimal electric field strength for electroporation of B. fragilis was also evaluated. As the electric field strength increased, the number of transformants increased (Fig. 3). Using the Gene Pulser II system (Bio-Rad Laboratories), an electric pulse at 12.5 kV/cm yielded the maximum number of transformants (4.02 × 105 ± 1.47 × 105 transformants/μg pLYL05).
FIG. 3.
Effect of electric field strength on the electrotransformation efficiency of B. fragilis strain NCTC9343 with pLYL05. B. fragilis strain NCTC9343 competent cells prepared from 48-h cultures were electroporated with 1.0 μg pLYL05 DNA at the electric field strengths indicated. After 3 h of postpulse incubation, cells were spread onto GAM agar plates containing 50 μg/ml Cfx. Transformant colonies were counted after 48 h of anaerobic cultivation at 37°C. The open and filled circles indicate the log10 numbers of viable cells/ml of culture and log10 numbers of transformants/μg pLYL05, respectively.
Electrotransformation of other sequenced intestinal Bacteroides strains.
Using the optimized electroporation parameters for B. fragilis strain NCTC9343, other sequenced Bacteroides strains, including B. fragilis strain YCH46, B. thetaiotaomicron strain VPI-5482, and B. vulgatus strain ATCC 8482, were transformed by pLYL05 DNA prepared from an E. coli strain. As B. thetaiotaomicron strain VPI-5482 is cefoxitin resistant, this strain was transformed with pNLY1. As summarized in Table 3, transformants were successfully obtained from all strains tested, although the transformation efficiencies of non-B. fragilis species were relatively low. To evaluate the effect of in vivo plasmid methylation on transformation, Bacteroides species were electroporated with homologously modified pLYL05 or pNLY1. In vivo methylation of the pLYL05 plasmid increased the transformation efficiencies for B. fragilis and B. thetaiotaomicron but not for B. vulgatus.
TABLE 3.
Electrotransformation of Bacteroides species using in vivo methylation
| Straina | Plasmidb | Log 10 no. of transformants/μg plasmid DNAc |
|
|---|---|---|---|
| Without in vivo methylation | With in vivo methylation | ||
| Bacteroides fragilis NCTC9343 | pLYL05 | 5.36 ± 0.04 | 7.23 ± 0.05 |
| Bacteroides fragilis YCH46 | pLYL05 | 4.42 ± 0.34 | 6.25 ± 0.08 |
| Bacteroides thetaiotaomicron VPI-5482 | pNLY1 | 3.43 ± 0.65 | 4.45 ± 0.25 |
| Bacteroides vulgatus ATCC 8482 | pLYL05 | 3.15 ± 0.16 | 2.56 ± 0.11 |
Competent cells were prepared from 48-h cultures and electroporated with 0.45 μg DNA of each plasmid indicated. After 3 h of postpulse incubation, cells were spread onto GAM agar plates containing selective antibiotics corresponding to the plasmid used.
All plasmids were purified from E. coli laboratory strain HB101. pNLY1 was used for B. thetaiotaomicron strain VPI-5482 as this strain is cefoxitin resistant.
The data are means ± standard deviations calculated from three independent experiments.
Construction of a B. fragilis deletion mutant.
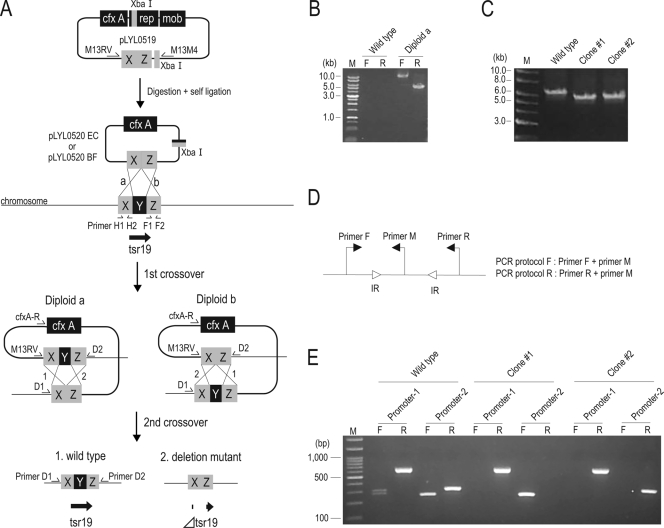
As our optimized electroporation protocol yielded relatively high numbers of transformed B. fragilis colonies using plasmid DNA prepared from an E. coli strain, we attempted to construct a deletion mutant of B. fragilis by electrically introducing a suicide targeted plasmid. We selected the gene encoding a tyrosine site-specific recombinase (Tsr19) as a target for gene disruption (6, 14, 16) as it mediates the on-off switching of two promoters that are located far apart. A schematic diagram of the expected two-step homologous recombination used to delete the internal portion of the Tsr19 gene is shown in Fig. 4A.
FIG. 4.
(A) Schematic diagram of attempted deletion of a B. fragilis gene. Homologous recombination between the target gene (Tsr19 gene) and the targeted suicide vectors pLYL0520EC and pLYL0520BF is shown. Plasmid pLYL0519 harboring 2-kb fragments homologous to the chromosomal target (X and Z) was digested with XbaI to remove the Bacteroides replication origin (rep). After self-ligation, pLYL0520 was introduced into B. fragilis and integrated into the target site by homologous recombination (diploid a or b). To obtain the deletion mutants, the diploids were screened by using loss of Cfx resistance. Y, region deleted; cfx A, cefoxitin resistance marker; rep, Bacteroides replication origin; mob, mobilization gene. (B) PCR to check integration of suicide plasmid pLYL0520BF into the target site on the B. fragilis strain NCTC9343 chromosome. The junctions of the expected integration were amplified using primers D1 and cfxA-R and primers D2 and M13RV (indicated by F and R, respectively, above the lanes). Lane M contained markers. (C) Confirmation of the Tsr19 gene deletion by PCR. The expected deletion was demonstrated by the 675-bp downshift of the amplicons produced using primers D1 and D2. (D) Orientation-specific primer sets used to detect the DNA inversions at two Tsr19 gene-regulated promoter regions (promoter 1 and promoter 2). Two primer sets were used for assessment of DNA inversions at promoter 1 (primer F, 5′-GCTTCAGAGAAGGTGACATACA-3′; primer M, 5′-TACTCAAACAGGCGTTTACA-3′; and primer R, 5′-CTCCATGCTTACGATAGGAC-3′) and promoter 2 (primer F, 5′-CTGAGTTTGCAGAGCTTCTG-3′; primer M, 5′-AGTTGAGATAATAGTCGCAT-3′; and primer R, 5′-AGCACTCTGTCTGCTAATGG-3′). Open arrowheads indicate inverted repeat sequences (IR). (E) Analysis of Tsr19 gene deletion mutants of B. fragilis strain NCTC9343, showing different promoter orientation patterns for the two distant Tsr19 gene-regulated invertible loci. The results of agarose gel electrophoresis of the amplification products from the regions indicated above the lanes are shown. F and R indicate the primers described above for panel D, as follows: F, primer F plus primer M; R, primer M plus primer R. Lane M contained 100-bp ladder DNA markers.
We constructed plasmid pLYL0519 in E. coli strain DH5α. The pLYL0519 plasmid contained 2-kb fragments (Fig. 4A) from regions flanking the region to be deleted (Fig. 4A). The Bacteroides replication origin was then removed by XbaI digestion, generating suicide plasmid pLYL0520EC in E. coli. Since in vivo methylation of the plasmid was effective for B. fragilis transformation as described above, suicide plasmid pLYL0520BF was also prepared; to do this, plasmid pLYL0519 was introduced into B. fragilis strain NCTC9343, modified, and then extracted to remove the Bacteroides replication origin.
Suicide plasmids pLYL0520EC and pLYL0520BF were designed to obtain diploid configurations at the target site on the NCTC9343 chromosome (Fig. 4A). B. fragilis strain NCTC9343 cells prepared from an early-exponential-phase culture (OD660, ∼0.2) or a 48-h culture were electroporated with the suicide plasmids. Although the transformant yields for simple replicative plasmid transfer were optimal when competent cells derived from the 48-h culture were used, suicide plasmid integration occurred at higher frequencies when early-exponential-phase competent cells were used along with a 12-h postpulse incubation step (data not shown). For each 0.1 μg of pLYL0520EC and pLYL0520BF, 136 to 217 and 242 to 4,257 Cfx-resistant colonies were generated, respectively. However, upon confirmation of integration by PCR using primers D1 and cfxA-R and primers D2 and M13RV (as shown in Fig. 4A), none of the 24 randomly selected pLYL0520EC-transformed colonies gave a pattern consistent with successful integration. In contrast, 48.7% of the Cfx-resistant pLYL0520BF-electroporated colonies yielded results consistent with the expected diploid (Fig. 4B). The remaining Cfx-resistant colonies were transformants from self-ligated pLYL0519 that had contaminated the preparation of pLYL0520BF.
Spontaneous resolution of the diploids was induced by cultivation in liquid media without Cfx. Appropriate cell dilutions were spread onto GAM agar plates, and nearly 4,000 colonies were then screened by replica plating to detect cells that had undergone deletion due to a second recombination event. Of the 12 Cfx-sensitive colonies identified, 5 were colonies of the expected deletion mutants, in which the internal region of the Tsr19 gene was deleted, as confirmed by PCR analysis (Fig. 4C). The disruption of the Tsr19 gene was further confirmed by performing a promoter inversion assay for the two invertible regions (Fig. 4E) using an orientation-specific primer set (Fig. 4D). Two types of locked mutants were generated. Together, our results showed that a B. fragilis genetic knockout was constructed by using an electroporation method.
DISCUSSION
Genetic knockout is an essential strategy for functional identification of genes of interest. However, in many bacterial species, genetic manipulation is often difficult due to barriers to introduction of foreign DNA. Diverse restriction-modification (R/M) systems in bacteria are partially responsible for the resistance to transformation by heterologous DNA (1). Due to the diversity of R/M systems in bacteria, genetic manipulation systems must be constructed and optimized for individual species or strains.
Bacteroides is a major constituent of the intestinal microflora in humans and animals. The metabolic activities of Bacteroides species can profoundly affect human physiology through nutrient degradation, synthesis of short-chain fatty acids, and immune system stimulation (2, 16). The recent release of whole-genome sequence data is expected to enhance the study of the molecular basis of human-microbe interactions in the gut. However, consistent with many R/M system genes observed in Bacteroides genomes, Bacteroides species show poor competence for heterologous DNA. For example, the genomes of two sequenced strains, B. fragilis strains NCTC9343 (5) and YCH46 (12), contain at least 14 R/M system genes (http://rebase.neb.com/rebase/index.html). Transconjugation using the filter mating procedure is usually employed for gene transfer in this group of bacteria (24), but this procedure is labor-intensive and time-consuming. Although Smith et al. previously described an electroporation method optimized for several Bacteroides species, the transformation efficiencies for plasmids purified from E. coli were quite low (20). In this study, we refined the electroporation parameters for transformation of Bacteroides species, especially for plasmids purified from E. coli. Furthermore, we disrupted a B. fragilis gene by homologous recombination using an electrically introduced suicide plasmid with our optimized electroporation parameters. The results obtained in our experiments indicated that for Bacteroides species, the optimized electroporation parameters for simple plasmid transformation are not necessarily optimal for insertional mutagenesis.
Consistent with the report of Smith et al. (20), the recovery of transformants was greatly influenced by the antibiotic resistance marker encoded on the transforming plasmid. Of the selective antibiotic markers tested in B. fragilis (Emr, Cmr, and Cfxr), cefoxitin resistance resulted in the highest level of recovery of transformants, and the plasmid encoding cefoxitin resistance yielded 10- and 700-fold more transformants than plasmids encoding chloramphenicol and erythromycin resistance, respectively, in strain YCH46. E. coli-Bacteroides shuttle plasmids pNLY1 and pLYL05 share the same backbone and differ only in the antibiotic resistance gene carried, yet their abilities to transform B. fragilis cells were considerably different. Similarly, the pVAL-2 shuttle plasmid, which was constructed by exchanging the Emr element of pVAL-1 with the Cfxr gene from pLYL05, yielded 10-fold-higher numbers of transformants in B. fragilis than the parent pVAL-1 plasmid. While the reasons for these differences remain unclear, a number of factors, such as the concentration of antibiotics used, the resistance mechanism, and the transcriptional level of the selective marker gene, may be involved. It is also possible that the Cfxr gene does not include a recognition sequence for the R/M system utilized by the B. fragilis strains used in this study.
Growth phase was the other important parameter that greatly influenced the recovery of transformants following electroporation in Bacteroides species. Competent cells used for transformation are usually prepared from early- to mid-exponential-phase cultures. Previous optimization of electroporation for Bacteroides species was also performed with competent cells prepared from early- to mid-exponential-phase cultures (20). Unexpectedly, we found that stationary-phase B. fragilis cells showed higher levels of competence than exponentially growing cells (Fig. 1). Of the culture phases tested for B. fragilis strain NCTC9343, competent cells prepared from 72-h cultures showed the highest transformation efficiency for pLYL05 when transformation indices (Ti) were compared. The highest absolute number of transformants was obtained when competent cells prepared from 48-h cultures were used. A recent report concerning the electroporation of Corynebacterium pseudotuberculosis also demonstrated that the highest level of competence was observed when stationary-phase cells were used for electroporation (9). The physiological changes associated with stationary-phase growth, such as alterations in membrane integrity, cell wall turnover, extracellular polysaccharide production, and R/M system activity, might explain the enhanced plasmid entry.
We obtained similar results for optimal electric field strength and postpulse incubation time for B. fragilis, as previously reported by Smith et al. (20). The number of transformants linearly increased with the electric field strength, and the maximum efficiency occurred at 12.5 kV/cm, which was the maximum electric field strength possible for the electroporation cuvettes with the 0.2-cm interelectrode distance used in the Gene Pulser II system (Bio-Rad Laboratories). At least 3 h of postpulse incubation was required for maximum transformation efficiency (Ti) when Cfxr was used as a selective marker. From these results, we concluded that for simple plasmid transfer into B. fragilis, the maximum transformant yield was obtained using competent cells prepared from 48-h cultures, plasmids harboring the Cfxr marker, an electric field strength of 12.5 kV/cm, and 3 h of postpulse incubation. When this protocol was used for other sequenced intestinal Bacteroides strains (B. vulgatus strain ATCC 8482 and B. thetaiotaomicron strain VPI-5482), plasmids pLYL05 and pNLY1 purified from E. coli were also successfully transferred, although the transformation efficiency was reduced compared to that for B. fragilis. As E. coli laboratory strains are superior for complex plasmid construction, our electroporation protocol described here, with improved accessibility of E. coli-purified plasmids in intestinal Bacteroides strains, greatly simplifies genetic manipulation of Bacteroides species. In addition, frozen competent cells in a 10% glycerol solution exhibited competence equivalent to that of freshly prepared cells for every Bacteroides species tested. This finding is particularly important for routine use of electroporation as a tool for genetic manipulation in Bacteroides.
To assess the applicability of our protocol to construction of genetic knockouts in B. fragilis, we attempted to disrupt the tyrosine recombinase gene (the Tsr19 gene) that mediates promoter inversions at two distant loci. Homologous recombination between the cloned fragments and the chromosomal target site occurred at higher frequencies when early-exponential-phase cells were used as the competent cells than when cells from a 48-h culture were used and following 12 h of postpulse incubation. This result indicated that the optimized electroporation parameters used for simple plasmid transformation were not necessarily optimal for insertional mutagenesis in Bacteroides. For integration of a suicide plasmid into the chromosome, it is possible that the cells need to be actively replicating for homologous recombination to occur. The copy number of the selective marker gene may also be related to the difference in optimal postpulse incubation time between simple plasmid transfer and insertional mutagenesis (i.e., multiple copies for replicative plasmid transfer versus a single copy for insertional mutagenesis), so that longer postpulse incubation times may be required for insertional mutagenesis. Our results also indicated that the electroporation parameters for insertional mutagenesis in Bacteroides may require separate optimization. Nonetheless, the efficient and reproducible plasmid transfer described here enabled B. fragilis to be transformed with in vivo-methylated (i.e., modified in isogenic hosts) targeted vectors, which led to generation of the expected genetic disruptants (Fig. 4). As shown in Table 3, in vivo methylation increased the transformation efficiency 100- and 10-fold in B. fragilis and B. thetaiotaomicron, respectively. However, in vivo methylation decreased the transformation efficiency in B. vulgatus. The reason for this is unclear, but it is possible that the extracted plasmid DNA may have been unsuitable for the R/M system of the recipient cells, as Bacteroides can interchange R/M system specificities via reversible DNA inversion (5). Thus, the in vivo methylation conditions may need to be optimized based on the recipient cells.
Random mutagenesis by certain transposons is an attractive tool for identification of genes responsible for interesting phenotypes. We introduced pEP4351 (7), which harbors Bacteroides transposon Tn4351, into B. fragilis strain NCTC9343. Electroporation employing parameters suitable for insertional mutagenesis produced 1,400 ± 831 transposon insertion mutants. Sequence analysis of 93 clones revealed that Tn4351 inserted at 65 independent sites distributed throughout the chromosome (data not shown). Construction of an in vivo-methylated pLYL05-based Tn4351 plasmid would be expected to increase the yield of mutants with Tn4351 inserted in B. fragilis.
In summary, we determined the optimal electroporation parameters for simple transfer of plasmids purified from E. coli into Bacteroides strains. Electroporation employing the optimized parameters successfully transformed the genomes of several sequenced intestinal Bacteroides species. Our electroporation protocol should enable enhanced genetic manipulation of intestinal Bacteroides species using complex plasmids constructed in E. coli hosts, including random insertional mutagenesis using certain types of transposons.
Acknowledgments
We are grateful to Nadja B. Shoemaker (University of Illinois) for providing E. coli-Bacteroides shuttle plasmids pVAL-1, pNLY1, and pLYL05.
This work was supported by grant-in-aid for scientific research 20510187 to T.K. from the Japan Society for Promotion of Science.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Ando, T., Q. Xu, M. Torres, K. Kusugami, D. A. Israel, and M. J. Blaser. 2000. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 37:1052-1065. [DOI] [PubMed] [Google Scholar]
- 2.Bäckhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 25:1915-1920. [DOI] [PubMed] [Google Scholar]
- 3.Bennion, R. S., E. J. Baron, J. E. Thompson, Jr., J. Downes, P. Summanen, D. A. Talan, and S. M. Finegold. 1990. The bacteriology of gangrenous and perforated appendicitis revisited. Ann. Surg. 211:165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brook, I. 1995. Bacteroides infections in children. J. Med. Microbiol. 43:92-98. [DOI] [PubMed] [Google Scholar]
- 5.Cerdeño-Tárraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 6.Chatzidaki-Livanis, M., M. J. Coyne, H. Roche-Hakansson, and L. E. Comstock. 2008. Expression of a uniquely regulated extracellular polysaccharide confers a large-capsule phenotype to Bacteroides fragilis. J. Bacteriol. 190:1020-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper, J., A. P. Kalinowski, N. B. Shoemaker, and A. A. Salyers. 1997. Construction and characterization of a Bacteroides thetaiotaomicron recA mutant: transfer of Bacteroides integrated conjugative elements is RecA independent. J. Bacteriol. 179:6221-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne, M. J., W. Kalka-Moll, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2000. Bacteroides fragilis NCTC9343 produces at least three distinct capsular polysaccharides: cloning, characterization, and reassignment of polysaccharide B and C biosynthesis loci. Infect. Immun. 68:6176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorella, F. A., E. M. Estevam, and P. G. Cardoso. 2006. An improved protocol for electrotransformation of Corynebacterium pseudotuberculosis. Vet. Microbiol. 114:298-303. [DOI] [PubMed] [Google Scholar]
- 10.Hooper, L. V., T. S. Stappenbeck, C. V. Hong, and J. I. Gordon. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4:269-273. [DOI] [PubMed] [Google Scholar]
- 11.Kelly, D., J. I. Campbell, T. P. King, G. Grant, E. A. Jansson, A. G. Coutts, S. Pettersson, and S. Conway. 2004. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat. Immunol. 5:104-112. [DOI] [PubMed] [Google Scholar]
- 12.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. U. S. A. 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindberg, A. A., A. Weintraub, D. L. Kasper, and J. Lönngren. 1982. Virulence factors in infections with Bacteroides fragilis: isolation and characterization of capsular polysaccharide and lipopolysaccharide. Scand. J. Infect. Dis. Suppl. 35:45-52. [PubMed] [Google Scholar]
- 14.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Patrick, S., S. Houston, Z. Thacker, and G. W. Blakely. 2009. Mutational analysis of genes implicated in LPS and capsular polysaccharide biosynthesis in the opportunistic pathogen Bacteroides fragilis. Microbiology 155:1039-1049. [DOI] [PubMed] [Google Scholar]
- 16.Peterson, D. A., N. P. McNulty, J. L. Guruge, and J. I. Gordon. 2007. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 15:328-339. [DOI] [PubMed] [Google Scholar]
- 17.Roche-Hakansson, H., M. Chatzidaki-Livanis, M. J. Coyne, and L. E. Comstock. 2007. Bacteroides fragilis synthesizes a DNA invertase affecting both a local and a distant region. J. Bacteriol. 189:2119-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salyers, A. A., N. B. Shoemaker, A. Cooper, J. D'Elia, and J. A. Shipman. 1999. Genetic methods for Bacteroides species. Methods Microbiol. 29:229-276. [Google Scholar]
- 19.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, C. J., A. Parker, and M. B. Rogers. 1990. Plasmid transformation of Bacteroides spp. by electroporation. Plasmid 24:100-109. [DOI] [PubMed] [Google Scholar]
- 21.Tzianabos, A. O., D. L. Kasper, R. L. Cisneros, R. S. Smith, and A. B. Onderdonk. 1995. Polysaccharide-mediated protection against abscess formation in experimental intra-abdominal sepsis. J. Clin. Invest. 96:2727-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzianabos, A. O., A. Pantosti, H. Baumann, J. R. Brisson, H. J. Jennings, and D. L. Kasper. 1992. The capsular polysaccharide of Bacteroides fragilis comprises two ionically linked polysaccharides. J. Biol. Chem. 267:18230-18235. [PubMed] [Google Scholar]
- 23.Tzianabos, A. O., A. B. Onderdonk, B. Rosner, R. L. Cisneros, and D. L. Kasper. 1993. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 262:416-419. [DOI] [PubMed] [Google Scholar]
- 24.Valentine, P. J., N. B. Shoemaker, and A. A. Salyers. 1988. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 170:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]