Abstract
The anaerobic, thermophilic cellulolytic bacterium Clostridium thermocellum is known for its elaborate cellulosome complex, but it also produces a separate free cellulase system. Among the free enzymes, the noncellulosomal enzyme Cel9I is a processive endoglucanase whose sequence and architecture are very similar to those of the cellulosomal enzyme Cel9R; likewise, the noncellulosomal exoglucanase Cel48Y is analogous to the principal cellulosomal enzyme Cel48S. In this study we used the designer cellulosome approach to examine the interplay of prominent cellulosomal and noncellulosomal cellulases from C. thermocellum. Toward this end, we converted the cellulosomal enzymes to noncellulosomal chimeras by swapping the dockerin module of the cellulosomal enzymes with a carbohydrate-binding module from the free enzyme analogues and vice versa. This enabled us to study the importance of the targeting effect of the free enzymes due to their carbohydrate-binding module and the proximity effect for cellulases on the designer cellulosome. C. thermocellum is the only cellulosome-producing bacterium known to express two different glycoside hydrolase family 48 enzymes and thus the only bacterial system that can currently be used for such studies. The different activities with crystalline cellulose were examined, and the results demonstrated that the individual chimeric cellulases were essentially equivalent to the corresponding wild-type analogues. The wild-type cellulases displayed a synergism of about 1.5-fold; the cellulosomal pair acted synergistically when they were converted into free enzymes, whereas the free enzymes acted synergistically mainly in the wild-type state. The targeting effect was found to be the major factor responsible for the elevated activity observed for these specific enzyme combinations, whereas the proximity effect appeared to play a negligible role.
Cellulose, the major plant cell wall structural polysaccharide, is an excellent potential energy source for microbial growth. The cellulases that hydrolyze the cellulose polymer chains can occur in two alternative states. “Free” (noncellulosomal) secreted cellulases usually contain a carbohydrate-binding module (CBM) as an integral part of the polypeptide chain for guiding the catalytic module to the substrate (11, 36, 47). Conversely, the “cellulosomal” cellulases are organized in a discrete multienzyme complex called the cellulosome (2, 17, 18, 35). The cellulosome complex is assembled by the high-affinity interaction between two complementary modules; a single dockerin module borne by each cellulase binds one of multiple cohesin modules which are located on a noncatalytic subunit called scaffoldin (8, 45). The targeting of the entire complex to the cellulose substrate is mediated by a CBM, which is also borne by the scaffoldin subunit. Otherwise, the free and cellulosomal enzymes contain very similar types of catalytic modules.
The cellulosome was first discovered in the anaerobic, thermophilic cellulolytic bacterium Clostridium thermocellum in 1983 (4, 34) and was studied and characterized extensively. This system is considered the most efficient of all cellulase systems for cellulose degradation due to the organization of the enzymes into a complex that “concentrates” them together on given sites of the cellulosic substrate and facilitates stronger synergism among the catalytic units. Indeed, the incorporation of dockerin-bearing cellulases into artificial designer cellulosomes (7) was shown to induce synergism between cellulases via targeting to the substrate or due to the proximity of the cellulases in the complex (14, 20, 22, 38). The designer cellulosome concept was used to bind specifically chimeric dockerin-bearing enzymes to artificial scaffoldin with matching cohesins from different cellulosomal species and a CBM (15, 16, 20-22, 38, 39).
The various free and cellulosomal cellulases are categorized into numerous families of glycoside hydrolases (GHs) based on their amino acid sequences, resulting three-dimensional structures, and modes of activity (13). Cellulases that attack the cellulose chain in different ways have the potential to degrade cellulose synergistically. A combination of family 48 and family 9 glycoside hydrolases results in “true” cellulolytic activity, i.e., solubilization of crystalline cellulose substrates. Moreover, these two families of cellulases are particularly well represented in the cellulosome systems of the known cellulosomal bacteria (6). Family 48 exoglucanases are essential cellulosomal components in bacterial cellulase systems, yet there is only a single member in nearly every cellulosomal microorganism. C. thermocellum is the only organism known so far with two members; one cellulosomal enzyme contains a dockerin (40, 48), and a second free noncellulosomal enzyme contains a CBM (10). Two of the most prominent C. thermocellum cellulosomal enzymes, Cel9R and Cel48S, have an important role in the C. thermocellum cellulosome. Cel48S is the most prominent enzyme in C. thermocellum, and Cel9R is a processive endoglucanase and is among the most prevalent processive family 9 GHs (19, 27, 43, 46, 52, 53). Both of these enzymes act in a processive manner to cleave cellulose. On the other hand, Cel9I and Cel48Y are two noncellulosomal cellulases of C. thermocellum and are analogues of the cellulosomal Cel9R and Cel48S enzymes, which have been shown to hydrolyze crystalline cellulose synergistically (10, 26, 29). These two noncellulosomal enzymes may be part of a second, “true” soluble cellulase system in C. thermocellum that complements the cellulosome, although they are both expressed at limited levels during growth of the bacterium (B. Raman, personal communication). These four analogues enabled us to compare cellulosomal and noncellulosomal cellulases which originate from the same microorganism.
In this study we created a set of wild-type and converted chimeric pairs of enzymes in order to examine the interplay of cellulosomal versus noncellulosomal cellulases in C. thermocellum. The converted chimeric enzymes were created by swapping the dockerin and the CBM of the cellulosomal and free enzymes, respectively, belonging to each family. Combinations of different pairs of enzymes were then assayed to determine their capacities to degrade a model crystalline cellulose substrate, as part of a designer cellulosome system or as soluble enzymes, in order to examine the advantage of each system over the other system, to determine whether there is a preferred mode of activity, and to address the feasibility of converting the enzymatic mode of action.
MATERIALS AND METHODS
Cloning of wild-type C. thermocellum enzymes.
The genes encoding wild-type enzymes (cellulosomal enzymes Cel48S and Cel9R and noncellulosomal enzymes Cel48Y and Cel9I) were cloned by PCR from C. thermocellum genomic DNA with primers that allow their insertion into either the pET21a or pET28a vector. PCRs were performed using ABgene ReddyMix ×2 (Advanced Biotechnologies Ltd., Epsom, United Kingdom). DNA samples were purified using a HiYield gel-PCR fragment extraction kit (Real Biotech Corporation, RBC, Banqiao City, Taiwan). All constructs were designed to contain a His tag for subsequent purification steps (see Table S1 in the supplemental material).
Cloning of the converted chimeric proteins.
Chimeric enzyme constructs were assembled from modules (catalytic modules, dockerins, CBMs) from the wild-type vectors by using PCR amplification. PCR was also used for insertion of restriction sites and preservation of reading frames in the new constructs (see Table S2 in the supplemental material). In most cases, the module (CBM or dockerin) together with its native adjacent linker was fused to the desired catalytic module. In the only exception, the CBM of Cel48Y was transferred to the catalytic module of Cel48S together with a truncated 20-residue linker instead of the original 102-residue linker. The shorter linker was used for construction of the converted chimera *48S-CBM due to technical difficulties in the expression and purification of the longer, native version of the particularly lengthy linker. For comparison, in the wild-type (cellulosomal) enzyme Cel48S, the catalytic module is separated from its C-terminal dockerin by a linker consisting of only 5 residues.
Protein expression and purification.
Escherichia coli BL21(DE3) cells were transformed with the desired plasmid and grown at 37°C in Luria-Bertani broth supplemented with 50 μg/ml kanamycin or 100 μg/ml ampicillin (Sigma-Aldrich Chemical Co., St. Louis, MO) and 2 mM CaCl2 for cultures used to prepare dockerin-containing enzymes to an A600 of ∼1. Isopropyl-1-thio-β-d-galactoside (IPTG) (Fermentas UAB, Vilnius, Lithuania) was added to a final concentration of 0 to 1 mM based on the results of predetermined optimization experiments. Cultures were then grown at 16°C overnight or at 37°C for 3 h.
Cells were harvested by centrifugation (4,000 × g, 15 min, 4°C), resuspended in Tris-buffered saline (TBS) (137 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl; pH 7.4) supplemented with 5 mM imidazole (Merck KGaA, Darmstadt, Germany) and protease inhibitor cocktail (1 mM phenylmethylsulfonyl fluoride [PMSF], 0.4 mM benzamidine, and 0.06 mM benzamide obtained from Sigma-Aldrich) and disrupted by sonication. The sonicate was heated for 15 to 30 min at 50 to 60°C and then centrifuged (20,000 × g, 30 min, 4°C). The supernatant fluids were mixed with ∼5 ml Ni-nitrilotriacetic acid (NTA) supplemented with 5 to 10 mM imidazole for 1 h on a 20-ml Econo-pack column on a rotator at 4°C (batch purification system). The column was then washed by using gravity flow with 50 to 100 ml wash buffer (TBS with 15 mM imidazole). Elution was performed by using TBS with 100 mM imidazole and then TBS with 250 mM imidazole.
Fractions (2 ml) were collected and analyzed by SDS-PAGE (10% acrylamide). The fractions containing the purified proteins were pooled, and CaCl2 (5 mM) was added to dockerin-containing enzymes, as was the protease inhibitor cocktail, which was added to all enzymes. The conditions used for overexpression of the wild-type and chimeric proteins are summarized in Table 1. Cel48S and *48S-CBM tended to form inclusion bodies. No attempts were made to renature these cellulases; the proteins were isolated directly from the supernatant phase using 10 liters of culture fluid.
TABLE 1.
Expression conditions, solubilities, and yields of the recombinant proteins used in this study
| Plasmid | Recombinant protein | Expression conditions |
Solubility | Final yield (mg protein/ liter culture) | ||||
|---|---|---|---|---|---|---|---|---|
| Temp (°C) | Time (h) | IPTG concn (mM) | Calcium (2 mM) | Length of heating (min)a | ||||
| pET21a | Cel9I (9I-CBM) | 37 | 3 | 0.2 | − | 30 | High | 24 |
| pET21a | Cel48Y (48Y-CBM) | 16 | 18 | 1 | − | 30 | High | 40 |
| pET21a | *9I-b | 37 | 3 | 0.2 | + | 30 | High | 20 |
| pET21a | *48Y-t | 37 | 3 | 0.2 | + | 30 | High | 10 |
| pET28a | Cel48S (48S-t) | 37 | 3 | 0.2 | + | 20 | Low | 0.5 |
| pET28a | 9R-b | 37 | 3 | 0.2 | + | 15 | High | 4 |
| pET21a | *48S-CBM | 20 | 18 | 0 | − | 30 | Low | 2.3 |
| pET21a | *9R-CBM | 37 | 3 | 0.2 | − | 30 | High | 17 |
Most proteins were heated at 60°C; the only exception was Cel48S, which was heated at 50°C.
Scaffoldins Scaf·B, Scaf·T, and Scaf·BT were expressed and purified using phosphoric acid-swollen cellulose (PASC) and a previously described method (28). In brief, following expression and sonication of each scaffoldin, the supernatant fluids were incubated with PASC for 1 h at 37°C to allow binding via the CBM. The matrix was washed three times with TBS (pH 7.4) containing 1 M NaCl and three times with TBS without added salt. The protein was eluted with 1% (vol/vol) triethylamine and neutralized with 1 M 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.5).
Protein concentrations were estimated by using the absorbance at 280 nm. The extinction coefficient was determined based on the known amino acid composition of each protein using the ProtParam tool on the EXPASY server (http://www.expasy.org/tools/protparam.html) (24, 25). Some proteins were concentrated using Vivaspin 5,000-molecular-weight cutoff concentrators. Proteins were stored in 50% (vol/vol) glycerol at −20°C.
Specificity of the enzyme-borne dockerins and their cohesin targets.
The procedure of Barak et al. (1) was used, with modifications to determine the specificity of the enzyme-borne dockerins and their cohesin targets. Maxisorp enzyme-linked immunosorbent assay (ELISA) plates (Nunc A/S, Roskilde, Denmark) were coated with each of the dockerin-containing enzymes used in this study and then interacted with 100 ng/μl of its single-cohesin scaffoldin (Scaf·B or Scaf·T) counterpart. Rabbit anti-CBM (diluted 1:3,000 in blocking buffer) was employed as the primary antibody for detection of the interaction.
Nondenaturing gel electrophoresis.
Samples (final protein concentration, 2 μM) were diluted in TBS supplemented with 10 mM CaCl2 and 0.05% Tween 20 and incubated at 37°C for 2 h. Nondenaturing sample buffer (192 mM glycine, 25 mM Tris) was added, and 15 μl/lane was subjected to PAGE (9% acrylamide gels), using a Bio-Rad power pack 300.
Sequence analysis.
Sequence pairwise alignment was performed using Align (global) (http://www.ebi.ac.uk/Tools/emboss/align/).
Enzyme assays.
Enzymatic activity was assayed using Avicel as a model insoluble crystalline cellulose substrate and was determined by measuring the reducing sugars released by the dinitrosalicylic acid (DNS) method (37). A typical assay mixture consisted of buffer (100 mM acetate buffer [pH 5.0], 24 mM CaCl2, 4 mM EDTA) plus 0.5 μM enzyme as determined by preliminary calibration of the linear range of enzyme activity. The reaction was initiated by addition of 40 μl of Avicel (Sigma-Aldrich) from a 10% (wt/vol) stock suspension in 100 mM acetate buffer to a 200-μl (total volume) reaction mixture (final concentration of Avicel, 2% [wt/vol]). The reaction was carried out in triplicate for 17 h in a 60°C shaker-incubator. The reaction was terminated by immersing the sample tubes in ice water. Samples were centrifuged at the maximum speed (20,800 × g, 10 min) to remove the substrate. DNS (150 μl) was added to 100 μl supernatant fluid, and the tubes were then boiled for 10 min. The absorbance at 540 nm was measured, and the specific activity was calculated using a glucose standard curve and was expressed in moles of glucose equivalents per mole of enzyme per minute. When the activity of a dockerin-containing enzyme with its matching scaffoldin was examined, 2 h of incubation at 37°C using equimolar quantities of the binding components (without the substrate) preceded the assay. Each assay was repeated at least three times.
Relative activity.
Due to the large number of systems that were tested in this study, separate experiments with selected common enzyme samples were performed to obtain between-experiment standards. Thus, the amount of soluble sugars released by the enzymes was normalized by comparison to the activity of the WT-free Cel9I (see below) or the activity of the WT-free pair (9I-CBM plus 48Y-CBM), as shown in Fig. 2 to 4.
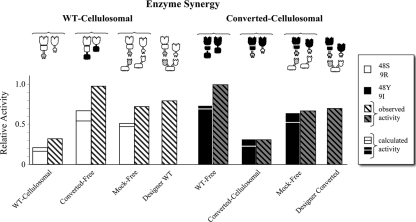
FIG. 2.
Comparative degradation of Avicel by individual family 48 and family 9 enzymes. Cellulosomal enzymes were converted to the free mode, and free enzymes were converted to the cellulosomal mode. The compositions of the reaction mixtures in this figure and Fig. 3 and 4 are indicated by diagrams above the bars. A white structure indicates a cellulosome-derived enzyme, and a black structure indicates a non-cellulosome-derived enzyme. Open bars, 48S and 9R catalytic modules in either “WT-cellulosomal,” “mock-free” or “converted-free” mode; filled bars, 48Y-CBM and 9I-CBM catalytic modules in either the “WT-free,” “converted-cellulosomal,” or “mock-free” mode. For definitions of enzyme systems see Table 2. The results of two independent experiments are shown. Triplicate reactions were carried out, and standard deviations are indicated by error bars. The relative activity was determined by comparison with the activity of the WT-free enzyme 9I-CBM, as described in Materials and Methods.
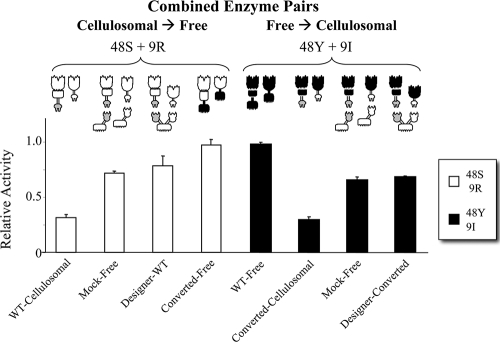
FIG. 4.
Synergism between combinations of family 48 and family 9 enzymes. (Left side) Synergy of the cellulosomal enzymes 48S and 9R for catalytic modules in “WT-cellulosomal,” “mock-free,” or “designer-WT” mode and converted into “converted-free” mode. The open bars indicate the calculated sums of activities of the individual enzymes; the contribution of the family 48 enzyme (top section) is considerably less than that of the family 9 enzyme (bottom section) for each of the pairs. The striped bars indicate the observed activity experimentally obtained for the combined enzyme systems. (Right side) Synergy of the free enzymes 48Y and 9I for catalytic modules in a “WT-free” mode and converted into cellulosomal modes, including “converted-cellulosomal,” “mock-free,” and “designer-converted.” The solid filled bars indicate the calculated sums of activities of the individual enzymes (top section, family 48 enzyme; bottom section, family 9 enzyme). For definitions of enzyme systems see Table 2. Values for single enzyme activities and pairs of enzymatic activities are shown in Table S3 in the supplemental material. Triplicate reactions were carried out. The relative activity was determined by comparison with the activity of the WT-free enzymes 48Y-CBM and 9I-CBM, as described in Materials and Methods.
RESULTS
Cloning and expression of recombinant proteins.
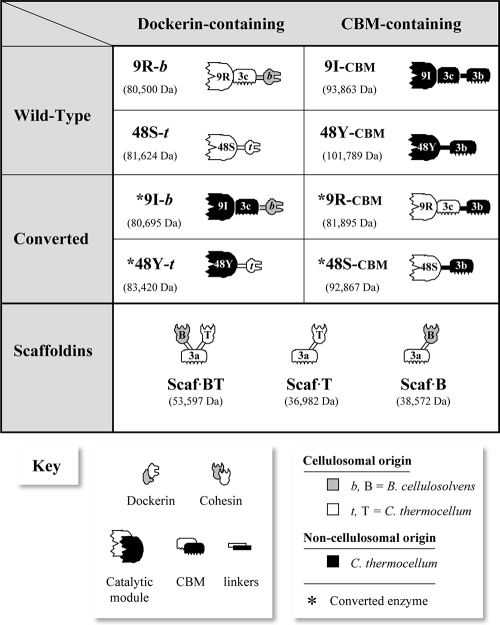
Genes encoding the following four wild-type enzymes were cloned and expressed from the genomic sequence of C. thermocellum: noncellulosomal CBM-bearing cellulases Cel48Y and Cel9I (designated 48Y-CBM and 9I-CBM, respectively) and the analogous cellulosomal dockerin-bearing cellulases Cel48S and Cel9R (designated 48S-t, where t indicates the origin of the appended dockerin module, C. thermocellum). Cel9R dockerin was replaced with a dockerin module from the divergent species Bacteroides cellulosolvens. This step was crucial for subsequent assembly of the designer cellulosome. To distinguish this enzyme from the authentic wild-type Cel9R (which contained the original C. thermocellum dockerin), it was designated 9R-b, where b indicates the origin of the dockerin (Bacteroides).
Next, the four chimeric “converted” enzymes were cloned and expressed. The noncellulosomal enzyme 48Y-CBM was thus converted into a cellulosomal enzyme and the cellulosomal enzyme 48S-t was converted into a noncellulosomal enzyme by swapping the C-terminal CBM and dockerin modules. The two converted enzymes were designated *48Y-t and *48S-CBM, respectively, where the asterisk indicates a converted enzyme (converted either from cellulosomal to noncellulosomal [in which the native dockerin was replaced by a cellulose-binding CBM] or vice versa). Similarly, the noncellulosomal enzyme 9I-CBM was converted into a cellulosomal enzyme and the cellulosomal enzyme 9R-b was converted into a noncellulosomal enzyme by swapping the C-terminal CBM3b and dockerin modules. The resulting converted enzymes were designated *9I-b and *9R-CBM.
The various proteins were expressed in E. coli and purified by batch purification on an Ni-NTA column using an added His tag. All of the purified recombinant proteins produced a single major band in SDS-PAGE (data not shown), and in each case the mobility was consistent with the expected molecular mass. Schematic diagrams of the constructs designed in this study and of the wild-type and converted cellulases are shown in Fig. 1. The purification yields and relative solubilities of the proteins are shown in Table 1. The family 9 glycoside hydrolases used in this study contain an additional accessory module, designated CBM3c, fused on the C-terminal side of the catalytic module. Unlike the CBM3b module, this module has no role in targeting the enzyme to the cellulose substrate. The role of the CBM3c module is to feed single carbohydrate chains of the substrate into the catalytic site of the enzyme, hence facilitating processive cleavage of cellulose (23, 26, 31, 44). Consequently, the catalytic module and the adjacent CBM3c module were considered a single unit for the purpose of designing converted chimeric enzymes.
FIG. 1.
Schematic diagrams of the wild-type enzymes, chimeric enzymes, and chimeric scaffoldins that were used in this study. The modular notation, structure, and molecular mass of each protein are indicated. White indicates a C. thermocellum cellulosome-derived component; black indicates a C. thermocellum noncellulosomal component; and gray indicates a B. cellulosolvens cellulosome-derived component. In the modular notation of the enzymes, the numbers indicate the family of the catalytic domain; R, I, S, and Y indicate the original names of the enzymes (Cel9R, Cel9I, Cel48S, and Cel48Y, respectively); and b and t indicate the source of the dockerin module (B. cellulosolvens from ScaB and C. thermocellum from Cel48S, respectively). B and T indicate the source of the divergent cohesins (B, the first cohesin from ScaA of B. cellulosolvens; T, the third cohesin from CipA of C. thermocellum). An asterisk indicates a converted enzyme (an enzyme converted either from cellulosomal to noncellulosomal, in which the native dockerin was replaced by a cellulose-binding CBM, or vice versa).
Phylogenetic relationship of the enzyme components.
Phylogenetic analysis of the family 48 glycoside hydrolases was previously described by Xu et al. (51), and the cellulosomal enzyme 48S-t and noncellulosomal enzyme 48Y-CBM from C. thermocellum were shown to be divergent. Nevertheless, the two catalytic modules have 53.7% identity and 67.2% sequence similarity (http://www.ebi.ac.uk/Tools/emboss/align/) and have the same exoglucanase mode of action (10).
The sequences of the family 9 catalytic modules cluster according to the four themes of the family 9 cellulases. Of other cellulases, 7 of the C. thermocellum GH family 9 enzymes are represented in the analysis and are distributed in themes B, C, and D, demonstrating their prevalence and diversity. The similar modular architectures of the cellulosomal enzyme 9R-b and the noncellulosomal enzyme 9I-CBM are reflected by the close association of their catalytic module sequences with each other and with other theme B GH family 9 cellulases (33). There is 60.6% sequence identity and 72.7% similarity between the sequences of the catalytic modules. The similarity of the sequences indicates that there are similar catalytic mechanisms, although one cellulase belongs to the cellulosomal system and the other belongs to the noncellulosomal system.
The free cellulases, 48Y-CBM and Cel9I-CBM, each contain a family 3b CBM module, which targets the enzyme and thus delivers the catalytic module to the insoluble crystalline substrate. For conversion of the enzymes, the CBM3b module derived from a member of the appropriate GH family (family 48 or 9) was used. The family 3a CBM of the cellulosomal scaffoldin subunit of C. thermocellum (CipA) also binds to crystalline cellulose and targets the entire complement of cellulosomal enzymes to the cellulose substrate. Owing to its particularly strong binding to cellulosic substrates, this CBM was used in the designer scaffoldins described in this study to target the dockerin-bearing enzymes to their substrate. Family 3b and family 3a CBM sequences are grouped on close branches on a phylogenetic tree (33), although the CBM3a module of CipA is closer to other scaffoldin-borne CBMs.
In contrast to family 3a and family 3b CBMs that interact with the surface of crystalline cellulose and target the enzymes to the substrate, the family 3c CBM binds a single cellulose chain and directs it to the active site of the catalytic module of family 9 cellulases. This CBM is unique in that it is always associated with GH family 9 enzymes downstream of the catalytic modules. This alternative mode of interaction with cellulose is reflected in the sequences of CBM3c, which diverge from those of CBM3a and CBM3b. CBM3c is fused tightly to the catalytic module by numerous intermodular contacts to ensure proper orientation of the two modules, so they can be considered a single functional unit (12, 23, 26, 31, 44).
Specificity of the interaction of the enzyme-borne dockerin with its matching cohesin.
The specificity of the dockerin-bearing cellulases and their matching cohesins was examined semiquantitatively by performing a sensitive enzyme-linked affinity assay in microtiter plates (1). The enzymes with the C. thermocellum dockerin module (derived from Cel48S), 48S-t and *48Y-t, interacted exclusively with the matching cohesin (Scaf·T) and did not interact with the divergent cohesin (Scaf·B). Likewise, the enzymes with the B. cellulosolvens dockerin, 9R-b and *9I-b, interacted exclusively with the matching cohesin (Scaf·B) and did not interact with the nonmatching divergent cohesin (Scaf·T) (see Fig. S1 in the supplemental material).
The stoichiometry of the interaction between the pairs of dockerin-bearing cellulases (wild-type 48S-t and 9R-b and the converted enzymes *48Y-t and *9I-b) and the matching scaffoldin (Scaf·BT) was determined by nondenaturing PAGE (see Fig. S2 in the supplemental material). Equimolar concentrations of the single components, scaffoldin and wild-type enzymes 48S-t and 9R-b (see Fig. S2A, lanes 1, 2, and 4, respectively, in the supplemental material), resulted in a single major band. Equimolar mixtures of the scaffoldin with a single cellulase, 48S-t or 9R-b (lanes 3 and 5, respectively), or with both of the cellulases (lane 6) resulted in a stronger major band that was shifted from the bands of the single proteins, indicating that a complex was formed with complete interaction between the dockerin-bearing enzyme and its matching cohesin. Essentially the same pattern was observed in the nondenaturing gel containing the converted enzymes *48Y-t and *9I-b (see Fig. S2B in the supplemental material). Residual banding patterns were also observed that indicated the presence of contaminating or noninteracting components (e.g., dockerins that did not fold properly) in each enzyme preparation.
Definitions of enzyme systems.
The comparative degradation of Avicel by different combinations of enzyme systems was tested. The different systems are defined in Table 2. The wild-type cellulosomal dockerin-bearing enzymes 48S-t and 9R-b are designated “WT-cellulosomal” enzymes. In a similar manner, the wild-type free CBM-bearing enzymes 48Y-CBM and 9I-CBM are designated “WT-free” enzymes. After the dockerin modules were swapped with CBM modules, the converted chimeras *48S-CBM and *9R-CBM were designated “converted-free” enzymes, and *48Y-t and *9I-b were designated “converted-cellulosomal” enzymes, reflecting their new mode of action based on their modular composition. An asterisk indicates that an enzyme was converted to a different mode. Since preattachment of a dockerin-bearing enzyme to a single-cohesin scaffoldin targets the cellulase to the substrate via the CBM, hence mimicking a free mode, we designated these systems “mock-free.” The “mock-free” enzymes used in this study comprised either 48S-t or *48Y-t attached to the corresponding Scaf·T, as well as 9R-b or *9I-b attached to Scaf·B. Designer cellulosomes were assembled from the “WT-cellulosomal” or “converted-cellulosomal” enzymes and designated “designer-WT” (48S-t plus 9R-b plus Scaf·BT) or “designer-converted” (*48Y-t plus *9I-b plus Scaf·BT).
TABLE 2.
Definitions of enzyme systems used in this study
| System | Components |
|---|---|
| WT-cellulosomal | 48S-t + 9R-b |
| WT-free | 48Y-CBM + 9I-CBM |
| Converted-cellulosomal | *48Y-t + *9I-b |
| Converted-free | *48S-CBM + *9R-CBM |
| Mock-free (dockerin- containing enzyme + | |
| single-cohesin scaffoldin) | 48S-t or *48Y-t + Scaf·T, 9R-b or *9I-b + Scaf·B |
| Designer-WT (WT- cellulosomal + designer | |
| scaffoldin) | 48S-t + 9R-b + Scaf·BT |
| Designer-converted (converted-cellulosomal + | |
| designer scaffoldin) | *48Y-t + *9I-b + Scaf·BT |
Catalytic activity assays of family 48 and family 9 cellulases.
The individual enzyme activities were examined, as were their activities in a “mock-free” state (combined with the corresponding single-cohesin scaffoldin) (Fig. 2). All of the cellulases (i.e., wild-type, chimeric, and converted enzymes) were active on crystalline cellulose (Avicel). The cellulosomal enzymes 48S-t and 9R-b were converted into free enzymes by swapping their dockerin modules with the CBMs of their free analogues. All of the family 48 glycoside hydrolases exhibited very low, yet detectable, catalytic activity, as expected from the previously reported activities of these enzymes (10, 32, 40, 49). The cellulosomal enzyme 48S-t displayed a low level of activity both in the “WT-cellulosomal” state and when it was bound to Scaf·T in the “mock-free” state. The “WT-free” 48Y-CBM and the “converted-free” *48S-CBM enzymes, both of which contained a CBM as an integral part of the polypeptide chain, also exhibited similarly low activities on crystalline cellulose substrates. This might indicate that the low activity is not necessarily a result of a lack of attachment of the cellulase to its substrate but rather is an intrinsic characteristic of the family 48 cellulases themselves, since the tunnel-like architecture of the catalytic module dictates an exoglucanase mode of action, whereas the endoglucanases can act on any part of the linear cellulose chain. The family 9 glycoside hydrolases displayed much higher levels of activity than the family 48 cellulases. The “WT-cellulosomal” enzyme 9R-b had the lowest activity, whereas the “mock-free” enzyme had activity that was almost 3-fold higher. The “converted-free” enzyme *9R-CBM and the “WT-free” enzyme 9I-CBM exhibited the highest level of activity.
In parallel experiments, we converted the free enzymes 48Y-CBM and 9I-CBM into cellulosomal enzymes by swapping their CBMs with dockerins of the corresponding cellulosomal analogues (Fig. 2). All of the family 48 glycoside hydrolases exhibited similarly low levels of activity. Among the family 9 glycoside hydrolases, the “converted-cellulosomal” *9I-b enzyme was more active than the “WT-cellulosomal” 9R-b enzyme that exhibited the lowest level of activity. The “mock-free” *9I-b enzyme (preattached to Scaf·B) exhibited improved activity due to the added substrate-targeting function, and its enzymatic activity was only about 1.3-fold lower than that of the “WT-free” 9I-CBM enzyme.
Catalytic activity of family 48 and family 9 combinations.
In the following experiments, the family 48 and family 9 enzymes that we produced were used in eight different combinations of enzyme systems, as described in Table 2. We tested the activities of the “WT-cellulosomal” enzymes (48S-t and 9R-b) converted to free enzymes (“converted-free”) and also compared their activities with those of the “WT-free,” “mock-free,” and “designer-WT” systems.
The chimeric “converted-free” enzymes were as active as the “WT-free” enzymes; the two pairs have the same modular architecture, but they have different catalytic modules. These free systems were the most efficient systems for cellulose degradation and were approximately 3-fold more active than the least active “WT-cellulosomal” enzymes. The “mock-free” and “designer-WT” pair (48S-t and 9R-b preattached to a matching single-cohesin scaffoldin and to a designer scaffoldin, respectively) were about 1.3-fold less active than the free systems. Nevertheless, these pairs were clearly about 2.2-fold more efficient than the “WT-cellulosomal” pair. There was no significant difference between the activities of the “mock-free” pair and the “designer-WT” pair. This implies that the observed improvement in the activity compared to that of the free enzymes was due to the targeting effect by the CBM and not to the proximity of the enzymes due to their inclusion in a chimeric scaffoldin (Fig. 3).
FIG. 3.
Comparative degradation of Avicel by combinations of family 48 and family 9 enzymes. Open bars, conversion from cellulosomal mode to free mode (the cellulosomal enzyme 48S and 9R catalytic modules in “WT-cellulosomal,” “mock-free,” and “designer-WT” modes were converted into the “converted-free” mode); filled bars, conversion from free mode to cellulosomal mode (the free 48Y and 9I catalytic modules in a “WT-free” mode were converted into a cellulosomal mode, either “converted-cellulosomal,” “mock-free,” or “designer-converted”). For definitions of enzyme systems see Table 2. The results of two independent experiments are shown. Triplicate reactions were carried out, and standard deviations are indicated by error bars. The relative activity was determined by comparison with the activity of the WT-free enzymes 48Y-CBM and 9I-CBM, as described in Materials and Methods.
The combined activity of the “WT-free” pair (48Y-CBM and 9I-CBM) was then compared to that of the “converted-cellulosomal” pair (*48Y-t and *9I-b). Replacement of the CBM with a dockerin module resulted in a level of activity that was 3-fold lower and was similar to the level of activity of the “WT-cellulosomal” analogues. This suggests that the lower efficiency for cellulose degradation is not due to a loss of function of the catalytic module but is due to a loss of the targeting effect. The preattachment of the “converted-cellulosomal” enzymes to a single-cohesin scaffoldin or to the designer scaffoldin Scaf·BT (“mock-free” or “designer-converted” state) increased the activity of the enzyme system so that it approached that of the original “WT-free” state (Fig. 3).
It has previously been reported that combinations of the “WT-free” enzymes, Cel48Y (48Y-CBM) and Cel9I (9I-CBM), synergistically increase the hydrolysis of crystalline bacterial cellulose by 2.1-fold with a 17-fold excess of Cel48Y over Cel9I (10). In the present study, however, equimolar ratios of the enzymes tested were used, as dictated by our current arrangement of the designer cellulosome. Nevertheless, our data demonstrated that there was significant synergistic activity of the enzyme pairs (Fig. 4). Both wild-type pairs exhibited a distinct synergism (about 1.5-fold). The cellulosomal pair continued to act synergistically in all of the states tested, including when enzymes were attached to a single-cohesin scaffoldin (“mock-free”) or to a designer scaffoldin (“designer-WT”) and also when the enzymes were converted to free enzymes (“converted-free”). Conversely, the free enzymes acted synergistically only in their wild-type state. The activities of the “converted-cellulosomal,” “designer-converted,” and “mock-free” enzyme pairs were essentially the same as the calculated sums of the individual activities (Fig. 4).
DISCUSSION
The multienzyme cellulosome complex from anaerobic bacteria is one of the major microbial enzyme paradigms currently being considered for conversion of plant cell wall biomass to bioenergy (30, 50). In this context, there is interest in employing designer cellulosomes for improving biomass degradation en route to second-generation biofuels as an alternative energy source and concurrent interest in obtaining an increased understanding of the structure-function relationship of cellulosome components (3, 5, 7, 9, 41, 42).
In previous studies we have shown that free cellulases from the aerobic bacterium Thermobifida fusca can be converted into cellulosomal enzymes and work in the designer cellulosome format by replacing their CBM with a dockerin module (14-16). In the C. thermocellum system used in the current study, the similarity of the cellulosomal and free family 48 and family 9 enzymes presents a unique opportunity to study their interconversion in a single bacterial species. Thus, we cross-converted both types of C. thermocellum cellulases into the alternative states (i.e., the cellulosomal enzymes were converted into the free mode and vice versa) and determined the comparative effects on cellulose degradation. As expected, the conversion of the dockerin-containing cellulosomal enzymes into the free mode increased their activity on cellulosic substrates due to the targeting effect of the appended CBM. The cellulose-degrading activity of a dockerin-containing enzyme was restored when the substrate-targeting function was reinstated by attaching the enzyme to a single-cohesin scaffoldin or a designer scaffoldin, indicating that the catalytic module was not significantly impaired by swapping the CBM with a dockerin. In view of the results described above, it is clear that the creation of a chimeric enzyme did not have a deleterious effect on the enzyme activity compared to that of the native enzyme analogue.
The catalytic activity of each individual enzyme and the catalytic activities of combinations of the family 48 and family 9 enzymes were tested using Avicel as a particularly recalcitrant microcrystalline cellulose substrate. It was generally observed that the activity of family 48 enzymes on Avicel was much lower than that of the family 9 glycoside hydrolases, in accordance with previous findings for these two families of enzymes. In addition, as discussed above, most cellulosomal enzymes that lack a module specialized for substrate binding were less active than their CBM-bearing analogues. Consequently, preattachment of a cellulosomal dockerin-bearing cellulase to a single-cohesin scaffoldin or a designer scaffoldin resulted in increasing the activity of the cellulase to a level comparable to that of its free analogue.
The studies described in this paper were initial attempts to produce designer cellulosomes using the cellulases of the primary cellulosome-producing bacterium C. thermocellum and thus complement previous studies in which Clostridium cellulolyticum and T. fusca enzymes were used (14, 20-22, 38, 39). The free C. thermocellum CBM-bearing enzymes (either “WT-free” or “converted-free”) were the most efficient enzymes for solubilizing microcrystalline cellulose (Avicel). It is thus clear that within the framework of the current experimental system, the targeting effect is the major factor responsible for the enhancement of activity observed for the family 48 and family 9 enzyme combinations, whereas the proximity effect appears to play little or no role, suggesting the possible significance of a second free cellulase system in C. thermocellum.
The lack of a proximity effect for these specific cellulases does not necessarily lessen the reported overall advantages of the cellulosomal system. This may imply that the current designer cellulosomes are rather primitive facsimiles of native cellulosomes, which require fine-tuning in the future. Thus, additional factors may be necessary, such as improved thermostability of the chimeric components, better understanding and design of the intermodular linkers, and interactions among the scaffoldin-borne modules. These considerations may indeed contribute to the overall synergism between two neighboring enzymes in a complex. Such considerations may also account for the greater proximity effect observed in previous studies of designer cellulosomes when either the C. cellulolyticum or T. fusca enzyme systems were used.
The disposition and organization of the microcrystalline cellulosic substrate used in our studies (Avicel) are very different at the molecular level from the disposition and organization of the microfibrillar cellulose rods that form the structural framework of the plant cell wall in its native state. In addition, the natural biological environment of C. thermocellum is anaerobic, and there are both competition with and assistance from other organisms. Moreover, in its native state, the cellulosome is attached to the bacterial cell surface, and the cell benefits immediately from the breakdown products of the cellulosic substrate. In comparison, the free cellulase system of C. thermocellum is soluble, and the enzymes may diffuse to areas that are distant from the cell, although we cannot exclude the possibility that the free enzymes also bind, at least transiently, either to the cell surface cellulosome or to other exocellular components. These factors, however, require additional studies beyond the scope of the present study. In this energy-limiting natural ecosystem, the cellulosome could thus have dramatic advantages over the free enzyme system, since it minimizes loss by diffusion of the cellulases and their products.
In conclusion, we exploited the special status of the C. thermocellum cellulase system, which contains multiple copies of the Cel48 exoglucanase and Cel9 processive endoglucanase, both in the free form and in the cellulosomal form. In general, the latter enzymes were functional in both states, and the individual catalytic modules, CBMs, and dockerins could replace their cellulosomal or noncellulosomal counterparts without significant consequences. The designer cellulosomes tested here comprised only two enzymes belonging to two enzyme families and were not attached to living cells. Moreover, the equimolar ratio of enzymes in the designer cellulosome used in this study may not represent the optimal ratio of the enzymes in the native system. Consequently, these designer cellulosomes might not possess all the advantages of the natural cellulosome. These differences between the native and artificial systems will be addressed in future work, as we continue to examine the feasibility and application of the designer cellulosome concept.
Supplementary Material
Acknowledgments
This research was supported by Brazilian friends of the Weizmann Institute of Science Alternative Energy Research Initiative and by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel, and from the Israel Science Foundation (grants 966/09 and 159/07). E.A.B. holds The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
Published ahead of print on 26 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barak, Y., T. Handelsman, D. Nakar, A. Mechaly, R. Lamed, Y. Shoham, and E. A. Bayer. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491-501. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., J.-P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multi-enzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, E. A., B. Henrissat, and R. Lamed. 2008. The cellulosome: a natural bacterial strategy to combat biomass recalcitrance, p. 407-426. In M. E. Himmel (ed.), Biomass recalcitrance. Blackwell, London, United Kingdom.
- 4.Bayer, E. A., R. Kenig, and R. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, E. A., R. Lamed, and M. E. Himmel. 2007. The potential of cellulases and cellulosomes for cellulosic waste management. Curr. Opin. Biotechnol. 18:237-245. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, E. A., R. Lamed, B. A. White, and H. J. Flint. 2008. From cellulosomes to cellulosomics. Chem. Rec. 8:364-377. [DOI] [PubMed] [Google Scholar]
- 7.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. [DOI] [PubMed] [Google Scholar]
- 8.Bayer, E. A., Y. Shoham, and R. Lamed. 2006. Cellulose-decomposing prokaryotes and their enzyme systems, p. 578-617. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 9.Bayer, E. A., Y. Shoham, and R. Lamed. 2008. Cellulosome-enhanced conversion of biomass: on the road to bioethanol, p. 75-96. In J. Wall, C. Harwood, and A. L. Demain (ed.), Bioenergy. ASM Press, Washington, DC.
- 10.Berger, E., D. Zhang, V. V. Zverlov, and W. H. Schwarz. 2007. Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyse crystalline cellulose synergistically. FEMS Microbiol. Lett. 268:194-201. [DOI] [PubMed] [Google Scholar]
- 11.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burstein, T., M. Shulman, S. Jindou, S. Petkun, F. Frolow, Y. Shoham, E. A. Bayer, and R. Lamed. 2009. Physical association of the catalytic and helper modules of a processive family-9 glycoside hydrolase is essential for activity. FEBS Lett. 583:879-884. [DOI] [PubMed] [Google Scholar]
- 13.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active Enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspi, J., Y. Barak, R. Haimovitz, D. Irwin, R. Lamed, D. B. Wilson, and E. A. Bayer. 2009. Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode. Appl. Environ. Microbiol. 75:7335-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caspi, J., D. Irwin, R. Lamed, H.-P. Fierobe, D. B. Wilson, and E. A. Bayer. 2008. Conversion of noncellulosomal Thermobifida fusca free exoglucanases into cellulosomal components: comparative impact on cellulose-degrading activity. J. Biotechnol. 135:351-357. [DOI] [PubMed] [Google Scholar]
- 16.Caspi, J., D. Irwin, R. Lamed, Y. Shoham, H.-P. Fierobe, D. B. Wilson, and E. A. Bayer. 2006. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocatal. Biotransform. 24:3-12. [Google Scholar]
- 17.Demain, A. L., M. Newcomb, and J. H. Wu. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 19.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth-rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fierobe, H.-P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Belaich, R. Lamed, Y. Shoham, and J.-P. Belaich. 2002. Degradation of cellulose substrates by cellulosome chimeras: substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 21.Fierobe, H.-P., A. Mechaly, C. Tardif, A. Belaich, R. Lamed, Y. Shoham, J.-P. Belaich, and E. A. Bayer. 2001. Design and production of active cellulosome chimeras: selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 22.Fierobe, H.-P., F. Mingardon, A. Mechaly, A. Belaich, M. T. Rincon, R. Lamed, C. Tardif, J.-P. Belaich, and E. A. Bayer. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined tri-functional scaffoldin. J. Biol. Chem. 280:16325-16334. [DOI] [PubMed] [Google Scholar]
- 23.Gal, L., C. Gaudin, A. Belaich, S. Pagès, C. Tardif, and J.-P. Belaich. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 26.Gilad, R., L. Rabinovich, S. Yaron, E. A. Bayer, R. Lamed, H. J. Gilbert, and Y. Shoham. 2003. CelI, a non-cellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gold, N. D., and V. J. Martin. 2007. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J. Bacteriol. 189:6787-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haimovitz, R., Y. Barak, E. Morag, M. Voronov-Goldman, R. Lamed, and E. A. Bayer. 2008. Cohesin-dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics 8:968-979. [DOI] [PubMed] [Google Scholar]
- 29.Hazlewood, G. P., K. Davidson, J. I. Laurie, N. S. Huskisson, and H. J. Gilbert. 1993. Gene sequence and properties of CelI, a family E endoglucanase from Clostridium thermocellum. J. Gen. Microbiol. 139:307-316. [DOI] [PubMed] [Google Scholar]
- 30.Himmel, M. E., Q. Xu, Y. Luo, S.-Y. Ding, R. Lamed, and E. A. Bayer. 2010. Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels 1:323-341. [Google Scholar]
- 31.Irwin, D., D.-H. Shin, S. Zhang, B. K. Barr, J. Sakon, P. A. Karplus, and D. B. Wilson. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 33.Jindou, S., Q. Xu, R. Kenig, Y. Shoham, E. A. Bayer, and R. Lamed. 2006. Novel architectural theme of family-9 glycoside hydrolases identified in cellulosomal enzymes of Acetivibrio cellulolyticus and Clostridium thermocellum. FEMS Microbiol. Lett. 254:308-316. [DOI] [PubMed] [Google Scholar]
- 34.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamed, R., E. Setter, R. Kenig, and E. A. Bayer. 1983. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol. Bioeng. Symp. 13:163-181. [Google Scholar]
- 36.Linder, M., and T. T. Teeri. 1997. The roles and function of cellulose-binding domains. J. Biotechnol. 57:15-28. [DOI] [PubMed] [Google Scholar]
- 37.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 31:426-428. [Google Scholar]
- 38.Mingardon, F., A. Chanal, A. M. López-Contreras, C. Dray, E. A. Bayer, and H.-P. Fierobe. 2007. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl. Environ. Microbiol. 73:3822-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mingardon, F., A. Chanal, C. Tardif, E. A. Bayer, and H.-P. Fierobe. 2007. Exploration of new geometries in cellulosome-like chimeras. Appl. Environ. Microbiol. 73:7138-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morag, E., I. Halevy, E. A. Bayer, and R. Lamed. 1991. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J. Bacteriol. 173:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordon, R. E., S. J. Craig, and F. C. Foong. 2009. Molecular engineering of the cellulosome complex for affinity and bioenergy applications. Biotechnol. Lett. 31:465-476. [DOI] [PubMed] [Google Scholar]
- 42.Ohmiya, K., K. Sakka, T. Kimura, and K. Morimoto. 2003. Application of microbial genes to recalcitrant biomass utilization and environmental conservation. J. Biosci. Bioeng. 95:549-561. [PubMed] [Google Scholar]
- 43.Raman, B., C. Pan, G. B. Hurst, M. Rodriguez, C. K. McKeown, P. K. Lankford, N. F. Samatova, and J. R. Mielenz. 2009. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4:e5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakon, J., D. Irwin, D. B. Wilson, and P. A. Karplus. 1997. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4:810-818. [DOI] [PubMed] [Google Scholar]
- 45.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 46.Stevenson, D. M., and P. J. Weimer. 2005. Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture. Appl. Environ. Microbiol. 71:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomme, P., R. A. J. Warren, R. C. Miller, D. G. Kilburn, and N. R. Gilkes. 1995. Cellulose-binding domains—classification and properties, p. 142-161. In J. M. Saddler and M. H. Penner (ed.), Enzymatic degradation of insoluble polysaccharides. American Chemical Society, Washington, DC.
- 48.Wang, W. K., K. Kruus, and J. H. D. Wu. 1993. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS), a major cellulosome component. J. Bacteriol. 175:1293-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, W. K., K. Kruus, and J. H. D. Wu. 1994. Cloning and expression of the Clostridium thermocellum cellulase celS gene in Escherichia coli. Appl. Microbiol. Biotechnol. 42:346-352. [DOI] [PubMed] [Google Scholar]
- 50.Wei, H., Q. Xu, L. E. Taylor II, J. O. Baker, M. P. Tucker, and S. Y. Ding. 2009. Natural paradigms of plant cell wall degradation. Curr. Opin. Biotechnol. 20:330-338. [DOI] [PubMed] [Google Scholar]
- 51.Xu, Q., E. A. Bayer, M. Goldman, R. Kenig, Y. Shoham, and R. Lamed. 2004. Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell-surface anchoring scaffoldin and a family 48 cellulase. J. Bacteriol. 186:968-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zverlov, V. V., J. Kellermann, and W. H. Schwarz. 2005. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5:3646-3653. [DOI] [PubMed] [Google Scholar]
- 53.Zverlov, V. V., N. Schantz, and W. H. Schwarz. 2005. A major new component in the cellulosome of Clostridium thermocellum is a processive endo-beta-1,4-glucanase producing cellotetraose. FEMS Microbiol. Lett. 249:353-358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.