Abstract
Five meilingmycins, A to E, with A as the major component, were isolated from Streptomyces nanchangensis NS3226. Through nuclear magnetic resonance (NMR) characterization, meilingmycins A to E proved to be identical to reported milbemycins α11, α13, α14, β1, and β9, respectively. Sequencing of a previously cloned 103-kb region identified three modular type I polyketide synthase genes putatively encoding the last 11 elongation steps, three modification proteins, and one transcriptional regulatory protein for meilingmycin biosynthesis. However, the expected loading module and the first two elongation modules were missing. In meilingmycin, the presence of a methyl group at C-24 and a hydroxyl group at C-25 suggests that the elongation module 1 contains a methylmalonyl-coenzyme A (CoA)-specific acyltransferase (ATp) domain and a ketoreductase (KR) domain. Based on the conserved motifs of the ATp and KR domains, a pair of primers was designed for PCR amplification, and a 1.40-kb expected fragment was amplified, whose sequence shows significant homology with the elongation module 1 of the aveA1-encoded enzyme AVES1. A polyketide synthase (PKS) gene encoding one loading and two elongation modules, with a downstream C-5-O-methyltransferase gene, meiD, was subsequently localized 55 kb apart from the previously sequenced region, and its deletion abolishes meilingmycin production. A series of deletions within the 55-kb intercluster region rules out its involvement in meilingmycin biosynthesis. Furthermore, gene deletion of meiD eliminates meilingmycins D and E, with methyls at C-5. Our work provides a more specific strategy for the cloning of modular type I PKS gene clusters. The cloning of the meilingmycin gene clusters paves the way for its pathway engineering.
Backbones of macrolides (e.g., erythromycin), polyenes (e.g., candicidin), polyethers (e.g., nanchangmycin), ansamycins (e.g., ansamitocin), and the polyketide portion of peptide-polyketide hybrids (e.g., oxazolomycin) are usually biosynthesized from simple carboxylic acids by the modular polyketide synthases (PKSs; type I PKSs) (6, 8, 30, 37, 38). Type I PKSs are multifunctional polypeptides and divided into modules, and each module is responsible for the incorporation of one carboxylic acid into the polyketide backbone. A minimal module is usually an ordered array of ketosynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP) domains, with the β-carbonyl group unreduced in its thioester products (16). KS catalyzes the carbon-carbon bond formation between the incoming extender unit and polyketide intermediate by the Claisen condensation, whereas ACP serves as the carrier for both the incoming extender units and the extended chains using a covalently bound phosphopantetheine arm. Selection and loading of extender acyl units is executed by acyltransferase (AT) domains, which contributes to the structural diversity of polyketides by recruiting different extender units such as acetate-derived malonyl coenzyme A (CoA), propionate-derived methylmalonyl-CoA, and glycerol-derived methoxymalonyl-ACP (5, 35). Deduced from sequence alignments of AT domains, the specificity for malonyl-CoA is usually determined by the presence of an HAFH motif in the active sites, whereas methylmalonyl-CoA is commonly selected by AT domains with a YASH motif in the active sites (7, 11, 24, 36). The fully extended polyketide chains bound to terminal enzymatic templates like acyl-ACP thioesters are usually released and cyclized by a type I thioesterase (TE) domain fused to the carboxyl terminus of the last elongation module.
Structure diversity of polyketides could also be attributed to the further modifications of β-ketothioester to hydroxythioester, α,β-unsaturated thioester, or the fully reduced β-carbon atom in the presence of ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) domains (27). KR domains usually contain a conserved motif, GGXGXXGXXXA, for NADP(H) binding and conserved residues of K2664, S2686, Y2699, and N2703 for catalysis like that in KR6 of 6-deoxyerthronolide B synthase (DEBS) (25). Moreover, different d-hydroxyl or l-hydroxyl stereoisomers regarding the chiral β-carbon are determined by the presence or absence of an LDD motif in the KRs (4, 25).
Antibiotic biosynthetic genes are often clustered together, which suggests successful cloning of a complete gene cluster through the localization of any essential structural, regulatory, or resistant genes (20). Since the cloning of the erythromycin biosynthetic gene cluster, numerous type I PKSs gene clusters have been cloned either through Southern hybridization using the DEBS genes as probes (12) or through PCR amplifications with degenerate primers of the erythromycin KS domains (21). However, since nearly each actinomycete genome contains many type I PKS gene clusters, this strategy will result in the cloning of dozens of clones from a genomic library. Further identification of the target PKS gene cluster requires additional strategies or is quite time-consuming. Therefore, a more straightforward strategy is required for the efficient cloning of a PKS biosynthetic gene cluster of interest.
Meilingmycin A, a 16-membered macrolide and identical to milbemycin α11, is the major product of Streptomyces nanchangensis (31). It has aglycone similar to and anthelmintic activity comparable to those of avermectins (3). However, meilingmycin A has a 5-carbon side chain on the methyl group of C-4, a saturated C-22—C-23 bond, and no sugar moiety attached (Fig. 1B). A 103-kb genomic region of S. nanchangensis was identified through the PCR amplifications of homologous fragments of aveE, aveF, and aveTE (14, 19), and its involvement in meilingmycin biosynthesis was confirmed by the deletion of an internal 24-kb genomic fragment (32).
FIG. 1.
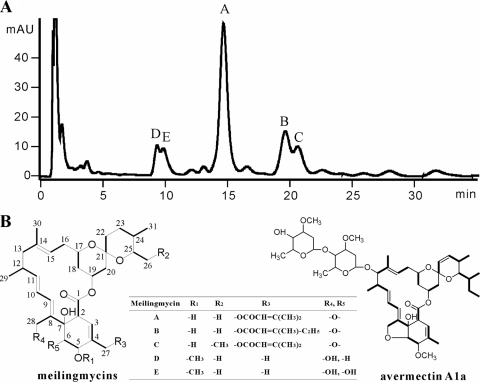
HPLC profile of meilingmycins and structure comparison between meilingmycins and avermectin. (A) HPLC profile of meilingmycins. (B) Structures of meilingmycins and avermectin A1a. The carbons of meilingmycins are numbered. The five meilingmycins are different at R1 to R5, and the differences are shown in the table. Meilingmycins A, B, and C contain a furan ring between R4 and R5 and a side chain at R3. No furan ring and side chain are identified in meilingmycins D and E, which are methylated at C-5. mAU, milli-absorbance units.
In this work, the previously cloned 103-kb region was sequenced and analyzed through gene deletions. However, the PKS genes encoding the loading and first two elongation steps for meilingmycin biosynthesis were not found in this region. Herein, a new strategy of acyltransferase-ketoreductase didomain PCR amplification was applied to localize this undefined segment, which was proved to be located 55 kb away from the previously identified gene cluster.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and general techniques.
Bacterial strains and plasmids used in this study are listed in Table S2 in the supplemental material.
S. nanchangensis NS3226, the wild-type producer for meilingmycin, was used for meilingmycin isolation and generation of mutant strains. DH10B (Gibco-BRL) and ET12567 (pUZ8002) (23) were used as Escherichia coli hosts for cloning and conjugation, respectively. Cosmid vector pHZ1358 (29) and fosmid vector pCC2FOS (Epicentre Biotechnologies) were used for constructing genomic library. Two pHZ1358-based cosmid genomic libraries, NS1 and NS2, and one pCC2FOS-based fosmid genomic library, NS3, had been constructed for this work. pJTU1278 and pJTU1289 derived from pHZ1358 were used for gene inactivation. pMD18-T (Takara) and pBlueScript II SK(+) (Stratagene) were used for DNA sequencing.
YMS solid medium (13) was used for S. nanchangensis sporulation, and tryptic soy broth (TSB) supplemented with 10.3% sucrose and 1% yeast extract was used for the growth of mycelia for the isolation of total DNA. SFM solid medium (17) was used for conjugation between E. coli and Streptomyces. Luria-Bertani medium was used for E. coli propagation. For Streptomyces, thiostrepton and apramycin were used at 12.5 μg/ml and 15 μg/ml, respectively, in solid and liquid media.
Recombinant DNA techniques used for E. coli were described by Sambrook and Russell (26), and those used for Streptomyces were described by Kieser et al. (17).
Fermentation, purification, and analysis of meilingmycins.
One 8-day-old YMS agar plate of the wild-type strain or mutants was extracted with absolute ethanol, and the extract was concentrated under reduced pressure to yield an oily substance, which was further extracted with 1 ml methanol. The methanol extract was analyzed by liquid chromatography-mass spectrometry (LC-MS) with the Agilent 1100 series LC/MSD Trap system. A high-performance liquid chromatographer (HPLC) was operated at a flow rate of 0.2 ml/min, with methanol-water (80/20, vol/vol) on an Agilent SB-C18 (2.1- by 50-mm, 2-μm) column. The ion trap mass spectrometer was operated with the electrospray ionization source in the positive-ion mode. The drying gas flow was at 8 liters/min, and nebulizer pressure was 30 lb/in2. The drying gas temperature used was 325°C, and the fragmentation amplitude was varied from 1.0 to 1.8 V.
For structure elucidation, the 8-day-old culture broth of the wild type (12 liters) was extracted with an equal volume of ethanol three times. The ethanol extract was concentrated to approximately 500 ml under reduced pressure and extracted further with an equal volume of ethyl acetate five times. The ethyl acetate extract was concentrated under reduced pressure to yield 4.34 g of an oily substance, which was chromatographed through silica gel (30 g, 300 to 400 mesh) and eluted with chloroform. Fractions possibly containing meilingmycins were combined and concentrated to give 0.6 g of crude sample. Methanol extract of the crude sample was applied to the preparative HPLC (Shimadzu) using Shim-pack (Prep-ODS, 20 by 250 mm; Shimadzu) and eluted with methanol-water (90/10, vol/vol) at 6 ml/min to yield 15 fractions. Detected by LC-MS, fractions 3, 4, and 6 probably containing meilingmycins were further purified by thin-layer chromatography (TLC) on HF254 silica gel with chloroform-methanol (10:1, vol/vol) as a solvent. Together with fractions 9 and 10, five different meilingmycins were purified by preparative HPLC for the second time with the same condition to yield meilingmycins D (4.7 mg), E (2.9 mg), A (12.4 mg), B (3.9 mg), and C (2.1 mg).
Nuclear magnetic resonance (NMR) spectra were recorded with a Bruker AM 500 NMR spectrometer in CDCl3 at 25°C, using the solvent as internal reference downfield of tetramethylsilane (TMS) at 0 ppm.
Sequencing and analysis of meilingmycin biosynthetic gene clusters.
Cosmids/fosmids 14G1, 2E4, 16B1, 9B8, and 20G1, a 17-kb ClaI fragment from 2G6, and an 8.5-kb SacI fragment from 26H12 were sequenced at CHGC (Chinese National Human Genome Center at Shanghai). In total, a 185,250-bp DNA sequence was obtained, including the previously sequenced region.
Sequence contig assembly and base editing were performed with the Phred/Phrap/Consed package (9). The sequence data were analyzed with the FramePlot-3.0beta online program (15) and GeneMark.hmm for Prokaryotes (version 2.4) using Streptomyces avermitilis as the model organism (18). DNA and deduced protein sequence homology searching was performed using BLAST (1). The PKS region was analyzed with NRPS-PKS (2).
Methylmalonyl-CoA-specific AT (ATp)-KR didomain PCR amplification.
A forward primer ATp1 (5′-CCGGTCGACTACGCCTCCCACTGC-3′) was designed for the conserved methylmalonyl-CoA-specific motif PVDYASHC of the AT domain. A degenerate reverse primer, KRp (5′-CCGGTGCCGCCGGTGATsAGbAyGGT-3′; “b” stands for C, G, or T; “s” stands for C or G; “y” stands for C or T), was designed for the NADP(H) binding motif T(V/I)LITGGTG of the KR domain. The resulting 1.4-kb amplified PCR product with primers ATp1 and KRp was cloned into the pMD18-T vector and sequenced. A pair of specific homologous primers, meiAT-P1 (5′-AAACCACCGACCGCACCA-3′) and meiAT-P2 (5′-ACCGGCTTCCAGATGTCCC-3′), was designed, according to the sequence results. Through PCR screening with this pair of primers, four fosmids (1F1, 14E7, 14G1, and 20G1) were cloned from the genomic library NS3, and one cosmid (10D8) was cloned from the genomic library NS2.
Gene deletion of the region containing the amplified ATp-KR fragment.
Complete digestion of cosmid 10D8 DNA by KpnI and religation resulted in the construction of pJTU4459, in which an internal 18.6-kb region of 10D8 was deleted and the 3.5-kb and 4.5-kb flanking fragments were connected. Then, a 1.50-kb KpnI fragment containing aac(3)IV was amplified from pHGF9827 (6) with primers SK-P1 (5′-AAAGGTACCTCGAGGTCGACGGTATC-3′, with engineered KpnI underlined) and SK-P2 (5′-AAAGGTACCGGCCGCTCTAGAACTAG-3′, with engineered KpnI underlined), which was inserted between the two flanking fragments to generate pJTU4460 for subsequent conjugation, as previously described (30). Allelic replacement of the 18.6-kb region in the mutant was confirmed by Southern hybridization. A pair of primers, 10D8-C1 (5′-CGCCGACTTCATGGAAACG-3′) and 10D8-C2 (5′-CGTGATGACAATGCGTGGTTT-3′), was designed for amplifying the probe. Comparison between the wild-type and mutant HYL23 was conducted through fermentation, extraction, and LC-MS analysis.
Gene deletion of meiD.
The 8.40-kb KpnI fragment containing meiD from fosmid 14G1 was firstly cloned into pBlueScript SK(+) to generate pJTU4465a. Then, the 8.40-kb KpnI fragment from pJTU4465a was inserted into pJTU1289 to generate pJTU4466. The oriT-aac(3)IV cassette was amplified from pIJ773 (10) with the primers MeiD-T1 (5′-ATCGGCAAACTCGGCGACATCGCCGGCCGCCGGGTCCTGtgtaggctggagctgcttc-3′) and MeiD-T2 (5′-TGATCAGCTCTCGGACGGACATCCCGGGCGCTTGGCGGTattccggggatccgtcgacc-3′) (pIJ773 homologous sequences are in lowercase). The resulting PCR product was used to replace the internal 615-bp region of meiD in pJTU4466 to generate pJTU4469 through ReDirect Technology (10), which was used to generate mutant HYL28 through conjugation. Allelic replacement of meiD in the HYL28 mutant was confirmed by PCR amplification with primers meiD-C1 (5′-TGAACGACGACACGCACGAA-3′) and meiD-C2 (5′-GCCGAAAGACACGCAGGACA-3′). Comparison between the wild-type and mutant HYL28 was performed through fermentation, extraction, and LC-MS analysis.
Supplemental material.
Detailed constructions for deletion of the pks1-pks2 region, deletion of meiC, deletion of meiF, deletion of the 55-kb intercluster region, determination of the left and right boundaries, NMR data for structural elucidation of meilingmycins, and chromosomal walking were described in the supplemental material.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was deposited in GenBank under accession number FJ952082 (Fig. 2A).
FIG. 2.
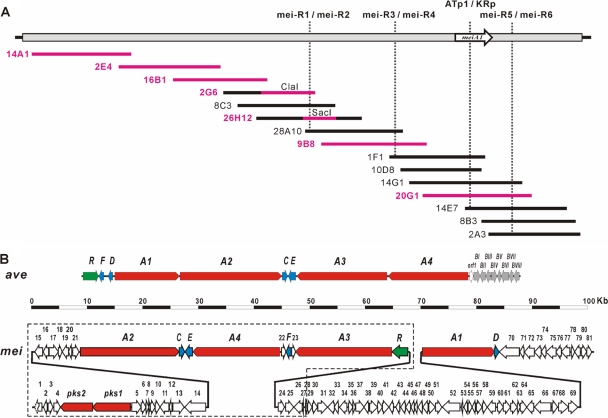
Physical map and gene organization of meilingmycin biosynthetic gene clusters. (A) Physical map of meilingmycin biosynthetic gene clusters. The vertical dashed lines indicate the positions of primers for chromosome walking. The horizontal lines indicate individual cosmid/fosmids, and those shown in pink are sequenced regions, including the 17-kb ClaI fragment from 2G6 and the 8.5-kb SacI fragment from 26H12. (B) Gene organization of meilingmycin biosynthetic gene clusters. ave, avermectin biosynthetic gene cluster (14); mei, meilingmycin biosynthetic gene cluster; red, PKS genes; blue, genes for post-PKS modifications; green, regulatory genes; blank arrows, genes probably not related to meilingmycin biosynthesis. The ORFs boxed with dashed lines are from the previously sequenced 103-kb region.
RESULTS
Structure elucidation of meilingmycins.
HPLC analysis of the fermentation broth of S. nanchangensis NS3226 identified five components of meilingmycins with similar UV absorbance, including the previously characterized meilingmycin A (Fig. 1A) (31). From a 12-liter liquid culture of the wild-type NS3226, meilingmycins A (12.4 mg, m/z 649.7 [M + Na]+), B (3.9 mg, m/z 663.7 [M + Na]+), C (2.1 mg, m/z 663.7 [M + Na]+), D (4.7 mg, m/z 567.8 [M + Na]+), and E (2.9 mg, m/z 583.7 [M + Na]+) were purified as amorphous powder. According to 1H-NMR, 13C-NMR, heteronuclear multiple-bond correlation (HMBC), heteronuclear multiple-quantum coherence (HMQC), and 1H,1H correlated spectroscopy (1H,1H COSY) analyses, the structures of A to E were proved to be identical with those of milbemycin α11, α13, α14, β1, and β9, respectively (Fig. 1B) (see the supplemental material). Meilingmycin A/milbemycin α11 is the main product in S. nanchangensis NS3226, whereas milbemycin β1/meilingmycin D is the major component in milbemycin producer Streptomyces hygroscopicus subsp. aureolacrimosus SANK 60286 (34).
Sequence analysis of the previously cloned 103-kb region.
Analysis of the available DNA sequence (103,513 bp) (Fig. 2B, encircled with dotted lines) led to identification of 37 open reading frames (ORFs), as shown in Fig. 2B and Table 1; see also Table S1 in the supplemental material. The overall gene organization between meilingmycin and avermectin biosynthetic gene clusters is different. However, homologous genes for aveC, aveE, aveF, and aveR were identified and named meiC (52% identity/66% similarity), meiE (62% identity/74% similarity), meiF (64% identity/78% similarity), and meiR (49% identity/60% similarity), respectively, in the meilingmycin biosynthetic gene cluster.
TABLE 1.
Deduced functions of mei biosynthetic genesa
| Protein | Amino acids | Proposed function | Homolog/origin | % Identity/% similarity | GenBank accession no. |
|---|---|---|---|---|---|
| Orf1 | 444 | Na+/H+ antiporter | Sare_1658/Salinispora arenicola CNS-205 | 57/72 | YP_001536537 |
| Orf2 | 225 | Intracellular protease/amidase | Tfu_2826/Thermobifida fusca YX | 64/75 | YP_290882 |
| Orf3 | 299 | LysR family transcriptional regulator | AbaB2/Saccharopolyspora erythraea NRRL 2338 | 58/72 | YP_001105667 |
| Orf4 | 398 | Cytochrome P450 | CypX/Streptomyces ambofaciens ATCC 23877 | 51/71 | CAJ88189 |
| MeiA2 | 5,987 | PKS: module 3 (KS-ATa-ACP)-4 (KS-ATa-KR-ACP)-5 (KS-ATa-KR-ACP)-6 (KS-ATp-DH-KR-ACP) | AveA2/S. avermitilis MA-4680 | 58/68 | NP_822114 |
| MeiCb | 330 | PKS modification | AveC/S. avermitilis MA-4680 | 52/66 | NP_822115 |
| MeiEb | 459 | Cytochrome P450 hydroxylase | AveE/S. avermitilis MA-4680 | 62/74 | NP_822116 |
| MeiA4 | 5,277 | PKS: module 10 (KS-ATa-KR*-ACP)-11 (KS-ATp-KRI-ACP)-12 (KS-ATa-DH-KR-ACP)-TE | AveA4/S. avermitilis MA-4680 | 66/75 | NP_822118 |
| Orf22 | 266 | Reductase | Orf-1/S. avermitilis MA-4680 | 70/80 | NP_822119 |
| MeiFb | 270 | C-5-ketoreductase | AveF/S. avermitilis MA-4680 | 64/78 | NP_822111 |
| Orf23 | 251 | Unknown | |||
| MeiA3 | 5,822 | PKS: module 7 (KS-ATp-DH-ER-KR-ACP)-8 (KS-ATa-DH-KR-ACP)-9 (KS-ATp-DH-KR-ACP) | AveA3/S. avermitilis MA-4680 | 56/66 | NP_822117 |
| MeiR | 964 | LuxR family transcriptional regulator | AveR/S. avermitilis MA-4680 | 49/60 | NP_822110 |
| MeiA1b | 4,330 | PKS: loading module (AT-ACP)-module 1 (KS-ATp-KR-ACP)-2 (KS-ATa-DH-ER-KR-ACP) | AveA1/S. avermitilis MA-4680 | 52/63 | NP_822113 |
| MeiDb | 286 | C-5-O-methyltransferase | MilD/S. griseochromogenes | 86/91 | AAR15334 |
| AveD/S. avermitilis MA-4680 | 58/69 | NP_822112 | |||
| Orf80 | 323 | Carbohydrate kinase | IolC1/S. avermitilis MA-4680 | 72/83 | NP_826515 |
| Orf81 | 300 | Unknown | ROP_10670/Rhodococcus opacus B4 | 59/73 | YP_002778259 |
Gene products not involved in meilingmycin biosynthesis, as confirmed through gene deletions, are listed in Table S1 in the supplemental material.
Gene products involved in meilingmycins biosynthesis, as confirmed through gene deletions or complementation to S. avermitilis mutant. KRI, inactive ketoreductase domain; KR*, nonfunctional ketoreductase domain deduced from the meilingmycin structure.
Five ORFs (pks1, pks2, meiA2, meiA3, and meiA4) encoding typical type I PKS subunits were identified, among which meiA2, meiA3, and meiA4 are highly homologous with aveA2, aveA3, and aveA4 of the avermectin biosynthetic gene cluster, respectively (Fig. 2B; Table 1). MeiA2 has the same domain organization as AVES2 (14), putatively encodes modules 3 to 6, and is responsible for the incorporation of C-13—C-20 to the meilingmycin polyketide backbone. MeiA3 putatively contains modules 7 to 9 and is responsible for the incorporation of C-7—C-12 to the backbone. There are KR, DH, and ER domains in module 7 of MeiA3, which catalyze the full reduction of the carbonyl group on C-13. However, the corresponding module in AVES3 contains a dysfunctional DH domain and lacks an ER domain, which leads to the retention of a hydroxyl group on C-13, serving as the attachment site for the oleandrose moiety (14). MeiA4 shows nearly the same domain organization as AVES4 for the incorporation and modification of C-1—C-6 (14), with the KR domain of module 10 (KR*) presumably active, as deduced from sequence analysis, but nonfunctional, as predicted from the meilingmycins structure. However, an inactive KR domain (KRI) is present within module 11 because it has a 16-residue deletion, including the required Lys residue, and also does not have the required Tyr and Asn residues (25).
pks1 and pks2 were 22.0 kb away from meiA2 (Fig. 2B). Pks1 contains two modules (see Table S1 in the supplemental material). One is aberrant (KSI-AT-ACP), with a shorter 123-amino acid (aa) KS domain and an RAFH motif in the AT domain, and the other is a malonyl-CoA-specific extension module (KS-malonyl-CoA specific AT [ATa]-KR-ACP). Pks2 has a methylmalonyl-CoA-specific extension module (KS-ATp-DH-KR-ACP) and a TE domain at the C terminus (see Table S1). A mutant with the pks2-orf15 region deleted maintains the same meilingmycin productivity as wild-type NS3226 (see Fig. S3 in the supplemental material), which rules out the involvement of Pks1 and Pks2 in meilingmycin biosynthesis.
There are seven ORFs (orf15-orf21) to the left of meiA2 with putative functions of regulation, transporter, or resistance. To determine whether they are related to meilingmycin biosynthesis, orf15-orf21 were deleted (see the supplemental material), and the resulting mutant HYL24 still produces meilingmycins at a level comparable to that of wild-type NS3226, as detected by LC-MS (see Fig. 4C), which clearly indicates that meiA2 is at the left boundary of the meilingmycin biosynthetic gene cluster. So far, the PKS portion responsible for the synthesis of C-21—C-26 was still missing, and new strategy was required to localize this portion.
FIG. 4.
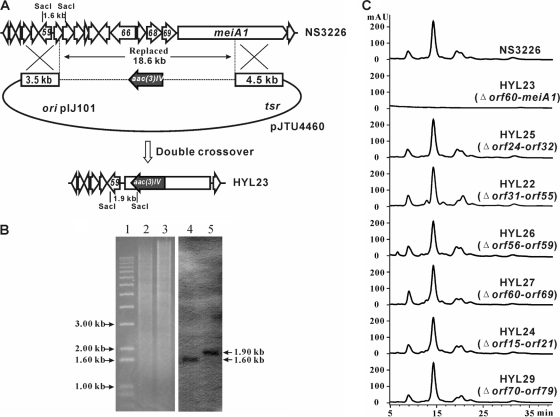
Gene deletion of meiA1 and upstream genes. (A) Schematic description of gene deletion. The 18.6-kb region, including most of meiA1, and 10 upstream ORFs were replaced by aac(3)IV through double crossover. Using a portion of the 1.60-kb SacI fragment at the left end of the deletion region as a probe for Southern hybridization, the SacI-digested genomic DNA from wild-type NS3226 will give a 1.60-kb signal, whereas the deletion mutant HYL23 will yield a 1.90-kb signal. (B) Confirmation through Southern hybridization. The 562-bp probe was amplified using primers 10D8-C1 and 10D8-C2 and cosmid 10D8 as a template. The Southern hybridization revealed that there is a 1.90-kb signal in mutant HYL23 and a 1.60-kb signal in wild-type NS3226. (C) Comparison between wild-type NS3226 and deletion mutants through LC-MS. HYL23, deletion (Δ) of orf60-meiA1 containing the amplified ATp-KR fragment; HYL25, deletion of orf24-orf32 within the 55-kb region; HYL22, deletion of orf31-orf55 within the 55-kb region; HYL26, deletion of orf56-orf59 within the 55-kb region; HYL27, deletion of orf60-orf69 within the 55-kb region; HYL24, deletion of orf15-orf21 to determine the left boundary of the mei gene clusters; HYL29, deletion of orf70-orf79 to determine the right boundary of the mei gene clusters.
ATp-KR didomain amplification.
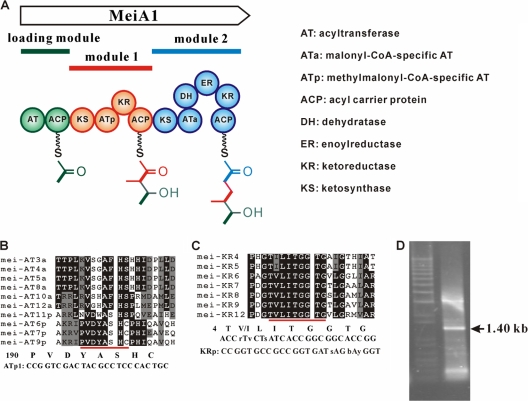
The presence of a methyl group at C-24 of meilingmycins predicts that the AT domain of the first extension module (module 1) is methylmalonyl-CoA specific (ATp), and the retention of a hydroxyl group at C-25 suggests the presence of the KR domain in the same module. Based on the canonical organization of PKS modules, module 1 was deduced to have a KS-ATp-KR-ACP assembly (Fig. 3A) (16). Therefore, a pair of degenerate primers was designed for amplifying the ATp-KR region using the total DNA of S. nanchangensis as template. Based on the alignment of available AT domains of MeiA2, MeiA3, and MeiA4, a forward primer ATp1 was designed, according to the conserved motif PVDYASHC (aa 190 to 197) of the ATp domain (Fig. 3B). Based on the alignments of available KR domains of MeiA2, MeiA3, and MeiA4, the reverse primer KRp was designed within the conserved NADPH-binding motif T(V/I)LITGGTG of the KR domain (Fig. 3C).
FIG. 3.
PCR amplification of the ATp-KR region. (A) Hypothetical domain organization of MeiA1. (B) Sequence alignment of the mei AT domains. The top six ATs are malonyl-CoA-specific AT domains (ATa). The rest are methylmalonyl-CoA-specific AT (ATp) domains. The conserved motif of ATp is underlined, and the primer ATp1 was designed according to this motif. (C) Sequence alignment of the mei KR domains. The conserved motif of KR for NADP(H) binding is underlined, and the degenerate primer KRp was designed according to this motif. (D) PCR amplification of the ATp-KR didomain. The expected 1.40-kb fragment was amplified with primers ATp1 and KRp.
Using the genomic DNA of S. nanchangensis NS3226 as the template, an expected 1.40-kb DNA fragment was amplified (Fig. 3D), cloned, and sequenced. The sequences of all the cloned 1.40-kb fragments are identical and show high homology with that of aveA1. Based on this newly obtained sequence, a pair of primers, meiAT-P1 and meiAT-P2, was designed and used to screen the whole genomic library. Four fosmids (1F1, 14E7, 14G1, and 20G1) and one cosmid (10D8) were identified from the genomic library (Fig. 2A).
In order to check whether the newly cloned region was involved in meilingmycin biosynthesis, an 18.60-kb DNA fragment containing ATp-KR was replaced by the apramycin-resistant determinant aac(3)IV, which generated the mutant HYL23 (Fig. 4A). Allelic replacement of the 18.6-kb region in the HYL23 mutant was confirmed by Southern hybridization (Fig. 4B), and LC-MS analysis of the fermentation broth of HYL23 revealed that meilingmycin production is completely abolished in this mutant (Fig. 4C).
Chromosome walking and sequence analysis of a newly cloned 80-kb region.
In order to connect the previously cloned 103-kb region with this ATp-KR-containing region, chromosome walking was initiated from the right end of 2G6 and resulted in the cloning of a continuous 180-kb region (Fig. 2A). In total, a 185,250-bp DNA sequence was obtained, including the previously sequenced region.
The amplified ATp-KR fragment was found to be located in a PKS gene named meiA1, whose encoded protein MeiA1 shows significant homology with the aveA1-encoded enzyme AVES1 in avermectin biosynthesis (52% identity/63% similarity). MeiA1 contains a loading module (AT-ACP) with an atypical IPAH motif in the AT domain, extension module 1 (KS-ATp-KR-ACP), and extension module 2 (KS-ATa-DH-ER-KR-ACP), with its deduced product similar to the C-21 to C-26 portion of meilingmycin backbone (Fig. 1B). Interestingly, it turns out that the distance between meiR and meiA1 is 55,169 bp, and 46 ORFs (orf24 to orf69) are located in this region (Fig. 2B; see also Table S1 in the supplemental material). Moreover, a C-5-O-methyltransferase gene, meiD, is localized downstream of meiA1 and homologous with the C-5-O-methyltransferase gene for milbemycin biosynthesis in Streptomyces griseochromogenes (86% identity/91% similarity) and with aveD (58% identity/69% similarity). The encoded product of meiD is probably responsible for the methylation of the hydroxyl group on C-5 of the minor components, meilingmycins D and E.
The 55-kb region between meiR and meiA1 is not involved in meilingmycin biosynthesis.
Functional analysis of orf24-orf69 in relation to meilingmycin biosynthesis was performed through an ordered series of large-fragment deletions (see the supplemental material). Genes orf24-orf32, orf31-orf55, orf56-orf59, and orf60-orf69 were replaced by oriT-aac(3)IV, and meilingmycin productivity is not obviously changed in the corresponding mutants of HYL25, HYL22, HYL26, and HYL27, respectively (Fig. 4C). Therefore, all 46 ORFs (orf24-orf69) of the 55-kb region were proved not to be required for meilingmycin production, which physically divides the meilingmycin biosynthetic genes into two separate gene clusters. Furthermore, sequence analysis reveals that the previous deletion of the 18.6-kb ATp-KR-containing region in HYL23 covers orf60-meiA1 (Fig. 4C), and the exclusion of orf60-orf69 in the meilingmycin biosynthesis confirms the unambiguous involvement of meiA1 in meilingmycin biosynthesis.
Gene meiD encodes a methyltransferase for the biosynthesis of meilingmycins D and E.
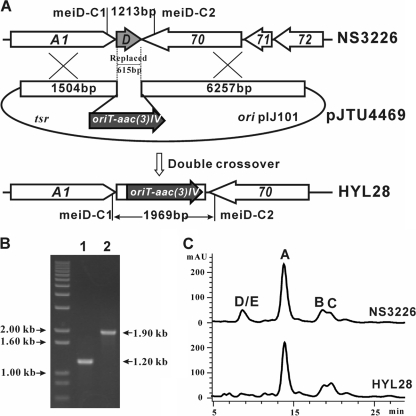
The involvement of meiD in meilingmycin production was proved through gene replacement. Using the ReDirect Technology (10), an internal 615-bp DNA region of meiD was replaced by the oriT-aac(3)IV cassette, which generated the mutant HYL28 (Fig. 5A). Allelic replacement of meiD in the HYL28 mutant was confirmed by PCR amplification with primers meiD-C1 and meiD-C2. An expected 1.90-kb product was amplified from the genomic DNA of HYL28, whereas a 1.20-kb expected product was amplified from the genomic DNA of wild-type NS3226 (Fig. 5B). Analyzed by LC-MS, the peaks of meilingmycins D and E disappeared in the fermentation extract of HYL28, while meilingmycins A, B, and C were still produced in this mutant (Fig. 5C). Thus, meiD proved to be responsible for the methylation of the hydroxyl group on C-5 of meilingmycins D and E.
FIG. 5.
Gene replacement of meiD. (A) Schematic representation of the replacement of a 615-bp internal fragment of meiD by the 1.40-kb oriT-acc(3)IV cassette. Wild-type NS3226 should yield a 1.20-kb PCR-amplified product, and mutant HYL28 should yield a 1.90-kb product by using primers meiD-C1 and meiD-C2. (B) Confirmation of the meiD-deleted mutant via PCR amplification. (C) HPLC analysis of wild-type NS3226 and meiD mutant HYL28. The peaks of meilingmycins D and E disappeared in mutant HYL28, and meilingmycins A, B, and C are still produced by the mutant.
Moreover, meiD also serves as the right boundary of the meilingmycin biosynthetic gene clusters, because the deletion of the 10 ORFs (orf70-orf79) downstream of meiD has no effect on the meilingmycin production in mutant HYL29 (Fig. 4C) (see the supplemental material).
DISCUSSION
Genes involved in the biosynthesis of single secondary metabolites are usually clustered, which could be considered a general principle and serves as the guideline for the cloning of new biosynthetic gene clusters. However, more and more biosynthetic genes are not found closely located, making it very difficult to clone all required genes and to perceive the complete biosynthetic mechanisms. It has been reported that the genes for ansamitocin (37), clavam (33), and moenomycin A (22) biosyntheses are separated into two or three parts on the chromosome. For example, cluster I and cluster II of the ansamitocin biosynthetic genes are separated by a 30-kb intercluster region. However, only three genes required for the synthesis of the 3-amino-5-hydroxy-benzonate (AHBA) starter unit are present in cluster II, and all other essential genes, including the PKS machinery, are located in cluster I (37). Through a series of large-fragment deletions, the meilingmycin biosynthetic genes, especially the PKSs, were proved to be divided into two clusters by a 55-kb region, with meiA1 encoding the first three modules located in one cluster and meiA2, meiA3, and meiA4 located in the other cluster. The separation of antibiotic biosynthetic gene clusters may have resulted from chromosome rearrangement and/or gene insertions.
Attempts to clone the missing part of the meilingmycin biosynthetic gene cluster with different strategies had been tried. First, four of the eight previously identified PKS contigs were deleted, and none of the four mutants lost meilingmycin productivity (data not shown) (31, 32). Second, routine chromosome walking was also performed on both sides of the previously cloned meilingmycin biosynthetic genes, and subsequent deletions on both sides failed to abolish meilingmycins biosynthesis in the mutants. Third, based on the saturated C-22—C-23 in meilingmycins, a pair of degenerate primers was designed, according to the conserved motifs of PKS ER domains. Unfortunately, too many positive cosmids were identified from the genomic library, making it impossible to identify the rest of the meilingmycin gene cluster through numerous deletions.
The strategy of amplifying the ATp-KR didomain through PCR utilizes the presence of a hydroxyl group at C-25, usually catalyzed by the KR domain of the downstream module, and the presence of a methyl group at C-24, reflecting the nature of the AT domain of the same module that recruits a methylmalonyl-CoA extender unit. Therefore, the first extension module of mei PKS is supposed to have ATp and KR domains. According to the evolutionary hypothesis on PKS, the KR domain is the first reduction domain inserted between AT and ACP to form a KS-AT-KR-ACP assembly (16). The predicted length between either the YASH motif of ATa or the HAFH motif of ATp and the NADP(H) binding motif of KR is 1.40 kb (Fig. 4). For modules with DH domains, the DH domains are usually present between the AT and KR domains to form a KS-AT-DH-KR-ACP assembly, and the amplified lengths between the AT and KR should be increased to about 2.10 kb. Furthermore, for modules with complete reductions, the ER domains were generally inserted between DH and KR domains as a late event in PKS evolution to form a KS-AT-DH-ER-KR-ACP assembly, which is supposed to give an amplified 3.0-kb large fragment with the pair of AT-specific and KR-specific primers. Through amplification of ATp-KR and ATp-DH-KR regions, the lasalocid A biosynthetic gene clusters was successfully cloned in our group (H. Wang and L. Bai, unpublished data).
Since most of the modules in one polyketide biosynthetic pathway have evolved from an ancestor module (16), the specificity for amplifying the correct ATp-KR region could be highly increased if the degenerate primers are designed based on available domains/modules from the same PKS machinery. In our experiment, a nondegenerate ATp-specific forward primer was designed according to the alignment of mei-AT6p, mei-AT7p, and mei-AT9p, and a degenerate KR-specific reverse primer was designed based on the alignment of mei-KR4 to mei-KR9 and mei-KR12 (Fig. 4).
Due to the presence of an atypical IPAH motif in the AT domain of the loading module, the meilingmycin biosynthesis is initiated with either malonyl-CoA (meilingmycins A, B, D, and E) or methylmalonyl-CoA (meilingmycin C) and proceeded by condensation of seven malonyl-CoA and five methylmalonyl-CoA extender units (Fig. 6). In the biosynthesis of avermectin, AveC was required for the dehydration of C-22—C-23 to yield the double bond between C-22 and C-23 with an unknown mechanism (28). MeiC, the homolog of AveC in the mei cluster, was proved to be essential for meilingmycin biosynthesis through gene inactivation (see Fig. S2 in the supplemental material) and may be involved in the complete reduction of the keto group on C-23 to generate the saturated bond between C-22 and C-23.
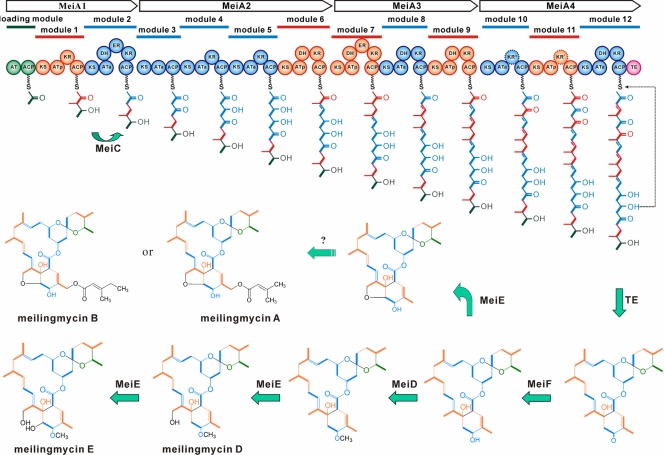
FIG. 6.
Proposed biosynthetic pathway to meilingmycins. R, H or CH3; green circles, loading module or starter units; orange circles, methylmalonyl-CoA-specific modules or acetate-derived extender units; blue circles, malonyl-CoA-specific modules or propionate-derived extender units; pink circle, the thioesterase (TE) domain; KR*, nonfunctional ketoreductase domain deduced from meilingmycin structure; KRI, inactive ketoreductase domain; AT, acyltransferase; ATa, malonyl-CoA-specific AT; ATp, methylmalonyl-CoA-specific AT; ACP, acyl carrier protein; DH, dehydratase; ER, enoyl reductase; KR, ketoreductase; KS, ketosynthase.
After being released and cyclized by the TE domain of MeiA4, the polyketide chain of meilingmycin is probably oxidized by the cytochrome P450 hydroxylase MeiE to generate a furan ring between C-8 and C-6 (meilingmycins A, B, and C), a hydroxyl group on C-8 (meilingmycin D), or two hydroxyl groups on both C-8 and C-6 (meilingmycin E). meiE could restore avermectin productivity in the aveE mutant (J. Qiu and L. Bai, unpublished data). However, AveE from the avermectin cluster catalyzes only the formation of a furan ring between C-8 and C-6 (13). Subsequently, the C-5 keto group is reduced to hydroxyl by the ketoreductase MeiF, an AveF homolog, after which the meilingmycin biosynthetic pathway is presumably diverged by either methylation or hydroxylation/acylation. Surprisingly, the inactivation of meiF abolishes meilingmycin productivity, and so far, no new derivative has been detected from the mutant through LC-MS (see Fig. S3 in the supplemental material). For the intermediates without the furan ring, the hydroxyl on C-5 was methylated by the methyltransferase MeiD to generate minor components (meilingmycins D and E). On the other hand, through hydroxylation/acylation at C-27 still to be characterized, major components of meilingmycins carrying a 3-methylbut-2-enoyl (meilingmycins A and C) or a 3-methylpent-2-enoyl side chain (meilingmycin B) are formed. However, the putative genes for the synthesis of these two side chains have not been found in the 185.0-kb sequenced region, which suggests that these genes are located in other regions of the chromosome.
Supplementary Material
Acknowledgments
We thank Tobias Kieser and Shuangjun Lin for critical reading of the manuscript and helpful discussion.
This work received financial support from the National Science Foundation of China, the Ministry of Science and Technology (973 and 863 Programs), the Shanghai Municipal Council of Science and Technology, and Shanghai Leading Academic Discipline Project B203.
Footnotes
Published ahead of print on 26 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ansari, M. Z., G. Yadav, R. S. Gokhale, and D. Mohanty. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405-W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burg, R. W., B. M. Miller, E. E. Baker, J. Birnbaum, S. A. Currie, R. Hartman, Y. L. Kong, R. L. Monaghan, G. Olson, I. Putter, J. B. Tunac, H. Wallick, E. O. Stapley, R. Oiwa, and S. Omura. 1979. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caffrey, P. 2003. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem 4:654-657. [DOI] [PubMed] [Google Scholar]
- 5.Chan, Y. A., A. M. Podevels, B. M. Kevany, and M. G. Thomas. 2009. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 26:90-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, S., X. Huang, X. Zhou, L. Bai, J. He, K. J. Jeong, S. Y. Lee, and Z. Deng. 2003. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem. Biol. 10:1065-1076. [DOI] [PubMed] [Google Scholar]
- 7.Del Vecchio, F., H. Petkovic, S. G. Kendrew, L. Low, B. Wilkinson, R. Lill, J. Cortes, B. A. Rudd, J. Staunton, and P. F. Leadlay. 2003. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 30:489-494. [DOI] [PubMed] [Google Scholar]
- 8.Donadio, S., and L. Katz. 1992. Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111:51-60. [DOI] [PubMed] [Google Scholar]
- 9.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 10.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haydock, S. F., J. F. Aparicio, I. Molnar, T. Schwecke, L. E. Khaw, A. Konig, A. F. Marsden, I. S. Galloway, J. Staunton, and P. F. Leadlay. 1995. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374:246-248. [DOI] [PubMed] [Google Scholar]
- 12.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2498. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda, H., H. Kotaki, and S. Omura. 1987. Genetic studies of avermectin biosynthesis in Streptomyces avermitilis. J. Bacteriol. 169:5615-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Omura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U. S. A. 96:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 16.Jenke-Kodama, H., T. Borner, and E. Dittmann. 2006. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis. PLoS Comput. Biol. 2:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Centre, Colney, Norwich, England.
- 18.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacNeil, T., K. M. Gewain, and D. J. MacNeil. 1993. Deletion analysis of the avermectin biosynthetic genes of Streptomyces avermitilis by gene cluster displacement. J. Bacteriol. 175:2552-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malpartida, F., and D. A. Hopwood. 1984. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature 309:462-464. [DOI] [PubMed] [Google Scholar]
- 21.Migita, A., M. Watanabe, Y. Hirose, K. Watanabe, T. Tokiwano, H. Kinashi, and H. Oikawa. 2009. Identification of a gene cluster of polyether antibiotic lasalocid from Streptomyces lasaliensis. Biosci. Biotechnol. Biochem. 73:169-176. [DOI] [PubMed] [Google Scholar]
- 22.Ostash, B., A. Saghatelian, and S. Walker. 2007. A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem. Biol. 14:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petkovic, H., A. Sandmann, I. R. Challis, H. J. Hecht, B. Silakowski, L. Low, N. Beeston, E. Kuscer, J. Garcia-Bernardo, P. F. Leadlay, S. G. Kendrew, B. Wilkinson, and R. Muller. 2008. Substrate specificity of the acyl transferase domains of EpoC from the epothilone polyketide synthase. Org. Biomol. Chem. 6:500-506. [DOI] [PubMed] [Google Scholar]
- 25.Reid, R., M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. R. Hutchinson, and R. McDaniel. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72-79. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schwecke, T., J. F. Aparicio, I. Molnar, A. Konig, L. E. Khaw, S. F. Haydock, M. Oliynyk, P. Caffrey, J. Cortes, J. B. Lester, et al. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. U. S. A. 92:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stutzman-Engwall, K., S. Conlon, R. Fedechko, F. Kaczmarek, H. McArthur, A. Krebber, Y. Chen, J. Minshull, S. A. Raillard, and C. Gustafsson. 2003. Engineering the aveC gene to enhance the ratio of doramectin to its CHC-B2 analogue produced in Streptomyces avermitilis. Biotechnol. Bioeng. 82:359-369. [DOI] [PubMed] [Google Scholar]
- 29.Sun, Y., X. He, J. Liang, X. Zhou, and Z. Deng. 2009. Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl. Microbiol. Biotechnol. 82:303-310. [DOI] [PubMed] [Google Scholar]
- 30.Sun, Y., X. Zhou, H. Dong, G. Tu, M. Wang, B. Wang, and Z. Deng. 2003. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 10:431-441. [DOI] [PubMed] [Google Scholar]
- 31.Sun, Y., X. Zhou, J. Liu, K. Bao, G. Zhang, G. Tu, T. Kieser, and Z. Deng. 2002. ‘Streptomyces nanchangensis,’ a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361-371. [DOI] [PubMed] [Google Scholar]
- 32.Sun, Y., X. Zhou, G. Tu, and Z. Deng. 2003. Identification of a gene cluster encoding meilingmycin biosynthesis among multiple polyketide synthase contigs isolated from Streptomyces nanchangensis NS3226. Arch. Microbiol. 180:101-107. [DOI] [PubMed] [Google Scholar]
- 33.Tahlan, K., H. U. Park, and S. E. Jensen. 2004. Three unlinked gene clusters are involved in clavam metabolite biosynthesis in Streptomyces clavuligerus. Can. J. Microbiol. 50:803-810. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, S., H. Miyaoka, K. Tanaka, R. Enokita, and T. Okazaki. 1993. Milbemycins alpha 11, alpha 12, alpha 13, alpha 14 and alpha 15: a new family of milbemycins from Streptomyces hygroscopicus subsp. aureolacrimosus. Taxonomy, fermentation, isolation, structure elucidation and biological properties. J. Antibiot. (Tokyo) 46:1364-1371. [DOI] [PubMed] [Google Scholar]
- 35.Walton, L. J., C. Corre, and G. L. Challis. 2006. Mechanisms for incorporation of glycerol-derived precursors into polyketide metabolites. J. Ind. Microbiol. Biotechnol. 33:105-120. [DOI] [PubMed] [Google Scholar]
- 36.Yadav, G., R. S. Gokhale, and D. Mohanty. 2003. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328:335-363. [DOI] [PubMed] [Google Scholar]
- 37.Yu, T. W., L. Bai, D. Clade, D. Hoffmann, S. Toelzer, K. Q. Trinh, J. Xu, S. J. Moss, E. Leistner, and H. G. Floss. 2002. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. U. S. A. 99:7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, C., J. Ju, S. D. Christenson, W. C. Smith, D. Song, X. Zhou, B. Shen, and Z. Deng. 2006. Utilization of the methoxymalonyl-acyl carrier protein biosynthesis locus for cloning the oxazolomycin biosynthetic gene cluster from Streptomyces albus JA3453. J. Bacteriol. 188:4142-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.